class: left, middle, inverse, title-slide # Data Visualization with R and ggplot2 ### Jessica Minnier, PhD & Meike Niederhausen, PhD<br><span style="font-size: 80%;"><a href="https://www.ohsu.edu/xd/research/centers-institutes/octri/education-training/octri-research-forum.cfm">OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop</a> </span> ### <span style="font-size: 80%;">2020/03/04 & 2020/05/20 <br><br><br> slides: <a href="http://bit.ly/berd_ggplot">bit.ly/berd_ggplot</a> <br> pdf: <a href="http://bit.ly/berd_ggplot_pdf">bit.ly/berd_ggplot_pdf</a></span> --- layout: true <!-- <div class="my-footer"><span>bit.ly/berd_tidy</span></div> --> <!-- <div class="my-footer"><a href="#visualtoc"> Visual TOC</span></div> --> --- # Load files for today's workshop .pull-left[ 1. Open slides [bit.ly/berd_ggplot](http://bit.ly/berd_ggplot) 1. Get project folder (detailed instructions: [bit.ly/berd_ggplot_instructions](https://bit.ly/berd_ggplot_instructions)) + Download zip folder at [bit.ly/berd_ggplot_zip](http://bit.ly/berd_ggplot_zip) + UNZIP completely (right click-> "extract all") + Open unzipped folder + Open (double click) `berd_ggplot_project.Rproj` + Inside RStudio 'Files' tab: click on file `00-install.R` and click "Run" to run all lines of code. 1. Open google doc for asking questions: [https://bit.ly/berd_doc](bit.ly/berd_doc) ] .pull-right[ <center><img src="img/horst_ggplot2_masterpiece.png" width="100%" height="100%"><a href="https://github.com/allisonhorst/stats-illustrations"><br>Allison Horst</a></center> ] --- # Learning objectives .pull-left[ - Understand the basic idea behind grammar of graphics - Be able to data to visual elements - Be able to customize plots in various ways - Use ggplot extensions to make even more plots! ] .pull-right[ <center><img src="img/ggplot-grammar-of-graphics-stack.png" width="100%" height="100%"><a href="https://www.sites.univ-rennes2.fr/mastersigat/Cours/Atelier%20Visualisation%20de%20donn%C3%A9es%20CERGY.pdf"></a></center> ] --- class: center, middle, inverse <center><img src="img/ggplotlogo.png" width="40%" height="40%"><a href="https://ggplot2.tidyverse.org/index.html"></a></center> --- # Grammar of Graphics - The "The Grammar of Graphics," is the theoretical basis for the ggplot2 package. + Much like how we construct sentences in any language by using a linguistic grammar (nouns, verbs, etc.), the grammar of graphics allows us to specify the components of a statistical graphic. In short, the grammar tells us that: >A statistical graphic is a mapping of data variables to aesthetic attributes of geometric objects. 3 **essential** components to a graphic: - data: the data-set comprised of variables that we plot - geom: this refers to our type of geometric objects we see in our plot (points, lines, bars, etc.) - aes: aesthetic attributes of the geometric object that we can perceive on a graphic. For example, x/y position, color, shape, and size. Each assigned aesthetic attribute can be mapped to a variable in our data-set. --- # Grammar of ggplot2 <center><img src="img/khealy_ggplot1.png" width="100%" height="100%"><a href="https://github.com/rstudio-conf-2020/dataviz"><br>Kieran Healy</a></center> --- # ggplot basics <center><img src="img/ggplot_basics_from_ppt.png" width="90%" height="100%"></center> --- class: center, middle, inverse # Tidy Data <img src="img/horst_tidyverse.jpg" width="70%" height="70%"> [Allison Horst](https://github.com/allisonhorst/stats-illustrations) --- ## Ggplot needs tidy data What are __tidy__ data? 1. Each variable forms a column 2. Each observation forms a row 3. Each value has its own cell  [G. Grolemond & H. Wickham's R for Data Science](https://r4ds.had.co.nz/tidy-data.html) See BERD workshop [Data Wrangling Part 1](https://jminnier-berd-r-courses.netlify.com/02-data-wrangling-tidyverse/02_data_wrangling_slides_part1.html#14) slides for more info. --- ## Gapminder data .pull-left[ [Gapminder](https://www.gapminder.org) is a foundation "fact tank" that collects reliable global statistics on many different measures, such as average life expectancy, population size, food supply, water source, etc. for individual countries <img src="img/gapminder_screenshot.png" width="90%" height="100%"> [Gapminder](https://www.gapminder.org/tools/#$chart-type=bubbles) ] .pull-right[ * `Gapminder_vars_2011.csv` contains select measures restricted to the year 2011. * `Gapminder_vars_2011_long.csv` is the same data as in `Gapminder_vars_2011.csv`, but in a _long_ format * Instead of individual columns for `CO2emissions`, `ElectricityUsePP`, ... `WaterSourcePrct`, * there is a column called `Measures` which contains these variables names and * a column called `Values` with the actual values for these measures. * This means the dataset contains multiple rows per country to account for each of these measures. ] --- ## A look at the `Gapminder_vars_2011.csv` dataset ```r gapminder2011 <- read_csv("data/Gapminder_vars_2011.csv") glimpse(gapminder2011) ``` ``` Rows: 195 Columns: 19 $ country <chr> "Afghanistan", "Albania", "Algeria", … $ CO2emissions <dbl> 0.412, 1.790, 3.290, 5.870, 1.250, 5.… $ ElectricityUsePP <dbl> NA, 2210.0, 1120.0, NA, 207.0, NA, 29… $ FoodSupplykcPPD <dbl> 2110, 3130, 3220, NA, 2410, 2370, 316… $ IncomePP <dbl> 1660, 10200, 13000, 42000, 5910, 1860… $ LifeExpectancyYrs <dbl> 56.7, 76.7, 76.7, 82.6, 60.9, 76.9, 7… $ FemaleLiteracyRate <dbl> 13.0, 95.7, NA, NA, 58.6, 99.4, 97.9,… $ population <dbl> 2.97e+07, 2.93e+06, 3.68e+07, 8.38e+0… $ WaterSourcePrct <dbl> 52.6, 88.1, 92.6, 100.0, 40.3, 97.0, … $ WaterSourcePrct_2011_quart <chr> "Q1", "Q2", "Q2", "Q4", "Q1", "Q3", "… $ geo <chr> "afg", "alb", "dza", "and", "ago", "a… $ four_regions <chr> "asia", "europe", "africa", "europe",… $ eight_regions <chr> "asia_west", "europe_east", "africa_n… $ six_regions <chr> "south_asia", "europe_central_asia", … $ members_oecd_g77 <chr> "g77", "others", "g77", "others", "g7… $ latitude <dbl> 33.00000, 41.00000, 28.00000, 42.5077… $ longitude <dbl> 66.00000, 20.00000, 3.00000, 1.52109,… $ world_bank_region <chr> "South Asia", "Europe & Central Asia"… $ world_bank_4_income_groups_2017 <chr> "Low income", "Upper middle income", … ``` --- name:visualtoc # Visual Table of Contents <a href="#barplot"><img src="figs/barplot_regions_out-1.png"width="150" height="150" title=figs/barplot_regions_out-1.png alt=figs/barplot_regions_out-1.png></a><a href="#histogram"><img src="figs/hist_LifeExp_out-1.png"width="150" height="150" title=figs/hist_LifeExp_out-1.png alt=figs/hist_LifeExp_out-1.png></a><a href="#density"><img src="figs/density_LifeExp_out-1.png"width="150" height="150" title=figs/density_LifeExp_out-1.png alt=figs/density_LifeExp_out-1.png></a><a href="#ridgeline"><img src="figs/ridges_LifeExp_out-1.png"width="150" height="150" title=figs/ridges_LifeExp_out-1.png alt=figs/ridges_LifeExp_out-1.png></a><a href="#boxplot"><img src="figs/boxplot_LifeExp_out-1.png"width="150" height="150" title=figs/boxplot_LifeExp_out-1.png alt=figs/boxplot_LifeExp_out-1.png></a><a href="#scatterplot"><img src="figs/scatter_FoodvsLifeExp_out-1.png"width="150" height="150" title=figs/scatter_FoodvsLifeExp_out-1.png alt=figs/scatter_FoodvsLifeExp_out-1.png></a><a href="#bubbleplot"><img src="figs/bubble_FemLitvsLifeExp_out-1.png"width="150" height="150" title=figs/bubble_FemLitvsLifeExp_out-1.png alt=figs/bubble_FemLitvsLifeExp_out-1.png></a><a href="#lineplot"><img src="figs/lineplot_YearLifeExp_out-1.png"width="150" height="150" title=figs/lineplot_YearLifeExp_out-1.png alt=figs/lineplot_YearLifeExp_out-1.png></a><a href="#ggmarginal"><img src="figs/margins_FoodvsLifeExp_out-1.png"width="150" height="150" title=figs/margins_FoodvsLifeExp_out-1.png alt=figs/margins_FoodvsLifeExp_out-1.png></a><a href="#correlation"><img src="figs/corrplotmix-1.png"width="150" height="150" title=figs/corrplotmix-1.png alt=figs/corrplotmix-1.png></a><a href="#facetdensity"><img src="figs/facet_density_all_out-1.png"width="150" height="150" title=figs/facet_density_all_out-1.png alt=figs/facet_density_all_out-1.png></a><a href="#facethist"><img src="figs/facet2x_density_all_out-1.png"width="150" height="150" title=figs/facet2x_density_all_out-1.png alt=figs/facet2x_density_all_out-1.png></a><a href="#volcano"><img src="figs/volcanoplot_out-1.png"width="150" height="150" title=figs/volcanoplot_out-1.png alt=figs/volcanoplot_out-1.png></a><a href="#heatmap"><img src="figs/heatmap_out-1.png"width="150" height="150" title=figs/heatmap_out-1.png alt=figs/heatmap_out-1.png></a><a href="#sidebyside"><img src="figs/ggpubr_out-1.png"width="150" height="150" title=figs/ggpubr_out-1.png alt=figs/ggpubr_out-1.png></a> Inspired by [EvaMaeRey (Gina Reynolds)](https://github.com/EvaMaeRey), author of the amazing [`flipbookr`](https://github.com/EvaMaeRey/flipbookr) package. --- name:barplot # Barplot .pull-left[ ```r ggplot(data = gapminder2011, aes(x = four_regions, fill = eight_regions)) + geom_bar() + labs(x = "World Regions", y = "Number of countries", title = "Barplot") + theme_bw() + theme( axis.text.x = element_text(angle = -30, hjust = 0), text = element_text(family = "Palatino")) + scale_fill_viridis_d(name = "Subregions") ``` See the last section of [ggplot1 Aesthetic specifications](https://ggplot2.tidyverse.org/articles/ggplot2-specs.html) for an explanation of `hjust`. <center><img src="img/hjust_vjust.png" width="50%" height="100%"></center> ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + * aes(x = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + * geom_bar() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + * aes(fill = eight_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + * scale_fill_discrete( * name = "Subregions" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + scale_fill_discrete( name = "Subregions" ) + * labs(x = "World Regions", * y = "Number of countries", * title = "Barplot") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + scale_fill_discrete( name = "Subregions" ) + labs(x = "World Regions", y = "Number of countries", title = "Barplot") + * theme_bw() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + scale_fill_discrete( name = "Subregions" ) + labs(x = "World Regions", y = "Number of countries", title = "Barplot") + theme_bw() + * theme(axis.text.x=element_text( * angle = -30, hjust = 0)) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + scale_fill_discrete( name = "Subregions" ) + labs(x = "World Regions", y = "Number of countries", title = "Barplot") + theme_bw() + theme(axis.text.x=element_text( angle = -30, hjust = 0)) + * scale_fill_viridis_d(name = "Subregions") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_9_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = four_regions) + geom_bar() + aes(fill = eight_regions) + scale_fill_discrete( name = "Subregions" ) + labs(x = "World Regions", y = "Number of countries", title = "Barplot") + theme_bw() + theme(axis.text.x=element_text( angle = -30, hjust = 0)) + scale_fill_viridis_d(name = "Subregions") + * theme( * text = element_text(family = "Palatino")) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/barplot_regions_auto_10_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:histogram # Histogram .pull-left[ ```r ggplot(data = gapminder2011, aes(x = LifeExpectancyYrs, fill = four_regions) ) + geom_histogram() + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + labs( x = "Life Expectancy (years)", title = "Histogram" ) + ggthemes::theme_economist() + theme(legend.position="bottom") ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + * aes(x = LifeExpectancyYrs) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + * geom_histogram() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_histogram() + * aes(fill = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_histogram() + aes(fill = four_regions) + * scale_fill_discrete( * name = "Regions", * labels = c("Africa", "Americas", * "Asia", "Europe") * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_histogram() + aes(fill = four_regions) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + * labs( * x = "Life Expectancy (years)", * title = "Histogram" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_histogram() + aes(fill = four_regions) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + labs( x = "Life Expectancy (years)", title = "Histogram" ) + * ggthemes::theme_economist() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_histogram() + aes(fill = four_regions) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + labs( x = "Life Expectancy (years)", title = "Histogram" ) + ggthemes::theme_economist() + * theme(legend.position="bottom") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/hist_LifeExp_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- # Legend position * "Generic" positions * `legend.position = "left"` * Other options: "top", "right", "bottom", "none" * Specificied by location * `legend.position = c(x,y)` * Specify x and y coordinates of position * Values should be between 0 and 1 * __c(0,0)__ corresponds to the __bottom left__ * __c(1,1)__ corresponds to the __top right__ --- name:density # Density Plot .pull-left[ ```r ggplot(data = gapminder2011, aes(x = LifeExpectancyYrs, fill = four_regions) ) + geom_density(alpha = 0.4) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + labs( x = "Life Expectancy (years)", title = "Density Plot" ) + hrbrthemes::theme_ipsum() + theme(legend.position=c(.2,.8)) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + * aes(x = LifeExpectancyYrs) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + * geom_density() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + * aes(fill = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + aes(fill = four_regions) + * aes(alpha=.4) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + aes(fill = four_regions) + aes(alpha=.4) + * scale_fill_discrete( * name = "Regions", * labels = c("Africa", "Americas", * "Asia", "Europe") * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + aes(fill = four_regions) + aes(alpha=.4) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + * hrbrthemes::theme_ipsum() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + aes(fill = four_regions) + aes(alpha=.4) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + hrbrthemes::theme_ipsum() + * theme(legend.position=c(.2,.8)) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + geom_density() + aes(fill = four_regions) + aes(alpha=.4) + scale_fill_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + hrbrthemes::theme_ipsum() + theme(legend.position=c(.2,.8)) + * labs( * x = "Life Expectancy (years)", * title = "Density Plot" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/density_LifeExp_auto_9_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:ridgeline # Ridgeline Plot .pull-left[ ```r library(ggridges) ggplot(data = gapminder2011, aes(x = LifeExpectancyYrs, y = four_regions, fill = four_regions) ) + geom_density_ridges(alpha = 0.4) + ggthemes::theme_clean() + theme(legend.position="none") + labs( x = "Life Expectancy (years)", y = "Regions", title = "Ridgeline Density Plot" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *library(ggridges) ``` ]] .column[.content[ ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) *ggplot(data = gapminder2011) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + * aes(x = LifeExpectancyYrs) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + * aes(y = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + * geom_density_ridges() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + geom_density_ridges() + * aes(fill = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + geom_density_ridges() + aes(fill = four_regions) + * aes(alpha = 0.4) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + geom_density_ridges() + aes(fill = four_regions) + aes(alpha = 0.4) + * ggthemes::theme_clean() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + geom_density_ridges() + aes(fill = four_regions) + aes(alpha = 0.4) + ggthemes::theme_clean() + * theme(legend.position = "none") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_9_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r library(ggridges) ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + aes(y = four_regions) + geom_density_ridges() + aes(fill = four_regions) + aes(alpha = 0.4) + ggthemes::theme_clean() + theme(legend.position = "none") + * labs( * x = "Life Expectancy (years)", * y = "Regions", * title = "Ridgeline Density Plot" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/ridges_LifeExp_auto_10_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:boxplot # Boxplot .pull-left[ ```r ggplot(data = gapminder2011, aes(x = LifeExpectancyYrs, # New! y = four_regions, fill = four_regions) ) + geom_boxplot(alpha = 0.3) + # add outlier.shape = NA # coord_flip() + theme_fivethirtyeight() + theme(axis.title = element_text()) + scale_fill_fivethirtyeight() + theme(legend.position = "none") + geom_jitter(width = .1, alpha = 0.3) + geom_violin(colour = "grey", alpha = .2) + labs( x = "World Region", y = "Life Expectancy (years)", title = "Boxplot" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + * aes(x = LifeExpectancyYrs) # New! ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! * geom_boxplot(alpha=.3) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + * aes(y = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + * aes(fill = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + * theme_fivethirtyeight() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + * scale_fill_fivethirtyeight() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + scale_fill_fivethirtyeight() + * theme(axis.title = element_text()) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + scale_fill_fivethirtyeight() + theme(axis.title = element_text()) + * theme(legend.position = "none") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_9_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + scale_fill_fivethirtyeight() + theme(axis.title = element_text()) + theme(legend.position = "none") + * geom_jitter( * width = .1, * alpha = 0.3 * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_10_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + scale_fill_fivethirtyeight() + theme(axis.title = element_text()) + theme(legend.position = "none") + geom_jitter( width = .1, alpha = 0.3 ) + * geom_violin( * colour = "grey", * alpha = .2 * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_11_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011) + aes(x = LifeExpectancyYrs) + # New! geom_boxplot(alpha=.3) + aes(y = four_regions) + aes(fill = four_regions) + theme_fivethirtyeight() + scale_fill_fivethirtyeight() + theme(axis.title = element_text()) + theme(legend.position = "none") + geom_jitter( width = .1, alpha = 0.3 ) + geom_violin( colour = "grey", alpha = .2 ) + * labs( * x = "World Region", * y = "Life Expectancy (years)", * title = "Boxplot" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/boxplot_LifeExp_auto_12_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- # Exercise Complete the third section of the `practice_ggplot.Rmd` file: "Bar plot". --- name:scatterplot # Scatterplot .pull-left[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha = 0.4) + geom_smooth(se = FALSE)+ geom_smooth(method = lm) + theme_minimal() + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + labs( x = "Daily Food Supply Per Person (kc)", y = "Life Expectancy (years)", title = "Scatterplot" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011, * aes(x = FoodSupplykcPPD, * y = LifeExpectancyYrs) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + * geom_point(alpha=.4) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + * aes(color = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + aes(color = four_regions) + * scale_color_colorblind( * name = "Regions", * labels = c("Africa", "Americas", * "Asia", "Europe") * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + aes(color = four_regions) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + * geom_smooth(se = FALSE) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + aes(color = four_regions) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + geom_smooth(se = FALSE) + * geom_smooth(method = lm) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + aes(color = four_regions) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + geom_smooth(se = FALSE) + geom_smooth(method = lm) + * theme_minimal() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs) ) + geom_point(alpha=.4) + aes(color = four_regions) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + geom_smooth(se = FALSE) + geom_smooth(method = lm) + theme_minimal() + * labs( * x = "Daily Food Supply Per Person (kc)", * y = "Life Expectancy (years)", * title = "Scatterplot" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/scatter_FoodvsLifeExp_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:bubbleplot # Bubbleplot .pull-left[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions, size = population) ) + geom_point(alpha = 0.4) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + scale_size( name = "Population Size (millions)", breaks = c(1e08,5e08,1e09), labels = c(100,500,1000) ) + hrbrthemes::theme_ipsum() + labs( x = "Daily Food Supply PP (kc)", y = "Life Expectancy (years)", title = "Bubbleplot" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011, * aes(x = FoodSupplykcPPD, * y = LifeExpectancyYrs, * color = four_regions) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + * geom_point(alpha=.4) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha=.4) + * aes(size = population) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha=.4) + aes(size = population) + * scale_color_colorblind( * name = "Regions", * labels = c("Africa", "Americas", * "Asia", "Europe") * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha=.4) + aes(size = population) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + * scale_size( * name = "Population Size (millions)", * breaks = c(1e08,5e08,1e09), * labels = c(100,500,1000) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha=.4) + aes(size = population) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + scale_size( name = "Population Size (millions)", breaks = c(1e08,5e08,1e09), labels = c(100,500,1000) ) + * hrbrthemes::theme_ipsum() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha=.4) + aes(size = population) + scale_color_colorblind( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + scale_size( name = "Population Size (millions)", breaks = c(1e08,5e08,1e09), labels = c(100,500,1000) ) + hrbrthemes::theme_ipsum() + * labs( * x = "Daily Food Supply PP (kc)", * y = "Life Expectancy (years)", * title = "Bubbleplot" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/bubble_FemLitvsLifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- # Exercise Complete the fourth section of the `practice_ggplot.Rmd` file: "Bubbleplot" --- name:lineplot # Lineplot .pull-left[ For the Lineplot example we are using the `gapminder::gapminder` dataset since it has longitudinal data across many years for life expectancy. ```r library(gapminder) ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent, group = country) ) + geom_point(alpha = 0.4) + geom_line(alpha = 0.7) + scale_color_colorblind(name = "Continents") + ggthemes::theme_clean() + labs( x = "Year", y = "Life Expectancy (years)", title = "Lineplot", subtitle = "Time series", caption = "Source: gapminder package" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## R package `gapminder` has a dataset called `gapminder` For the Lineplot example we are using the `gapminder::gapminder` dataset since it has longitudinal data across many years for life expectancy. ```r library(gapminder) head(gapminder,15) ``` ``` # A tibble: 15 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 2 Afghanistan Asia 1957 30.3 9240934 821. 3 Afghanistan Asia 1962 32.0 10267083 853. 4 Afghanistan Asia 1967 34.0 11537966 836. 5 Afghanistan Asia 1972 36.1 13079460 740. 6 Afghanistan Asia 1977 38.4 14880372 786. 7 Afghanistan Asia 1982 39.9 12881816 978. 8 Afghanistan Asia 1987 40.8 13867957 852. 9 Afghanistan Asia 1992 41.7 16317921 649. 10 Afghanistan Asia 1997 41.8 22227415 635. 11 Afghanistan Asia 2002 42.1 25268405 727. 12 Afghanistan Asia 2007 43.8 31889923 975. 13 Albania Europe 1952 55.2 1282697 1601. 14 Albania Europe 1957 59.3 1476505 1942. 15 Albania Europe 1962 64.8 1728137 2313. ``` --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder, * aes(x = year, * y = lifeExp, * color = continent) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + * geom_point(alpha = .4) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + geom_point(alpha = .4) + * geom_line(alpha = .7) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + geom_point(alpha = .4) + geom_line(alpha = .7) + * aes(group = country) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + geom_point(alpha = .4) + geom_line(alpha = .7) + aes(group = country) + * scale_color_colorblind( * name = "Continents" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + geom_point(alpha = .4) + geom_line(alpha = .7) + aes(group = country) + scale_color_colorblind( name = "Continents" ) + * ggthemes::theme_clean() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder, aes(x = year, y = lifeExp, color = continent) ) + geom_point(alpha = .4) + geom_line(alpha = .7) + aes(group = country) + scale_color_colorblind( name = "Continents" ) + ggthemes::theme_clean() + * labs( * x = "Year", * y = "Life Expectancy (years)", * title = "Lineplot", * subtitle = "Time series", * caption = "Source: gapminder package" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/lineplot_YearLifeExp_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name::ggmarginal # `ggmarginal` https://cran.r-project.org/web/packages/ggExtra/vignettes/ggExtra.html .pull-left-40[ ```r # library(ggExtra) p <- ggplot(data = gapminder2011, aes(x = FoodSupplykcPPD, y = LifeExpectancyYrs, color = four_regions) ) + geom_point(alpha = .4) + scale_color_discrete( name = "Regions", labels = c("Africa", "Americas", "Asia", "Europe") ) + theme(legend.position="bottom") + labs( x = "Daily Food Supply PP (kc)", y = "Life Expectancy (years)", title = "Scatterplot" ) ``` ] .pull-right-60[ ```r ggMarginal(p, type = "density", margins = "both", groupColour = TRUE, groupFill = TRUE ) ``` <img src="04_ggplot_slides_files/figure-html/margins_FoodvsLifeExp_out-1.png" width="720" style="display: block; margin: auto;" /> ] --- class: inverse, middle, center # Corrolelograms --- name:correlation ## Correlation matrix ```r M <- cor(gapminder2011 %>% select(FoodSupplykcPPD:WaterSourcePrct), use = "complete.obs" # specified since there are missing values ) M ``` ``` FoodSupplykcPPD IncomePP LifeExpectancyYrs FoodSupplykcPPD 1.0000000 0.48221951 0.64233437 IncomePP 0.4822195 1.00000000 0.51567562 LifeExpectancyYrs 0.6423344 0.51567562 1.00000000 FemaleLiteracyRate 0.4816309 0.44804036 0.64921874 population 0.0768498 -0.07838737 0.05467681 WaterSourcePrct 0.6980454 0.53687914 0.80693858 FemaleLiteracyRate population WaterSourcePrct FoodSupplykcPPD 0.48163092 0.07684980 0.69804539 IncomePP 0.44804036 -0.07838737 0.53687914 LifeExpectancyYrs 0.64921874 0.05467681 0.80693858 FemaleLiteracyRate 1.00000000 -0.05188109 0.74980282 population -0.05188109 1.00000000 0.05559188 WaterSourcePrct 0.74980282 0.05559188 1.00000000 ``` --- ## `corrplot::corrplot()` https://cran.r-project.org/web/packages/corrplot/vignettes/corrplot-intro.html .pull-left[ ```r library(corrplot) corrplot(M, method = "number") ``` <img src="04_ggplot_slides_files/figure-html/unnamed-chunk-6-1.png" width="504" style="display: block; margin: auto;" /> ] .pull-right[ ```r corrplot(M, method = "ellipse") ``` <img src="04_ggplot_slides_files/figure-html/unnamed-chunk-7-1.png" width="720" style="display: block; margin: auto;" /> ```r corrplot.mixed(M) ``` <img src="04_ggplot_slides_files/figure-html/unnamed-chunk-8-1.png" width="504" style="display: block; margin: auto;" /> ] --- name:ggcorr ## `GGally::ggcorr()` https://ggobi.github.io/ggally/index.html ```r # library(GGally) gapminder2011 %>% select(FoodSupplykcPPD:WaterSourcePrct) %>% # specifying which columns to use ggcorr() ``` <img src="04_ggplot_slides_files/figure-html/unnamed-chunk-9-1.png" width="720" style="display: block; margin: auto;" /> --- name:ggpairs ## `GGally::ggpairs()` https://ggobi.github.io/ggally/index.html ```r # library(GGally) gapminder2011 %>% select(FoodSupplykcPPD:WaterSourcePrct) %>% # specifying which columns to use ggpairs() ``` <img src="04_ggplot_slides_files/figure-html/unnamed-chunk-10-1.png" width="720" style="display: block; margin: auto;" /> --- class: inverse, middle, center # Faceting --- name:facetdensity # Faceted Density Plot .pull-left[ ```r ggplot(data = gapminder2011_long, aes(x = Values, color = four_regions) ) + facet_wrap(~ Measures, scales = "free", ncol = 2 ) + geom_density() + ggthemes::theme_few() + theme(legend.position="top") + labs( x = "", title = "Faceted Density Plots", # Add a figure number! tag = "Fig 1", # note that color is being # specified inside labs! color = "Regions" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- # Wide vs. long data - __Wide__ data has one row per subject, with multiple columns for their repeated measurements - __Long__ data has multiple rows per subject, with one column for the measurement variable and another indicating from when/where the repeated measures are from .pull-left[ wide <img src="img/SBP_wide2.png" width="73%" height="73%"> See BERD workshop [Data Wrangling Part 2](https://jminnier-berd-r-courses.netlify.com/02-data-wrangling-tidyverse/02_data_wrangling_slides_part2.html#26) for slides on how to make wide data long. ] .pull-right[ long <img src="img/SBP_long2.png" width="30%" height="30%"> ] --- # Dataset `Gapminder_vars_2011_long.csv` (1/2) * This is the same 2011 Gapminder data we've been using thus far, but in a __long format__ instead of wide. * Instead of individual columns for `CO2emissions`, `ElectricityUsePP`, ... `WaterSourcePrct`, * there is a column called `Measures` which contains these variables names and * a column called `Values` with the actual values for these measures. * This means the dataset contains multiple rows per country to account for each of these measures. ```r gapminder2011_long <- read_csv("data/Gapminder_vars_2011_long.csv") glimpse(gapminder2011_long) ``` ``` Rows: 1,365 Columns: 8 $ country <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanist… $ population <dbl> 29700000, 29700000, 29700000, 29700000, 29700000, 29700… $ four_regions <chr> "asia", "asia", "asia", "asia", "asia", "asia", "asia",… $ eight_regions <chr> "asia_west", "asia_west", "asia_west", "asia_west", "as… $ six_regions <chr> "south_asia", "south_asia", "south_asia", "south_asia",… $ WorldRegions <chr> "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia",… $ Measures <chr> "CO2emissions", "ElectricityUsePP", "FoodSupplykcPPD", … $ Values <dbl> 4.12e-01, NA, 2.11e+03, 1.66e+03, 5.67e+01, 1.30e+01, 5… ``` --- # Dataset `Gapminder_vars_2011_long.csv` (2/2) ```r gapminder2011_long %>% select(country, population, four_regions, Measures, Values) %>% head(15) ``` ``` # A tibble: 15 x 5 country population four_regions Measures Values <chr> <dbl> <chr> <chr> <dbl> 1 Afghanistan 29700000 asia CO2emissions 0.412 2 Afghanistan 29700000 asia ElectricityUsePP NA 3 Afghanistan 29700000 asia FoodSupplykcPPD 2110 4 Afghanistan 29700000 asia IncomePP 1660 5 Afghanistan 29700000 asia LifeExpectancyYrs 56.7 6 Afghanistan 29700000 asia FemaleLiteracyRate 13 7 Afghanistan 29700000 asia WaterSourcePrct 52.6 8 Albania 2930000 europe CO2emissions 1.79 9 Albania 2930000 europe ElectricityUsePP 2210 10 Albania 2930000 europe FoodSupplykcPPD 3130 11 Albania 2930000 europe IncomePP 10200 12 Albania 2930000 europe LifeExpectancyYrs 76.7 13 Albania 2930000 europe FemaleLiteracyRate 95.7 14 Albania 2930000 europe WaterSourcePrct 88.1 15 Algeria 36800000 africa CO2emissions 3.29 ``` --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011_long, * aes(x = Values) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + * geom_density() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + geom_density() + * facet_wrap(~ Measures, * scales = "free", * ncol = 2 * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + geom_density() + facet_wrap(~ Measures, scales = "free", ncol = 2 ) + * aes(color = four_regions) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + geom_density() + facet_wrap(~ Measures, scales = "free", ncol = 2 ) + aes(color = four_regions) + * ggthemes::theme_few() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + geom_density() + facet_wrap(~ Measures, scales = "free", ncol = 2 ) + aes(color = four_regions) + ggthemes::theme_few() + * theme( legend.position="top") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + geom_density() + facet_wrap(~ Measures, scales = "free", ncol = 2 ) + aes(color = four_regions) + ggthemes::theme_few() + theme( legend.position="top") + * labs( * x = "", * title = "Faceted Density Plots", * tag = "Fig 1", * color = "Regions" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet_density_all_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:facethist # Faceted 2x Histogram .pull-left[ Note that a __long__ dataset is being used in this example! See next slide for an explanation. ```r ggplot(data = gapminder2011_long) + geom_histogram(aes(x = Values), fill = "darkorange") + facet_grid( four_regions ~ Measures, scales = "free_x" ) + ggthemes::theme_igray() + theme( strip.text.y = element_text(size=10, angle=45, face = "bold"), strip.text.x = element_text(size=6), axis.text.x = element_text(angle=45, hjust=1) ) + labs( x = "", title = "Faceted Density Plots" ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = gapminder2011_long, * aes(x = Values) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + * facet_grid( * four_regions ~ Measures, * scales = "free_x" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + facet_grid( four_regions ~ Measures, scales = "free_x" ) + * geom_histogram(fill = "darkorange") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + facet_grid( four_regions ~ Measures, scales = "free_x" ) + geom_histogram(fill = "darkorange") + * ggthemes::theme_igray() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + facet_grid( four_regions ~ Measures, scales = "free_x" ) + geom_histogram(fill = "darkorange") + ggthemes::theme_igray() + * theme( * strip.text.y = * element_text(size=10, * angle=45, * face = "bold"), * strip.text.x = element_text(size=6), * axis.text.x = element_text(angle=45, * hjust=1) * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = gapminder2011_long, aes(x = Values) ) + facet_grid( four_regions ~ Measures, scales = "free_x" ) + geom_histogram(fill = "darkorange") + ggthemes::theme_igray() + theme( strip.text.y = element_text(size=10, angle=45, face = "bold"), strip.text.x = element_text(size=6), axis.text.x = element_text(angle=45, hjust=1) ) + * labs( * x = "", * title = "Faceted Density Plots" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/facet2x_density_all_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class:inverse, center, middle # Gene Expression --- # Pasilla Data ```r glimpse(pasilla_data) ``` ``` Rows: 8,377 Columns: 15 $ gene <chr> "FBgn0000008", "FBgn0000017", "FBgn0000018", "FBgn0000… $ baseMean <dbl> 95.144292, 4352.553569, 418.610484, 6.406200, 989.7202… $ fc <dbl> 1.0015792, 0.8467929, 0.9300151, 1.1573681, 0.9383590,… $ log2FoldChange <dbl> 0.002276441, -0.239918944, -0.104673912, 0.210847792, … $ lfcSE <dbl> 0.2237287, 0.1263369, 0.1484891, 0.6895876, 0.1477518,… $ stat <dbl> 0.01017501, -1.89904084, -0.70492676, 0.30575928, -0.6… $ pvalue <dbl> 9.918817e-01, 5.755911e-02, 4.808558e-01, 7.597879e-01… $ padj <dbl> 9.972108e-01, 2.880017e-01, 8.268337e-01, 9.435011e-01… $ treated1 <dbl> 7.607917, 11.938311, 9.143372, 6.479135, 10.187606, 6.… $ treated2 <dbl> 7.834912, 12.024557, 9.011505, 6.577240, 10.024965, 6.… $ treated3 <dbl> 7.595052, 12.013565, 8.944883, 6.475226, 10.017728, 6.… $ untreated1 <dbl> 7.567298, 12.045721, 9.315269, 6.565256, 10.450060, 6.… $ untreated2 <dbl> 7.642174, 12.284647, 9.098290, 6.479802, 10.080550, 6.… $ untreated3 <dbl> 7.844603, 12.455939, 8.966546, 6.422196, 10.069559, 6.… $ untreated4 <dbl> 7.669147, 12.077404, 9.066286, 6.395509, 9.996554, 6.4… ``` --- name:volcano # Volcano Plot .pull-left[ ```r # Create subset for labeling pasilla_data_top = pasilla_data %>% filter(-log10(padj) > 10, abs(log2FoldChange) > 2.5) ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + ggrepel::geom_text_repel( data = pasilla_data_top, aes(label = gene), color = "black", box.padding = 0.5, min.segment.length = 0) + xlim(c(-7,7)) + geom_vline(xintercept = c(-2.5, 2.5), lty = "dashed", color="grey") + ggthemes::theme_clean() + labs( x = bquote(~Log[2]~ "fold change"), y = bquote(~Log[10]~adjusted~italic(P)), title = "Volcano Plot", subtitle = "Gene Expression of Pasilla Data" ) ``` <img src="04_ggplot_slides_files/figure-html/volcanoplot_nice-1.png" width="504" style="display: block; margin: auto;" /> ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: split-40 count: false .column[.content[ ```r *ggplot(data = pasilla_data, * aes(x = log2FoldChange, * y = log10(padj))) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_1_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + * geom_point() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_2_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + * scale_y_reverse() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_3_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + * aes(color = padj < 0.05) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_4_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + * ggrepel::geom_text_repel( * data = pasilla_data_top, * aes(label = gene), color = "black", * box.padding = 0.5, * min.segment.length = 0) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_5_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + ggrepel::geom_text_repel( data = pasilla_data_top, aes(label = gene), color = "black", box.padding = 0.5, min.segment.length = 0) + * xlim(c(-7,7)) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_6_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + ggrepel::geom_text_repel( data = pasilla_data_top, aes(label = gene), color = "black", box.padding = 0.5, min.segment.length = 0) + xlim(c(-7,7)) + * geom_vline(xintercept = c(-2.5, 2.5), * lty = "dashed", color="grey") ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_7_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + ggrepel::geom_text_repel( data = pasilla_data_top, aes(label = gene), color = "black", box.padding = 0.5, min.segment.length = 0) + xlim(c(-7,7)) + geom_vline(xintercept = c(-2.5, 2.5), lty = "dashed", color="grey") + * ggthemes::theme_clean() ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_8_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- class: split-40 count: false .column[.content[ ```r ggplot(data = pasilla_data, aes(x = log2FoldChange, y = log10(padj))) + geom_point() + scale_y_reverse() + aes(color = padj < 0.05) + ggrepel::geom_text_repel( data = pasilla_data_top, aes(label = gene), color = "black", box.padding = 0.5, min.segment.length = 0) + xlim(c(-7,7)) + geom_vline(xintercept = c(-2.5, 2.5), lty = "dashed", color="grey") + ggthemes::theme_clean() + * labs( * x = bquote(~Log[2]~ "fold change"), * y = bquote(~Log[10]~adjusted~italic(P)), * title = "Volcano Plot", * subtitle="Gene Expression of Pasilla Data" * ) ``` ]] .column[.content[ <img src="04_ggplot_slides_files/figure-html/volcanoplot_auto_9_output-1.png" width="504" style="display: block; margin: auto;" /> ]] --- name:heatmap # Heatmap with `pheatmap::pheatmap()` It's possible to make heatmaps in ggplot2 with `geom_tile()`, but there are many other better functions using base R that cluster and annotate the data. This is using `pheatmap` package. .pull-left-60[ We need to create the data: ```r # select expression data pasilla_heat <- pasilla_data %>% select(treated1:untreated4) # subtract off gene-specific means pasilla_heat <- pasilla_heat - rowMeans(pasilla_heat) # calculate standard deviation of each centered gened sd_gene <- apply(pasilla_heat,1,sd) # select top 500 most variable pasilla_heat <- pasilla_heat[order(sd_gene, decreasing = TRUE)[1:500],] # create annotation data pasilla_col <- data.frame( trt = factor(c(rep("trt",3), rep("untrt",4))), id = 1:7, row.names=colnames(pasilla_heat)) ``` ] .pull-right-40[ ```r head(pasilla_heat, n = 3) ``` ``` treated1 treated2 treated3 untreated1 untreated2 untreated3 untreated4 2390 -1.5997691 0.8713581 1.568570 -1.500656 -2.103343 1.338488 1.4253512 521 -1.3218267 0.9954861 1.278523 -1.521073 -1.425689 1.040472 0.9541077 7886 -0.5901012 0.8225366 1.339219 -1.762754 -1.701829 1.155933 0.7369965 ``` ```r pasilla_col ``` ``` trt id treated1 trt 1 treated2 trt 2 treated3 trt 3 untreated1 untrt 4 untreated2 untrt 5 untreated3 untrt 6 untreated4 untrt 7 ``` ] --- # Heatmap with `pheatmap::pheatmap()` .pull-left[ ```r pheatmap::pheatmap( mat = pasilla_heat, show_rownames = FALSE, annotation_col = pasilla_col ) ``` ] .pull-right[ <img src="04_ggplot_slides_files/figure-html/heatmap_out-1.png" width="504" style="display: block; margin: auto;" /> ] --- name:sidebyside # Side by side plot with [`ggpubr`](https://rpkgs.datanovia.com/ggpubr/) .pull-left[ ```r p1 <- ggplot(data = pasilla_data, aes(x = log2FoldChange, y = -log10(padj), color = log10(baseMean))) + geom_point() + geom_vline(xintercept = c(-2.5, 2.5), lty = 2, color="grey") + theme_few() + scale_color_viridis_c() + labs(x = bquote(~Log[2]~ "fold change"), y = bquote(~Log[10]~adjusted~italic(P)), title = "Volcano Plot") p2 <- ggplot(data = pasilla_data, aes(x = baseMean, y = log2FoldChange, color = log10(baseMean))) + geom_point() + scale_x_log10() + geom_hline(yintercept = 0, color = "red") + theme_few() + scale_color_viridis_c() + labs(y = bquote(~Log[2]~ "fold change"), x = bquote(~Log[10]~ "mean expression"), title = "MA Plot") ``` ] .pull-right[ ```r ggpubr::ggarrange(p1, p2, labels = "AUTO", common.legend = TRUE, legend = "bottom") ``` <img src="04_ggplot_slides_files/figure-html/ggpubr_out-1.png" width="80%" height="80%" style="display: block; margin: auto;" /> Other options: [cowplot](https://wilkelab.org/cowplot/articles/index.html) and [patchwork](https://github.com/thomasp85/patchwork). ] --- name:ggplotly # Interactive `plotly` graphs with `ggplotly()` .pull-left-40[ ```r # Save ggplot p1 <- ggplot( data = pasilla_data, aes(x = log2FoldChange, y = -log10(padj), color = log10(baseMean), * key = gene) ) + geom_point() + geom_vline( xintercept = c(-2.5, 2.5), lty = 2, color="grey") + theme_few() + scale_color_viridis_c() ``` ```r *plotly::ggplotly(p1) ``` ] .pull-right-60[ <div id="htmlwidget-5d8caa45ccaf8ea53375" style="width:504px;height:504px;" class="plotly html-widget"></div> <script type="application/json" data-for="htmlwidget-5d8caa45ccaf8ea53375">{"x":{"data":[{"x":[0.00227644122547208,-0.239918943537598,-0.104673911941356,0.210847791831904,-0.0917880713589005,0.463023709880682,0.314627344900028,0.619680103735074,0.41502490975807,0.152672420616915,0.604264329564366,0.682236716768903,-0.135407850172273,0.296146667073232,-0.119806201003982,-0.0234143270793296,0.205784496280956,0.299639996888025,2.6797251699955,-0.055813761871667,-1.02029911580308,-1.06713405242892,-0.00820169402913157,-0.1485475370658,0.753767415582456,0.453019639894436,0.00715683327611612,-0.161124087468721,0.206114987105945,-0.239771649083013,0.0641408816970531,1.29088453773643,0.156830618374471,-0.105262432930006,0.0820616573270056,-0.403216783547897,0.0720127650571099,-0.0985671054536023,0.490249416620732,0.419777842046993,0.277616965391449,0.136160862041411,-0.195018810593225,-0.121531672439517,-0.015918802680835,-0.0293008600721702,0.153811726744196,0.39757515244875,-0.166650339887359,-0.0133228281438921,0.0342052686397548,0.0995483976003433,-0.252272312199305,-0.104844641419382,-0.114240487585443,-0.0278354104374264,0.0515732599071316,0.114449795842143,-0.397699090272753,0.168356566630008,-0.162758455612891,-0.101434526828003,0.0798131712193889,-0.45071377754458,0.0573740618309733,0.060429389448582,-0.0470845802773908,-0.162671177583536,-0.125440528350665,0.152238218762925,-0.486883424212599,-0.669627363948953,0.489440890907982,-0.120069193864542,-0.531414894259549,-0.18074713290145,0.636562833544618,-0.0541038568445091,0.037972057248998,0.0178116750259057,-0.0722090723685616,0.254352828695105,0.0247973031914333,0.160415721317664,-0.174135638501346,0.0069671832230966,0.192194671127609,0.00856675455892629,0.641532763800809,0.384500422267955,0.0416551846349744,-0.0940377336776901,0.0894979701852478,0.0887113088997418,-0.125557666848428,0.0213072125609429,0.465970979895667,-0.188342771362734,-0.376081342771834,0.063814579877409,0.526752737355074,-2.03311982892339,-0.10302494618338,0.233969368971857,-0.00727239928231366,-0.241816873964233,-0.370406241317787,0.440449216219173,0.195500080751918,0.527257917863026,0.366826549722377,-0.00825508550483659,-0.162897009726981,-0.0762214261473141,-0.136898385381501,0.0978319172412978,-0.223925620285606,-0.0448618448161468,0.12632086351839,0.863214437536151,0.0241025677920427,0.688237460188684,0.107699220319637,0.331399462210467,-0.578807129598283,0.339622111849695,-0.712007148713957,0.0441241381142444,-0.121980939708785,0.0412635856578165,0.172411798971452,-0.127428929050497,0.531569450835937,-0.192004046453173,0.308436072291278,0.0170079831432651,-0.133110950647438,0.743796424503186,0.144187903968657,-1.22632935218755,-0.208976806517168,0.312065458488467,-0.32556265193855,0.468971003488432,-0.156012812968561,-0.171984015513582,-0.395691062242729,-0.211053327986248,-0.0487691613477529,0.0646771946433001,0.305655840452116,-0.135923717192483,-0.122447347920827,1.06714253945902,0.377097216442365,0.462596577072445,-0.283880245625566,-0.132123985754295,-0.107940412939322,-0.157595416782751,0.0494658748725619,-0.118259085266693,0.0840230538479547,-0.0737073544779353,-0.0224874183625884,-0.0373281082413931,0.598492764500546,0.276758466745779,0.470133176319484,0.0468080402486816,-0.229092177347556,-0.00992596177978916,-0.00617870432881503,-0.0243260820937582,0.0915323549982684,0.375923760045338,0.579774412130785,0.00158346218900298,0.0855354616724839,0.164307073678188,0.644457755594917,0.708013227223587,0.375070279697661,-0.278533211342548,0.0728440077464996,0.0785344869129412,-0.142030914605124,-0.522620677092203,0.672049002464056,-0.0277738708754679,0.680543883536111,-0.157374821628296,-0.349697883060372,-0.0604963395616626,-0.337000176087171,-1.25430213972342,-0.127589253331118,0.0628927715912921,0.188984042467867,-0.0660552872829662,-0.311522035612713,0.0762504063336782,-0.0373235727637371,0.179617761849453,0.0366824239422272,0.0726149746117479,0.266609472009272,0.193723880559883,0.34654859446205,-0.0696177618548092,0.136676741150534,0.00737179811759932,0.479578391646047,-0.155262864184357,-0.00708795952699272,0.177246755056381,0.0369689130187189,-0.086104248214125,1.48584691288193,1.34749359578934,1.71871276322136,0.494411589371809,0.0967307185213786,0.172137249214725,-0.238814156287248,0.174102650663092,0.2087320433577,-0.0333336194575512,-0.0962125191375694,0.665569087340211,0.938455305354959,0.279216553573339,-0.229328696447637,-0.0864807324763389,0.239281841470231,-0.100204276552528,0.0406680071936135,-0.39149173580883,0.146778575886616,-0.139184934649015,0.720279131431292,0.151141051276528,0.515612907099692,-0.0437326242950905,-0.130822402528658,-0.512919290672798,0.125007704239398,-0.0439236537300564,0.193623772496906,-0.143558772408809,-0.127944121591958,0.140370431128182,-0.204982922106306,-0.244039451550992,-0.219650122019594,-0.0380487199093277,-0.0788190663206405,-1.87469624645493,-0.165110075642968,-0.0430071544208684,0.479282815377848,0.0174814904716664,-0.19966533348418,0.287817786998464,-0.184848940031489,-0.118274144694849,0.273075678265196,0.0761105871064977,-0.0472725305545267,0.154794035150833,-0.0393665636230119,0.222884777711839,-0.149600559805767,-0.100798308179324,0.159097281610265,-0.254900299052761,-0.0132755047782184,-0.0327571452555086,-0.10360494870708,0.210252895754403,-0.195487825101382,0.136922777993764,-0.0823520323146857,-0.108623049638787,0.144859803244416,-0.0720747734429808,0.0625719822377154,-0.108718901645322,-0.0438743628800586,0.0760703944176457,0.00433928639471237,-0.747749696427826,0.0142504219199082,-0.534733366328332,0.025893305057006,0.771387292270866,-0.0401334720258357,0.411107893341524,-0.0787446107848092,1.01053946981169,0.0312711238281912,-0.0724050103091901,-0.142202405238983,-0.0981183280226116,0.116482868242301,-0.115152530348234,-0.10916001534704,-1.51317096779296,-0.0433770801274454,-0.156916114202028,-0.176615729450692,0.232703779255794,-0.19663034271745,0.100548491678535,0.143552618983331,-0.202468060448102,0.108138026970402,0.349141815694925,-0.130907713205666,0.0222113797869765,-0.195569128079435,0.139010943392506,-0.530602331378846,-0.0900449959937367,-1.15632228016232,-0.00259013721989301,-0.231334909875608,-0.667041939911446,-0.344781805431193,-0.021963493251632,-0.0456981574355427,0.121222100483504,0.203491959773895,-2.1316844758972,0.251488589710459,0.219074811042964,-0.174952355803762,0.349686372061708,-0.00625304555105879,0.0552962762502903,0.0496953984755119,-0.272579078599242,-0.0923470496200162,0.107825486366306,0.170315570222073,-0.872875538564755,0.112777173686055,0.116029834653025,-0.255411182334242,-0.0269612836304957,-1.17710932493337,-1.02656736496906,0.129776675049358,0.125314054800297,-0.11719458402863,-0.0897271884864987,-0.0945143314620591,0.0416630604291866,0.0732162427180959,-0.0419929428426119,0.162021542827904,0.110481539560505,0.144466771359269,-0.0492023723019648,-0.10960884719187,0.411904568124252,-0.0951335541054474,-0.0124856734919455,0.149983929867273,0.928203434808945,0.0744872673132483,-0.139071849045409,-0.0602658749127277,-0.0253746499384273,0.641611140394603,0.712552611239184,0.09477161051701,0.0982567001055591,0.308592047177623,-0.428690470128323,-0.230781621005786,0.0560458349320821,-0.361977237027318,0.0386114277447523,0.232232734990997,-0.0705335774526275,0.918494504357451,-0.0111173105782719,0.164706668653248,-0.101849153741547,0.792558747212014,0.100956867513895,0.410264473581822,0.204033109497061,-0.210799641925476,-0.156139189471765,0.249866988727186,0.0914645247784538,1.01576550018695,-0.147041667449602,0.191349303989207,-0.206640691258551,-0.0664202196896826,0.0235641256336705,0.137813798317082,0.127456063134976,-0.0633471377719746,0.283511380595693,-0.173489723208269,0.557226700416612,0.218540006512487,-0.189229812538684,0.0521424611793421,0.0666654543085484,0.0231340320188958,0.051620520025166,-0.0298786827858912,-0.100380725627409,-0.226378300445985,0.560486137770777,0.0194500018255768,-0.343026359047099,-0.468667111511018,0.0719216010512966,0.538498867915147,0.285320926387168,-0.188612033135106,-0.363353913756798,0.0958975566285064,-0.160803150218091,-0.0342977281141243,0.0444974339741277,0.266933017401816,-1.79952704829131,-0.111907884009318,-0.0447591014076425,0.0676517796331192,0.78746225148451,-0.164100721350985,-0.168130949654539,-0.30255529788074,0.339285373146778,0.00858680701537477,-0.110822222889902,0.0396232504335719,-0.0050331185070688,-0.050253016712492,0.00314624329409809,0.130415421721941,1.98177497294926,0.070414840533073,-0.0734925463125775,-0.0141758594787541,-0.057836262021313,0.0307001163288055,0.142154008351458,0.127466968843638,0.00917972561302497,0.192495766249285,0.06473765040331,0.0869310313457575,0.131853650049591,2.15369363558488,0.729521023472057,-0.407623119780023,0.14733115546917,-0.0342008367303126,-0.0526868639308813,0.124907101543429,0.0027190838193616,0.0396936122615657,-0.688948463162318,-0.459817284732983,-0.102052221208218,-0.342829151498821,-0.115591789475343,-0.320511112600439,0.00584759359826043,0.0718073734216663,-0.0404147337999117,-0.0882775964355114,-0.229948180680616,-0.306066332502422,-0.383977122000155,0.124824902321778,0.305125748501814,-0.136793267416485,0.216281889073241,0.0370165596119904,-0.10631097644967,-0.31413415419259,0.222291490936621,-0.00602327869512555,-0.0308294350117739,-0.128988886046473,-0.0647848090977099,0.216901907384619,-0.516841006006912,0.160904406494662,-0.0311860200234071,-0.224404379488547,0.134920836027455,1.07326663089927,0.286650146633057,-0.46231490830822,-0.164019809411793,-0.289449887215874,-0.0845800867456743,0.516091658155508,0.206678982917207,0.759127640080204,0.421235747808706,0.465665256265404,-0.49716031355779,-0.435282035006355,-0.248799424018415,0.000944170015803771,0.920766845701198,-0.102446107575214,-0.0902102469787988,0.455461376692379,-0.283426598757453,0.158908279774889,-0.097250495191655,0.0295367998460793,-0.0189199768981334,-0.0897755528145305,-0.34100488930964,0.834069036071159,-0.435912757537887,0.439547090509414,0.576068918262024,-0.04268908174961,-0.129289769562901,0.123961871464354,-0.0369493003026962,-0.215939484989812,0.406538934223614,0.0353275361991336,0.043175376429001,-0.142059842134494,0.0257004496709498,0.0839717666340494,0.841696618493023,-0.0190775003269182,0.0435519709593388,-0.288026227047818,0.0212271503713136,-0.129794813510837,0.541873114895258,-0.00994409166956245,-0.0432431020833011,-0.197370476920204,-0.445857188081994,-0.320189640155586,-0.153366660349855,-0.0352670296879148,0.644977245323191,-0.242622758022187,0.108148567724824,0.0937162228621596,0.55904364900478,0.217596910734393,0.219833882175417,-0.787164339663074,0.0198068556671739,0.0299018185752471,-0.0485443989955841,0.0165147358133088,0.135388691483051,-0.0636498791995527,0.0677430145404785,-0.334070890745318,0.27753451874563,0.0829574483498366,-0.188701061105654,1.0786533427867,-0.204176459778429,-0.0981500245722108,0.432409013641907,0.11024537801735,-0.0460153529017471,-0.196387403274663,0.150210257813825,-0.159024696301153,0.0480995480550795,0.558704363591123,0.213952121083505,0.244581120151493,-0.282444923135763,-0.170245928343441,-0.325032250378228,-0.134640060439457,0.0358798204300643,0.0671969905364612,0.913795076355343,-0.0271872080699709,-0.0252254411261143,0.324029998028436,0.405004605205452,-0.212349346597363,-0.209959883455319,-0.15275648536572,-0.153643592713406,0.026425508164365,-0.00423592814325173,0.403005512541168,0.0401653942344626,0.76963495453521,0.512071338285459,-0.11695102115112,0.0578558147626392,-0.334727617807191,-0.473734637191236,0.0115489638299034,-0.0247158630955737,0.0353053831254702,0.143406570896685,-0.330028971887597,0.118699497652938,0.0070787569148182,0.249648944722804,0.704218410389086,-0.12846342336279,0.0229362096380166,-0.738145622798422,-0.674573142778526,-0.00946289752480699,-0.149440178210411,-0.125905413384926,-0.177363845497006,-0.276194573668911,0.0884138018916251,-0.579613030563586,-0.0281803734127188,0.100296133143514,0.049035904521855,0.135163977210609,-0.209243869773489,-0.104067115829065,-1.42134255839859,-0.211622831652762,0.0422616058474335,0.076903946581721,-0.621680971652643,-0.365054436928713,0.118586063112651,-0.297572524916891,-0.0924528396025743,-0.89655952573601,0.00906126177080744,-0.187742590679524,0.73491609188242,0.409616679230825,-0.0834813669095003,-0.0666466889535452,0.145374745469035,-0.45990349189237,-0.184174601215762,0.07961453057247,0.0887235933174426,-0.818673741352393,-0.677588206426555,-0.0713370300970556,0.00917326569366217,-0.0695004216982163,0.0953828827874405,-0.158977149940068,-0.15051285213725,0.12526010139699,0.438090576898145,-0.189456111112475,-0.197457127539847,-0.0426100656401202,0.0259097094715988,0.0864261957097102,0.137331239324141,0.0176912689416046,0.032353790441972,-0.149773252003318,0.223233735932551,-0.263135037285257,-0.109672897741487,0.0528867744469383,0.307526869956057,-0.129638419789524,-0.0298537731824548,0.358236570169164,-0.347118834170974,0.087383569380345,0.274733477749459,0.0809172791996673,-0.61095911944209,-0.0428952950346654,0.0955168132663633,-0.212770295416745,-0.162472216957366,-0.0193957433395908,-0.259587729411739,-0.330548126897192,-0.166216488384759,-0.124699747404675,0.0612607580202686,0.109033743751149,-0.0783576283531024,0.116437613438027,-0.632110483356312,0.66742470351208,-0.0729517278279335,0.00437544353114819,0.142117506790122,0.20030781407628,1.04946071619985,0.212359318648962,-0.173607742085926,-0.00916168808313849,-0.0668517286439928,-0.0733064970442448,0.017701758021694,-0.333696884885658,0.0788639550618823,-0.193749752627287,-0.672919764002828,-0.0742453609237802,0.0137794474306713,0.50698217092416,-0.238189046811646,-0.184657568765109,0.0693210540552149,0.176486224762815,-0.710189064393179,0.425290729497923,-0.300208541964797,-0.103765869711503,-0.0320195950432755,-0.519025780951642,-0.0496542843443175,0.106917678247913,1.04772360713707,-0.190496293144424,-0.0481854547670108,0.0207255877097407,0.0468012969836973,0.297086518340918,0.00433477271867942,0.121117475317804,0.0075830820317023,-0.0386052892340013,-0.119609159548339,-0.540658406011973,-1.43796776132563,-0.489407867174956,0.22718518697554,0.451586859060363,-0.133572817837837,0.28870656075708,0.0789125298552669,-0.0542387165448331,-0.360545544663641,-0.153642793713079,0.560083816945634,-0.138532982784666,-0.0959756190896534,0.130453368236029,0.129846222942513,-0.175650925296594,0.24141208467538,-0.071959758399816,0.0737909681608495,-0.225935539417416,-0.382676501951427,-0.0082074907014438,0.195363875272098,0.22707450411376,0.583314474280141,0.0248994445107773,-0.759644436583624,-0.161915091553129,0.387013966256901,-0.0379839694193347,0.139893714913318,-0.285298885829616,-0.0633455148733615,0.0398360875858034,0.0446868573863507,0.0598628624699902,-0.157550899983394,-0.00831295759878562,-0.042019087309501,-0.0473758579978174,-0.254728279658654,-0.0546742291204253,0.170585527386525,-0.565230755512567,-0.635771050681603,0.0161265149537542,-0.495940579813673,0.150621763917892,0.892877046219088,0.109804603537408,0.0219008372045817,0.134034611849148,0.410235406949008,-0.11925854437607,-0.0928837398817691,0.0941490564978543,-0.0825845281781106,-0.123816739530725,0.0838010108003058,0.0762685171114439,0.052772568333255,-0.00529197159619943,0.0867836467018705,0.0625100273126355,0.101370305951158,-0.22724147844732,-0.0406249102986885,-0.220948610277625,0.502656791692075,-0.743434063741567,1.65833408756444,-0.158617782036792,0.290099366450885,0.31290057259419,0.0450691666402124,-0.227429287312655,-0.451854349734859,0.236051019255315,-0.195834157387752,-0.515717501216615,0.300455993279255,-0.0915954507579407,-0.233925949405875,0.0714065506174753,-0.659148436890218,-0.0671080210127576,-0.178638536177241,-0.087143005529307,0.0789623187291068,0.096639585979757,0.636798716324416,0.149726200964056,0.269743965244014,0.196490877241668,0.0214777014145808,0.00515872715461912,0.0423710439818559,-0.173531110006109,0.240361722347671,-0.0969154885204523,-0.028511637377066,-0.0198726939150267,-0.472427648098633,-0.213475333556036,-0.202037519834076,-0.0971283303074626,0.445696482132746,0.0571539309328769,-0.103898164672864,0.221249613851741,0.207311898638664,-0.0669215442670967,-0.213469122235827,0.863952929754299,0.225248294177716,-0.271901689560813,-0.157606048442757,0.462588657142522,0.118571039488947,0.0441108931903427,-0.184901225532028,-0.260942838126872,-0.690320826101334,0.0605626451980772,-0.0633745574573211,-0.0336850136051658,-0.156823793598552,-0.167828454979907,0.109604384851634,0.0573129814467114,-0.169328342837953,0.177348129828096,-0.267484006494957,0.361764956634125,0.758191870284167,2.04874033063349,-0.191763527861238,-0.756604401228491,0.032693188149663,0.565965878817802,-0.0883083487587348,-0.134763365575745,0.266135723343561,0.674133291498302,-0.186201794460805,0.0933892582381892,-0.0698774808120101,0.0132458063071897,-0.241336718162612,0.356584963257779,0.00120171068238756,-0.219219325137263,0.0552562088880677,0.00882769714677802,-0.0604741370277369,-0.0768147645689498,0.0214056424520583,-0.18212798215983,-0.0264510615938768,0.229363631127813,-0.136475818545267,0.214133569899151,0.142454490001038,-0.182151949909549,0.129549792304218,-0.165241518285416,-0.0748448553831468,1.26026744338738,0.602683422971357,0.49155069770637,0.311190142584345,0.0591031172509821,0.449926911885098,-0.108424005641451,0.00778550956513394,-0.0449143179198596,-0.0633913129651337,-0.738015190363945,-0.033913825121527,-0.145813180887719,-0.313967357267895,-0.0899231771431792,0.123268548041811,0.0548351492329455,-0.199433009868537,0.92130258407855,0.619636121538752,0.210438218090365,-0.373538560992209,-0.169671793152693,0.100588788461762,0.0729537283167931,0.69984290241995,-0.146198274025214,0.400205436749256,0.0609871135012563,-0.579698394490877,-0.063855838291327,0.117783424551034,0.126459816134725,-0.213820605173365,0.0571188204653573,0.135467774685212,0.165864450522101,0.231766569319013,0.0803979570283129,-0.336422795577571,-0.0766195887022146,0.435747254150721,-0.0825229502676273,-0.222696661075226,-0.0635229273384202,0.0163674637438169,-0.242977189315513,0.160152902531294,0.414242681646083,0.37900435442067,0.00059705748484159,0.141417797569271,0.0582516643755901,0.0228702201848614,0.129452837060222,0.437603155010036,-0.038183697308638,0.433606956182216,0.112317175274174,-0.125792213414189,-0.0179426375133924,0.44372150305529,-0.0486785657922247,-0.078588986110324,-0.348268867669048,-0.141136215891088,0.245613490980622,0.38393839383787,0.023547138863847,0.0567100942614934,-0.0807328170184689,-0.229754653010074,-0.0064605264696973,0.0251056478917382,-0.490792399488406,0.112164454160149,-0.244137810122878,-0.00797469524427933,-0.219491717436165,0.199418542039683,0.402607154201337,0.2341783245962,0.188897855556067,-0.135992381974552,0.395500275405311,0.00758819856368173,-0.452887296654503,-0.0751709082151723,-0.484894366558172,0.0874182490221023,-0.0350097370710017,-0.241507396462437,0.444674575265236,0.0401382635378429,0.382498293065537,-0.266317818407969,-0.0668009169603145,-0.175739550058378,-0.138629235726622,0.114316762401186,0.00796457228953585,-0.0225403578240079,0.121800074101732,-0.632257333044175,-0.070550127124317,0.496344506085659,-0.292332522536506,0.636939847586474,0.630794210966737,0.401380139620367,-0.0881549899504031,-0.439426628485963,0.375835195753448,-0.870117276536815,-0.262978984858127,0.563187560011385,0.415997796696377,-0.435282635093765,0.185323115881387,-0.0318401708716558,-0.830662083712955,-0.131523540187729,0.133262415919859,-0.0734632794942002,-0.0478131617348246,-0.31182186628501,0.373737477521028,0.38400437007897,-0.00370023705991753,-0.0685013506266281,0.155327302905413,0.0589291607162988,-0.094262706721016,-0.262684240569188,-0.198281524730364,0.163889234222372,-0.120980121642881,-0.111798919774845,-0.0118237947367044,0.105703848159836,-0.0818722478494703,-0.0938824733793286,-0.0956847685294328,0.486726756312355,0.722025606451277,0.210168933081975,-0.517371514499645,0.0768472289353735,-0.05689605875675,0.184762525605849,0.270592806051303,-0.146150149921535,0.212939186106253,-0.353520542224905,0.598740436975375,-0.137247639305056,-0.128669808046342,-0.119755098038065,-0.00390017154310366,0.250460913025776,-1.1873154134583,-0.0023887418594898,0.0437367567455533,-0.0520588203383684,-0.186206995742792,0.182247026908635,-0.205111718117564,-0.0258838287022916,-0.3587590799928,-0.0942087622668796,-0.00538187415655972,-0.0140962796540882,-0.32052354552402,-0.188424117096569,0.331604452134627,0.0678020865077751,0.483646266102181,0.0376624173339386,0.560905224542734,-0.0363726219662155,0.0516459300844757,0.185301145876116,-0.434105638042688,0.113477996308776,-0.350628980076537,-0.141136878487718,-0.074233657767547,0.223887931446195,0.00630734829845848,-0.0636546028677679,-0.0119780491577503,0.233533854518378,0.21019286405635,0.11887589403172,0.0101153854147673,0.660713840357273,0.13176958225821,-0.148642343920676,0.212180160561969,0.0326651240552448,0.15208247668046,-0.037775111268991,-0.297238369051831,0.251713691894676,-0.0395176305508545,-0.226928619459004,0.0124180190950857,-0.219935615641596,-0.154132271444339,-0.148882034175925,-0.0150820260551246,0.00457668760361989,0.0735843062965342,0.0677757974342343,-0.261049873311346,-0.390004440109051,0.190368700596163,0.14112554780373,0.060843232022839,-0.050366197979574,0.157214761598217,-0.0111231039142006,0.108641882561392,-0.119292420274754,-0.00101282742895375,0.419511637005429,-0.0509289791402798,-0.0815940010286738,-0.288444317102543,-0.994260540252165,0.0311929801274435,0.0639650487070924,-0.410668093883977,0.227953924564934,0.0355678842976743,-0.15931566579038,0.212986070263948,0.276594269033003,0.0222767673250116,-0.245756280440818,0.208447848161172,0.0106588602127708,0.380932202557967,-0.174629894065345,-0.889615679465129,-0.110672285928989,0.531342280618101,0.200030007976761,0.0476443551951508,0.0836193422888099,0.0372018766550319,-0.168491257079681,-0.440196540232498,0.303895901710792,0.06838647636624,0.057086820504575,1.06673264075735,0.29380785732056,-0.102962278539435,0.551186312816521,0.391715208397667,-0.924081869769849,-0.297788328449889,0.208538948173655,-0.081567425243777,-0.375932561657832,-0.13291630949899,0.623748597894524,-0.152057431235314,-0.07044036053938,-0.143728218753575,0.348422367744022,0.00385016201030365,0.107036909254409,-0.151076288159106,0.00802600051268362,-0.260242230963579,0.221141571043903,-0.273745110622496,-0.266760858867366,-0.172106796190128,-0.287988315827927,0.217081957216896,0.0434771902503871,-0.0636753315802037,0.0401252410856642,-0.866944882829282,-0.139221745320675,-0.293979110582086,0.385055000650678,-0.141762512707266,0.525199537542111,0.232035437446841,0.0419413124486692,0.0115395962744783,-0.239716672941975,-0.0990066150613232,-0.322498374810246,-0.0715292025473925,-0.48698490531548,-0.107338433547157,-0.153078198888942,-0.0262259458984721,-0.424777830892647,-0.129218572017163,0.163827180448873,-0.234281259952505,0.134078179231379,0.09310939582474,0.106220182140646,-0.103420339365064,0.100659200377547,-0.150068834756144,-0.0521288565720542,0.162482263276885,-0.667344239446303,-0.478416756115669,-0.0308099740617827,-0.111385747915631,-0.153771435271347,0.121839609146129,0.37327718740694,0.118385049493091,-0.206874195865559,0.0142431010996507,0.526773222139802,-0.0748834537742943,-0.0778073533136653,0.238930279981555,-0.109864142864319,0.00815726659213836,-0.206135451022623,0.529638839792423,0.11428934832073,-0.0207658396328644,0.673315786331327,-0.21368744879307,0.0783044569048753,-0.0614152369083038,-0.0560130704451487,0.426703112540128,-0.0198776628212268,-0.0221355513624719,-0.11359171661835,0.331101860656707,-0.0316415068128156,0.0528818625181344,0.04952789614731,0.162927449169547,-0.270520171499011,-0.553254195696124,0.0568994789276077,0.309045726222226,0.153658264164522,0.0606725341421008,-0.113298226926203,-1.97364564178577,-0.19686573085662,-0.29638386521994,0.0432809876620039,0.0891655772445324,-0.314852904771119,0.0213553080540592,0.292005499962153,-0.179644655650553,-0.336405185833585,-0.439237117007696,0.0254508163178749,-0.0658993062811248,0.275519500094105,-0.164994692159479,-0.217893053622903,-0.207387823893695,0.0483377180021685,-0.0268951013928223,0.116801500241961,0.482133706869175,-0.127651826429824,0.0118899472731272,-0.0143524940960999,-0.104700798544207,0.62435143459728,-0.227590607892881,-0.227005481303037,0.00098079544733495,0.229965766765709,-0.210895702640229,0.472596771369417,0.0442192037963237,-0.131576417026513,-0.0799287718213816,-0.131137948175491,0.286930286907236,-0.274618048331895,-0.107653099745292,-0.0799632780529776,0.0833586167726463,0.110406000867179,0.471487315694418,0.12510361980489,-0.157714862130156,0.153238526332059,0.0505078845528633,-0.183194028398082,0.110822720033095,0.000510246026508002,0.136729513246548,-0.0634107853273338,0.047040526064619,-0.218966875553474,0.00045310670708472,-0.230409416010215,-0.0170902602818057,-0.201264128367049,0.091243421392844,-0.32535755526086,-0.361277023188176,-0.0675989694746189,0.0106915442107702,-0.0945919996596328,0.0251902376996544,-0.270177591605771,-0.196320406840047,0.289034510664705,0.0273882357247411,-0.0265598536630298,-0.146315985113231,0.103853746248492,0.638526639596514,-0.0709355441903936,2.12297184804257,0.390396237128291,-0.157027086966031,0.330653954381873,-0.679623832402112,0.103206849931078,-0.185528106028996,0.259490914562216,0.014028189863014,0.0736352851926536,-0.137506813153918,1.01964031285675,-0.432600137864503,0.00725173511945848,-0.0217613708590119,-0.123265364573531,-1.15438608513958,0.257285381980408,-0.0146563569848312,0.157084908001789,-0.0140492329805238,0.18127626439411,-0.150895749805243,-0.171284791270144,-1.48961963307351,-0.110461880391552,0.303976308485477,-0.353024666341859,-0.387103451257025,0.0503632757982173,-0.05679345815292,0.643809775801642,0.0729079133164419,-0.116741492559159,0.0256657779689111,-0.428514444391559,-0.205158959903789,-0.350594996372284,-0.086915302192106,-0.0825203520437123,0.438303576052918,-0.775659707235641,0.281165241740919,-0.0873581119049615,-0.200124351322015,-0.0273635780888044,0.089485895457568,-0.0381068511023609,-0.234611994098953,-0.0604874798670203,-0.129861064089432,-0.372197746099503,-0.162010818918797,0.0364756685609209,-0.194427447778915,-0.212515905600906,-0.0717541448945138,0.365818254014609,-0.0108848605904444,0.255260797700713,-0.0682948019055557,-0.215618158615473,-0.366756444006301,-0.376685331340298,-0.141069082717937,-0.109516201812024,-0.312750504850788,-0.246340053342539,-0.0751185104524109,-0.0348488510722151,0.163696036780637,-0.0796616247580436,-0.032562008124143,0.0348356812507574,0.0357224968810741,-0.293046844598906,0.0578966451789488,0.137889054017997,0.435593152045021,0.11966298865416,0.0475329767608453,-0.120889161609164,-0.0117533029641729,0.0780584101046448,-0.0677488381371736,0.203613592576059,-0.0677686035179718,-0.0355644458752147,-0.122242750328786,0.194172598022396,-0.0146800831327006,-0.155202973129316,-0.11411848132417,0.173385729710487,0.0254350429376615,0.00803046616611324,-0.138621836081798,-0.116126105078468,0.00963190488688758,0.159996587685907,-0.0931948495854092,-0.130192221655232,-0.0966311571117085,-0.252638731741847,0.261380846368416,0.0271526638556033,0.0120698939814855,-0.856608963064505,-0.187315798144074,0.251543004783621,0.0781323588071178,-0.0438140092332004,0.220668922842196,0.0143025369287205,-0.213712752626129,-0.471337352082843,0.0231277777098433,0.00208638145821978,-0.235560603233298,0.986768288868347,0.351885455886685,-0.11075288360342,-0.527000015295608,0.00667983739925366,0.727647025041747,0.0367085869939905,0.291848138639882,-0.193193666414952,0.00366626852723903,0.0826822139616456,-0.194430167987428,-0.186448958464771,0.768151800131636,0.119677521381125,0.298251136382648,0.101227968471779,0.11925576178123,-0.00289772643667846,0.213276404098061,-0.0652811338312847,0.118537176122273,0.0817217192145218,0.0932700997465027,-0.0181894350797911,0.11834604189532,0.06137136304182,0.195291933837909,-0.0638238345729498,-0.0882513792873526,0.0485511035795212,0.0794899919369078,0.0394951081311321,-0.0357102862770193,0.473562726024879,1.85878880737386,-0.0431583869451886,-0.0433166692293455,0.14034844237401,-0.149662001502148,-0.454028906242895,-0.00670713991466573,-0.00555457569076347,0.127503667365476,-0.300049403531638,0.238846687631019,0.16801192798249,-0.0406569642262192,0.160602737323255,0.193617632389518,0.078069934452714,-0.383773962352726,-0.326261927568396,0.0939031299591168,-0.122856069989631,-0.231582522261085,0.337407742400987,-0.121177882043191,-0.0803711808198887,-0.121338047059823,-0.0768748501390029,0.0500997441008571,0.224947614394687,0.00517091617698925,-0.423968014005759,-0.0839833616394793,-0.0624209885002283,0.484842851816115,-1.52534930097457,-0.164868967075975,-0.381828644225184,-0.0729237540077598,0.10062297721029,-0.0109165286504091,0.0826751537991106,0.266121960568699,0.00898674486434761,0.0302542465297291,0.147567572584528,-0.0652722490043435,0.21405640012863,-0.122425950841053,0.0773670609542034,0.320673020159239,0.266876292362978,0.196683034419736,-0.117180637311157,0.792450875351244,0.157671008042037,-0.117427879108489,-0.179253235189113,-0.0195198680654404,-0.127842872623166,0.172810114939285,0.133908668181228,0.517406099063479,0.0483864583145193,-0.323047706093767,0.0307193214646145,-0.396075998784127,-0.0980520486801726,0.31058485066322,-1.57163653259637,-0.599143822992328,0.489996939628936,-0.0370085934281994,-0.109065598708573,0.106555952299017,0.0391239052620507,0.108430721057109,0.0305741800799599,-0.213608109869058,0.000869256772275875,-0.210811341256547,0.249041968387955,-0.152121837005607,-0.230649834955303,-0.0180258102357406,-0.324455679245205,-0.714181042137659,-0.0943951776425708,-0.328438321555001,0.254606359839378,-0.00122635928557263,-4.5858334065302,-0.250051156555414,-0.326816355129984,0.0727790924206477,-2.28669846362272,-0.287137523489686,-0.0996114844043616,-0.0366275899860513,0.0766847187586033,-0.125980239496164,0.159840217568859,0.0399878938352561,0.147408935311258,-0.113077791408812,0.0241310466248073,-0.0769413104157704,-0.00349129802416512,0.115621375586854,-0.241074992813331,-0.182915696857406,0.620826432359485,0.117447540728677,0.240075781203302,0.0775989969971388,-0.0878850235309588,0.0745449254861948,0.0333782128262588,0.123449500750725,-0.275224541954593,-0.19899734181035,0.185743543115145,0.158614181771064,-0.241464650807905,-0.0679396491871126,-0.0573118305656462,-0.208362979552793,0.0584727416043617,-0.108929591766428,-0.0244818287181972,-0.0726866764102653,-0.142381180630735,1.50345632459605,0.370415754365999,-0.108573078861161,-0.0662891378919184,0.0691422714543312,0.381334158074579,-0.103599267112772,-0.15884116852725,0.207586048007828,-0.226458274138404,-0.0918629477645724,-0.546314643937838,0.0285777910579217,-0.0709175797935897,0.107580535403972,-0.0131413522656212,0.0837426712376444,0.0483449093785433,0.273819299537856,0.105223297737595,1.06780241354292,-0.191021493249318,0.0681224738916538,1.48298474872701,-0.109567800500185,-0.175808373706927,-0.0415509860644062,0.141189908782023,0.275593080809207,0.199801783844818,-0.190366447579931,0.173588458357045,0.120432796825364,-0.0150068696995496,-0.0542730728679102,-0.12269474114966,-0.124613402346142,0.261789919213505,0.00354738997688111,-0.145834181813181,-0.166393188692327,0.00491363369938687,-0.243676896533113,-0.224645126983578,-0.349051097068961,0.0209683506591565,-0.0201644179556432,0.0782702850381341,-0.841045656468268,-0.11514773802314,-0.0265019724368275,0.64278915781179,-0.0656953186285606,-0.153207599844641,0.806254382143483,0.0246199271958718,-0.122120451157353,-0.00820095127690691,0.153259813273746,0.220897339671474,0.717773633567037,-0.061516313873677,-0.0476191547249565,-0.232043005397568,0.464713184637045,-0.0781330573905063,-0.0156956505125736,1.84661581409003,-0.0664929508183642,-0.0794100860800435,-0.780974148507986,0.022684552225464,-0.0511642586532319,-0.261381951082575,0.142540880317012,-0.0758585686566349,-0.503365282354722,-0.0480525034754183,0.208687951514302,-0.146120851689137,0.214524417540184,0.0383037094691722,-0.172855354100996,0.129262736428735,0.0163436837656029,0.255308394274465,-0.0774251167274215,0.0383457850594921,0.395695650661144,0.168743806635996,0.371679081577316,0.0611153502076041,0.0684283880566439,0.547854764959116,-0.49450961247938,-0.330309152979494,-0.297152256946962,-0.0793241799220187,-0.373523575523975,0.30978247015228,-0.386161346224974,-0.344193841729399,-0.183391402893322,0.268220390941433,-0.221401610254454,0.434393945834911,-0.0450554921196202,-0.00168325388937576,-0.109469162383756,-0.00741057827716443,-0.0573181806479332,0.239492596950465,0.39445437713934,-0.571482211979631,-0.0337789413045622,0.103227841326689,0.0290094825467117,-0.0819632966865882,0.160040370750011,-0.0907591717220929,-0.165252904826079,-0.0879193973739085,0.0411171775061354,-0.0611673757312302,-0.158364386119265,0.240552173652906,0.264678662796932,0.0415841923108643,0.151447637084095,0.469521532255654,0.106551837788595,-0.0615757348831501,-0.125174253521432,-1.12522735975382,0.0593370133581705,0.250711398564347,0.065108313478759,0.333396848561548,-0.021064260918896,0.0961308410338994,-0.524958837120895,0.325490045528279,0.0867883132047226,0.151271109101355,0.0627305777669743,0.0664439294757698,0.102719869955143,-0.179905684603878,0.077590822640909,-0.067395142507949,0.263113756887842,0.434097575695499,0.0270254399650005,-0.129720296503728,0.282865699793528,-0.0193096387198509,-0.23317518806915,-0.394683528622889,-0.55477329209655,-0.0940333048005935,0.299084879767658,0.228551907698009,0.391183841967426,-0.274269724121489,-0.045509921148503,0.550054364590538,0.237229242811339,-0.206903574633095,0.684885204250761,-0.067076299130344,0.39487793586968,0.119982088977212,-0.0867390963140259,0.0115335826204851,-0.22134959518394,0.0783379591262813,0.075571048344305,0.081579874248537,-0.0278294757545383,-0.0596732673984027,-0.00783122579290206,-0.0724912974035992,0.753232980594171,0.170478142535863,-0.0632155524924807,0.0985109163462801,-0.234809814419162,0.221902356177976,-0.0106592641404878,-0.00416943196724867,-0.211750580870408,-0.0181291566990933,0.110442713995622,0.104138646930001,0.428836764342625,-0.248404527783056,0.0193796288023799,0.00865218740123153,0.228104887919919,-0.211732695765429,-0.127072562902285,0.0472053984597926,-0.288352198475278,0.102626480863794,0.134923471017414,0.0430552359204078,-1.00613962942262,0.221210783883476,0.131049821829189,0.343427627805179,0.19253940202362,0.0756639711619601,0.0999410411227116,-0.0962441268160283,0.0432262530198074,-0.283949595542806,1.15610100361786,0.370057923639593,-0.215812554706061,-0.107209891710763,-0.0324182966947136,0.308635176306625,0.168192743721873,0.0914223526609238,0.304526305232252,-0.425626179620746,-0.112458729207987,0.112441279298915,-0.0703200126777791,-2.36245863367408,0.17952016524022,0.161133096964597,-0.00389718689604192,-0.106586760481192,-0.0466020457053416,-0.428046695492086,-0.0769365367959904,-0.344683647736504,0.534150172216047,-0.907407541273841,-0.0310400155178234,-0.026170285886948,0.345208318567025,0.226169683362364,-0.246487045247283,0.136842047964484,0.0183822072106818,-0.014095747312905,-0.134953115072752,-0.0432482292496397,0.187183770844298,0.0169353624676038,-0.0432928001940328,0.0422662329033678,0.342689283254569,-0.676409486459506,0.11474127217381,0.178879117520886,-0.0141864967258294,-0.144898429404513,-0.011338236675148,0.0309843248086637,0.182619612034602,0.085891541180188,-1.2691201926889,-0.222926206602472,-0.106925174392094,-0.329812845734919,-0.189623423053226,-0.357833939801918,0.255421634367937,-0.41723254086787,0.156854793785961,0.0429344311076857,0.0180610523061692,-0.0131903519329608,0.19569528247016,0.0512850083781052,-0.0429382394913854,-0.00306136836468292,0.0252046502041168,0.640107603735958,0.159521131965995,-0.0806303129597269,-0.00675320019068008,0.018291695897588,-0.0726398600646109,0.0194684626997219,0.0552886561632121,-0.257806196199918,0.130305190103229,0.190605267507575,0.0139643353094545,-0.659336551046707,-0.0323197043349806,0.0790068155160438,0.214989810931617,-0.116599471827999,0.141658637590007,-0.327370109978179,-1.04369213169322,0.159135422272696,-1.19202209332487,-0.050176475884094,-0.163015545310798,-0.0327380529774354,-0.161773972742726,0.0853909406753669,-0.0434805946780982,-0.291546277670847,0.00740495199490419,-0.0837210143569565,-0.354273582537947,-0.0394441270288849,-1.18422117596218,-0.0924903616199637,0.0535124211883474,-0.096061928432517,-0.546918777659046,-0.0662751532552345,-0.0125031020017738,-0.172545532263608,0.457616576722396,-0.101703114924625,0.134494313287149,-0.05257378026679,-0.0087834210497792,0.134580165630928,0.295196824997518,-0.125718278065034,0.0628014406495232,-0.0179919557641824,0.0400445743561551,0.0520436724821029,-0.294334831587166,-0.179492110599819,0.0130708428059182,0.0845163007114809,0.0227978811751326,-0.207342095992265,0.321903197225229,0.900861688726965,-0.190417690075087,0.196771337156556,0.18494059040419,-0.365845060064594,0.315250655677667,0.249893625242298,-0.135696583979295,-0.118370381026931,-0.0733628579711635,-0.0729482431351239,-0.106312667948141,0.134193592280121,-0.0247405997288592,-0.0226629587259671,-0.119290469072398,-0.197467510370637,-0.388218273961427,-0.499308937784775,-0.147340309515291,-0.044239135817889,0.118309025124212,-0.0841775194011405,-0.113970446324183,0.507972657441811,-0.442802574803888,-0.110528281905307,0.181274876739348,-0.109260533264169,0.0293014640254034,-0.0256949160552008,-0.182447496425686,-0.897127227307733,0.259941571443932,0.025133162672884,-0.1595095744774,-0.0680801519504688,-0.507207913480128,-0.267058320853029,-0.844877551867883,0.0528119507856907,-0.12317372413368,0.121293115179192,-0.793556444539473,-0.231267188104525,-0.181479618466859,-0.552593526507504,-0.178021501039311,-0.52496594495236,-0.834962587203258,0.648549109010067,0.253394895925924,0.10271550233371,-0.334861399750752,-0.00426886129761274,-0.124680248760534,-0.580852483503268,-0.090885586523123,-0.228503470820991,-0.116004224546283,-0.227199460213429,0.0634239867848848,0.0678800048378501,0.0374258053537113,0.0722855742333091,-0.0112251759497801,-0.306978503309445,-0.371777683784633,-0.215594284352156,-0.0728471937075477,-0.470472714414822,-0.16152212362067,-0.0944883811640601,0.0115827146390909,-0.142252435380445,0.149571322022842,0.118801522538489,0.0999903960059035,-0.135988620056666,0.0878693937715594,0.0103269056644539,0.159743054600649,-0.0640425993949749,0.579684482171346,0.235061011325787,-0.22586574471312,0.594199108191117,-0.348843052323794,0.633144935349817,-0.104054812166298,-0.134084227327411,0.26535194429697,0.0171692300104881,0.287431576373445,0.21428474893369,-0.0660698374469799,0.240491687684572,0.465713971557006,0.112677713073547,0.273267681079469,0.18322179816406,0.554335273545512,0.260148672770212,0.646223207879439,0.176869193323694,-0.127072333798484,-0.0434332834774624,-0.0499156399827355,-0.326327551336694,0.607474226798236,-0.362939900156844,0.0907253834045126,-0.115259710883497,0.14885670305823,0.117046394276161,0.309876784181272,0.163154749095243,1.29662932206517,0.22296183066854,-0.0183449847611973,-0.143188744425533,0.0409430631201088,-0.390454732644576,-0.0914487183312447,-0.67351639593546,-0.745409124094284,-0.4159133207839,-0.24648187033316,-0.0787900726463766,0.0344472143131651,-0.0302933803767149,-0.349931364399924,0.185957622921761,0.744317692878629,-0.0250912505421918,-0.345656278316918,0.0478727570678805,0.169333264950662,-0.223184531925622,-0.202567748728889,0.154760317769463,0.460642084254584,-0.101105609600322,-0.248280576524272,-1.34812694374434,0.984318354974083,0.0207660289234148,-0.1508969229929,-0.0478549940844651,-0.0586843163764223,-0.0100579073874419,-0.529237267325373,-0.0498746359784804,0.0684425657633654,-0.0156435545893722,-0.219433818997958,-0.15028535268835,-0.525370438949568,0.0149502438032403,-0.269858235255546,-0.23426930600758,-0.405710550296727,0.652711468031132,-0.154079806187892,-0.0916954949667723,0.264137232256323,-0.19643818603052,-0.0407720565839532,-0.00361766435293032,0.203977905530679,-0.135250444902379,-0.0528229258920291,-1.05116614253274,-0.099757797920491,0.539159308588971,0.145710820418634,0.233983540088989,0.0467661231500903,-0.0348968420004559,-0.0693440435201079,-0.834814328167957,0.00139759871113931,-0.956172455141243,-0.539709374712136,0.125737624469668,0.0146843636152419,-0.02129924104635,0.0730814607592324,-0.479939241689157,-0.608974781114114,-0.185980232177864,0.604731278919297,-0.139426288117367,0.011991475232639,-0.0828141761751479,0.630194704212559,-0.309341297259279,0.137741026682316,0.0321324107203146,0.253883955693922,-0.0120892430891401,-0.0343470317541331,-0.375498865941116,0.011288584538331,0.249390033342214,-0.222373752137512,0.14627924435171,-0.0671588179374321,0.293086602842643,0.0841477175479958,0.216900301742503,0.100387244236311,0.0852173365210735,0.0461586611990569,0.235679020106074,0.208476689080459,0.22848150835124,0.142958356687671,0.281795426819105,0.166458095419694,-0.125287736644773,-0.144266881011549,-0.255160329396432,0.210279661592595,-0.164020444398727,-0.0577890161158801,-0.257145853969142,0.0345638250738605,-0.0664716839745641,0.0514144999311548,0.430050718559904,-0.109543715871367,-0.228292310362634,0.114358537746618,0.102385645746099,0.0558606138938846,0.429697164600385,0.244034687033103,0.0858049021285471,-0.201240857843675,-0.101497839336253,0.0442916946046283,0.855345658923386,1.24948276930731,-0.302639872919672,-0.395875944714942,-0.31408530064039,0.184766533419235,-0.115259454195467,-0.095188489836336,0.0916956738500154,0.124762983784435,-0.433510900343167,-2.40448162483983,-0.34873990030436,0.98129901618736,0.210673855544475,-0.159014285782049,0.350245454121088,-0.706271417191606,-0.194752034158094,-0.139050463269308,-0.0410549796477176,-0.078208636963284,0.00286706832785589,-0.431809832752077,-0.0925427476139046,0.258778222694899,-0.126512702798467,0.0800427031804993,-0.0872507151846829,0.0971920886010302,-0.00854467646421774,0.230584183555126,0.55477037227227,-0.715199236258017,0.101119713453742,0.127247226325364,-0.184816598550539,0.135434679629697,1.4377905090844,-0.244882960807432,-0.195212901017111,-0.029472259848246,0.0913178677569026,-1.46998633641686,-0.44987106371244,-0.0927232698670287,-0.126467276085149,-0.452460773347549,0.11422878456114,-0.439217158852246,-0.0147048329745991,0.254622059647286,0.226180445707098,-0.666291907582822,-0.259061573078863,0.0198498090797088,0.198340182859035,0.12830990028284,0.532077402855524,0.135382068592234,0.0268840969618435,-0.222502845543569,-0.0291988793283265,-0.0374017242009615,0.0618907697153146,0.0920168258940781,-0.0856381445659494,0.0649909260902902,-0.133983212122929,-0.403288388711233,0.399979775028191,0.0321891680131113,0.187898503336945,0.0363843659704883,0.263455658864457,0.559248335780878,0.449129088856876,0.388262641226639,0.859451496807118,0.0209807999678695,0.655063829796887,0.263419244097102,-0.0606521539459448,0.544763812223927,0.329837733491121,0.192525433130916,0.00856944491906458,0.463813512461222,0.342442074428501,0.505447095216189,-0.541563112077685,-0.0173620937406072,0.259664631542658,0.111780069707062,0.217647086393002,0.853080406980717,0.00770428744015967,0.228439114856833,0.203338723568487,0.261226102877525,0.545400623242556,0.155703933838417,0.0665937674722927,0.4511307079904,0.211196791326589,-0.154179853486871,0.454070924129275,-0.263527804926392,0.13206495465318,0.0461391468933189,-0.121953886704795,-0.0326807196657509,0.282036247127051,0.754975374883986,0.0159490414247626,-0.201354113251966,0.0734981711073056,-0.134447533270784,-0.0118146360758563,0.209680531226111,0.319353910274846,-0.0417645686020791,-0.100185707723899,0.00901050215066617,0.40483384764783,-0.264099830679543,0.0935672834127923,0.0200334572896065,-0.105301610962876,-0.257124181103722,-0.488762054221844,-0.0597048994949487,-0.107452586216931,1.00993402791943,-0.306212971521267,-0.00287698923311658,0.0782704308675452,0.490727556795569,-0.559883653326467,0.16488365944098,0.280475692896646,-0.128345919500582,-0.0319424745534415,-0.00299428891083919,-0.403951040324067,-0.322634370020388,-0.323475278913186,0.162814833957278,-0.403136061117728,-0.809925141715351,-0.111556647014506,0.304437909897174,-0.159434343690411,0.000535049205694727,0.0776238921159156,0.0695534392848689,0.0337463204019682,-0.247404679035518,-0.0728031829173963,-0.061954766210091,0.424887863151632,0.153121379866698,0.583886257763159,0.810391850034791,-0.026662170018478,0.0439572892116312,0.129409182943327,-0.216984874173816,0.167592912649453,1.33885355593197,-0.279554116588955,0.238749380005316,-0.0127617570367219,0.149717200940463,0.301608042501623,0.252527389588854,-0.117167486727045,0.120153142263907,0.451922405447955,-0.169044961854709,-0.134678660100954,-0.0920329762546334,-0.271818158244424,0.0421935640934547,0.1348146094132,-0.160404479413192,0.126067336907551,0.173479545131844,0.290279121852249,-0.505635073271329,0.0280105773970403,-0.158387904842788,0.0068586131475036,-0.0162021599540538,0.0822349081328117,-0.233520005868601,0.404914540112989,-0.314781509543143,-0.226784288177697,0.628041125720677,-2.44494493880418,-0.166760254309334,-0.201395759008961,0.0413551960222562,0.0949201508372307,-0.086934762109671,0.953915367925426,-0.0914833911356906,-0.159919531127313,0.231192837787229,0.0258819866365537,0.421966397136084,0.27572419810139,0.132634130226651,0.269614018542998,-0.0692929190267449,-0.278916623775639,0.0717035190119937,-0.0720554314768256,-0.462175186617045,0.154157141618777,0.0467525164472968,-0.0581415268874746,0.316895680527292,0.702272217089568,-0.567918463377769,-0.723633123468066,0.162873569368216,0.0518606385790853,-0.159183140377861,-0.338767003327966,-0.2205672481279,0.617658344039805,-0.0730478429500575,0.265015760194591,0.35918965086325,-0.221126685482241,0.329444990144259,-0.124045706184732,-0.198450944108457,0.77796431725985,-0.104844000962499,-0.509493690588704,0.0921131132350104,0.0563828650313837,-0.339693224851274,-0.116354948220537,0.206249085361749,-0.196325875204011,0.657187008484821,-0.0251354299971282,0.174736037881956,0.0101011094561065,0.0721822846534931,0.0804035790515925,0.0638266715625199,0.674930629823593,0.305346147075539,-0.124233975853691,0.0192905204420219,-0.0999093468435502,0.198600598414982,0.566191440154065,0.245167210640329,0.711438077712318,-0.0894721755226626,-0.182188671452925,0.177327419167131,0.727790360280809,0.167172711273646,-0.21396773176111,0.380705953972074,-2.84321905155454,-0.110849119025061,-0.139790750967987,1.13554785820568,-0.331832518642109,0.338338252180765,-0.321480506817598,-0.193121362169214,0.227086685925101,0.322637100348353,0.0465297015094548,-0.122157051791021,-0.202293169712881,0.00844156470150791,-0.26896100059689,-0.224322391961987,0.242731602451792,-0.180489244731582,-0.339772800777257,0.382872913621528,0.341512550763334,-0.107041973390047,0.00931488679149229,0.106254414006769,0.0830955661310035,0.0785752013207165,0.000885409482622947,-0.764265498977557,0.0488870914897344,0.0780248176791402,0.00985246312614527,-0.0652828538449803,0.0953870272173652,0.173314850640324,0.300134580887121,-0.0407440715257682,-0.285424183219689,-0.857275792975472,-0.104029411751336,0.0411331503490867,-0.151102923921117,-0.208980957528656,0.111083151057692,0.203978938901,-0.0781556691080251,0.662097374246436,0.205838682203501,0.91669559016781,-0.237327055120489,-0.210009254175422,-0.17633208347233,-0.0565747065899988,-0.0568290720040444,0.3566599241343,-0.362505419162644,0.312583897560394,0.0630291122207927,0.00598347343564404,0.0724472439894598,0.0886695609953269,0.0446225134883552,-0.0601107591846403,-0.13602886766069,0.270234621780978,-0.0336282995890377,-0.943959096191354,0.128031187587454,-0.25237507184366,0.350524874682653,0.110168391032592,-0.277693224853457,1.61418370912914,-0.216988248104803,0.0648489295050173,-1.32205535248023,-0.21513039529826,-1.11752791218953,0.145902889629313,-0.0379863199295899,0.147492489699306,-0.0639033763046049,-0.568145910087898,-0.475694541822315,0.528031719104462,-0.0293404424430511,-0.112104979876376,-0.0864645016705586,-0.00476139744604157,-0.296872623192354,-0.0322677209049873,-0.0379806231774314,0.0194006521341121,-0.111110392102828,-0.390134885279131,-0.0545339516219018,0.479028498918599,-0.0533377808353141,-0.147848543452462,0.167462707885578,-0.0468046553867191,0.0479209714683859,0.128729761398125,-0.0402893664980137,0.398832820883895,0.236445731603948,0.162578981506288,0.0190239676896192,0.513936685972696,-0.00597660380999067,-1.2395582579374,-0.0827965187003737,0.0369310289055855,-0.054804900965142,0.0722319891560495,-0.0309557345426973,0.00281899253217973,-0.00498033113125524,0.0912316160593152,-0.0686367567809069,0.171993158954385,-0.0991976919481037,0.494393437164729,0.254846239602421,0.153461277560479,0.12493253651757,-0.312893769230284,0.789075163643455,-0.586661764294131,0.119201375165472,0.405590777050101,0.0318252242892029,-0.148720723647161,-0.107279692139906,-0.227319576266574,0.340828254510624,0.627364096034527,-0.403035854110021,0.321768532118106,0.046936101518802,0.00198579508104035,0.141813472409631,0.117137921654692,0.159456234430691,-0.0503163335496582,0.225152753520963,-0.694355499937267,0.570835170244067,0.204446200558413,-0.0029403185943914,-0.261410177989357,0.0125191163302431,-0.146332846414559,0.191043915215214,0.200598407590303,-0.317352905759652,-0.090348680881815,-0.0099805452486362,-0.246587623196265,-0.140556284567913,0.304922411985483,-0.121688900502453,0.321173340551667,0.365067936276073,-0.149859397126626,0.0417114994886649,0.16729646465343,0.18259188541264,-0.191157493723128,-0.70360055776737,0.316491440188806,0.0546355182247922,0.134858100886131,0.174226622884763,-0.198268110251223,0.34060582175789,0.285187345684843,-0.0743709105250579,0.219849121601015,0.166176839157653,-0.144259319252898,-0.302368363991129,0.185805483493567,0.114484315973572,-0.24913936781871,-0.00910184549772046,0.499999780974623,0.0817688498797505,0.06823296674107,0.110409916202379,-0.2719391342862,-0.240848560346948,0.198681456689279,-0.078995418081556,-0.265037512227031,0.112361193772548,-0.251607597842864,0.417456916150319,0.204414205941339,0.104478632663477,-0.00596989435154336,-0.147546641524503,-0.0119909050023841,-0.203566047275688,0.150601811724705,0.00102948779740751,-0.048638848495008,-0.414020585134462,-0.262349963512899,-0.563456545070998,0.0586903317997338,-0.278442218432875,0.42359249704971,0.0641072054449928,-0.275919494635412,0.031985824880849,-0.0560372645209289,-0.0566790995789637,0.149206022783999,-0.08530661526741,0.159355405651897,0.710453910271232,0.368888646421629,-0.542424662702387,-0.0448425694817057,0.00200574160203806,0.523871166789626,-0.0149393272423636,0.563577030091515,0.568366565212868,0.724381316903831,0.237327494745598,-0.521386989586636,0.110223323734301,-0.458761144071906,0.0221664433216325,2.087459118946,0.0132584722386546,-0.0261774094949267,-0.11725947205071,0.215546822198128,0.607174257326092,-0.00715236022864047,-0.16430279016772,-0.502414172792391,0.0418061248269313,-0.135175566762631,-0.193132762818786,0.0900857803495302,-0.0473491887900418,-0.00435321927249283,0.0700596288759504,0.47117387129773,0.133775745268833,-0.158678261332259,-3.21321218267998,-0.63256321418903,0.212239239538106,0.122369297060789,-0.586432878869111,0.0105243454670955,0.271770269609535,-0.323969681665727,-0.0442912970953477,0.42903507885864,-0.297837356694539,-0.13482326179169,-0.039615607092069,-0.120647807950924,-0.0388702482852726,0.360094452828027,0.589056256958391,0.585294994022724,0.398188572577407,-0.525448057502533,-1.04510186941508,0.320107725767387,-0.0325509848214258,-0.0485485769355412,-0.277689287884684,-0.0655102027438728,-0.363327247113111,-0.0575625917706132,-0.0150828620673744,0.192975306534303,0.377202888085403,-0.0707076025885705,-0.178957594441907,-0.0691494814931183,-0.183770055741603,-0.322686934100166,-0.144083198382539,-0.0094407421041703,0.26096766786254,-0.0217153092888991,-0.106400812089406,-0.0495438522981858,-0.158045957053599,0.072510443827038,-0.182618969341395,-0.0256664059012344,-0.162001331265851,-0.103594545843478,-0.00207561031196027,-0.534955690328297,0.215339660441497,-0.353134952618776,0.246233349603938,-0.058217049823133,0.143051198861141,0.0237393652228424,-0.180496136787861,-0.0178492774284928,0.359171208513773,0.626378899620211,0.029562615817708,-0.401048737007961,-0.0761244419320967,-0.128770486602555,-0.18427527164457,0.181818226484696,-0.241896888952526,-0.020019162916145,-0.084621771788939,0.178805824515044,0.0193965085013591,-0.196879894839443,0.0574276085938533,-0.451658871353098,0.0882686528940645,-0.128362465222731,-0.0041469203542141,-0.406024823607143,-0.0558317315949482,0.0645166214891699,0.0909920312002701,-0.247596956109007,0.080244003198568,-0.0168603342517416,-0.378753590660128,0.137424090958499,-0.0449403558606307,0.421640229017227,-0.1979733230363,0.111969991711717,-0.107872085219655,0.0957414337031562,0.0266660214911348,-0.12098022555575,0.150272375509166,0.0358411456325785,0.110615453928462,0.0430952822647609,0.213152271263138,-0.197672910208928,0.506905344480827,-0.153612961809605,-0.1217004023897,0.0737176132485559,-0.0670674777637058,1.07582893205392,0.165979941819713,0.432304642614239,-0.114684878859086,-0.062122118199569,-0.135887563431414,0.208820488904619,-0.325506189667661,0.464568145322608,-0.0743503097434337,-0.0261762926336384,-0.0694837059816987,-0.126683400716291,-0.554931964744786,0.00320142129163785,0.0940597837138587,0.0259970722104739,0.141980453518868,0.236601383281535,0.0384077034985382,0.244512185504703,-0.0274483530837471,0.0963981184979113,0.617622852612657,0.350307833389985,0.249417389504406,0.0332147053863359,0.361155086733347,0.181951799090319,-0.0822558234231929,0.0557093900087957,-0.0679046385739607,0.129444647991082,0.0123401763673853,0.0493108674316528,0.761901083718493,0.0908990974752071,0.0975219898913122,-0.159076151885328,0.187196694372047,-0.0832312540463442,-1.48487397988785,0.122633120112041,1.00113288643165,0.382893664562722,0.0739456700198308,0.721747526996317,0.102383096826037,0.115862544531303,0.0795326957849767,0.421101628904347,0.251894076087514,0.957644979565222,-0.179465934826203,0.071872808912715,0.0856692226538973,-0.0798160060197913,0.140851178615421,1.58539633990618,0.0615966228172125,0.189920662351533,-0.532765320582401,0.313531121906869,-0.476212130465327,0.538137477626537,-0.0960306101554345,0.656291976083613,0.264114027422932,0.0531785024138741,0.0630100156446574,0.068182978746292,-0.266737677047572,0.558389078083435,-0.0341405951121808,-0.141870488624378,0.165039456358788,-0.144690157345998,-0.357483932446212,-0.00269726040562039,0.0492513163254627,0.133272097853376,0.606462838702477,0.178028129699914,0.305735396229294,-0.132154028368494,-0.0380984101265492,-0.937857552384866,-0.0973162930611493,-0.473939510333023,-0.500953286429548,0.949672254183312,-0.0379146936044828,-0.294505652756963,0.0867540906063456,0.153924338755303,-0.0685371203998192,-0.102507729922763,-0.138259542371044,0.242139917621868,-0.358276054326131,-0.240362106044894,-0.188018253207776,0.163118186965385,-0.546860766864398,-0.288681694089405,-0.380994675405526,-0.0942401183837501,-0.192199210827504,-0.412688155894531,0.372121188236749,0.409343200044232,0.692866517489378,-0.0133620135776896,0.0714216636085764,0.101556828558057,0.239604167484217,-0.117284282045302,-0.550218272282288,-0.0837510387368071,-0.0879381829329323,0.305356514147817,0.0105385823282068,0.00461465046160945,-0.174709368437246,0.0472760688092419,0.364681425650668,0.046084043042909,0.5440456266907,-0.315810992418285,0.0695167608151379,0.0834759165964806,0.390682357096311,0.220384459378703,-0.257273168886134,0.193309838494238,-0.319276763047748,-0.0734624644549909,0.131550140292623,-0.0488338088264735,0.101848957026593,0.25678806563764,-0.602746408443816,-0.0912917333061909,0.213544463420735,-0.0937409202108139,0.174823543823049,0.182347850827829,0.144798005272228,-0.221645440842356,1.02267248867506,-0.110686596777343,0.249550149882547,-0.472070246695019,-1.6059594570587,-0.202705346185996,0.37653598577874,0.354711979752988,0.0740280478320542,-0.121838779207209,0.291541646130767,0.559680637870277,0.158434163104955,1.18134994674718,0.00879947642482386,0.282409150048565,0.196675467068904,-0.504372589806965,-0.00823411171276568,-0.391289248334597,-0.0772269858325875,0.130533060302539,-0.347146750978017,0.447642588843636,-0.137314239681409,0.0784278276247827,-0.0526832398118242,0.0533721519237669,-0.14465070392657,-0.109264291022125,-0.0639377043997432,0.14683590495253,0.256079842240674,-0.154154689786634,-0.0143482535334996,0.230658973507062,-0.128213520883505,0.0173036373023856,-0.287986679489672,0.282720130455911,0.0820187804275142,-0.206900694407644,0.0477378560319373,-0.118832106849269,0.158260039025084,-0.143399453422729,0.113677303323645,-0.0638723694497363,0.361087589366436,-0.00936520058598573,0.684820148701866,0.4311320785438,-0.11897722056072,-0.147638226830937,0.0498125019998203,-0.260904791570155,-0.70582798747197,0.0794870360288935,0.138940745127702,-1.32158664503131,0.0580632053339791,-0.593883388913548,0.357561842054641,-0.724090436056004,0.0434270727144383,-0.102206498791478,-0.212479716092894,0.826219739721627,-0.0390779550245262,0.161636553024229,-0.17062945499919,-0.384489704777777,0.133287519157833,-0.438877485990887,0.0486624838506282,-0.0650466306391142,0.189626792287702,0.345797281817407,0.0901033377208192,-0.0690739052535603,0.0436192062262907,0.419706593492887,-0.244037526779017,0.810196343254096,0.0089691055840256,-0.0069669705343473,0.105156060030088,0.0373476180987119,-0.671404601175495,-0.0180492898018433,0.111522960801516,0.145721041396365,0.0724290493444159,0.0966525875836859,0.0635837628539902,-0.188707156212954,-1.4114919639955,1.22592408277182,-0.18345586080081,0.28372713305322,-0.0434261110631536,0.0275544226395644,-0.855150225035779,0.183281076894317,-0.0479115385383456,0.271938969996986,-0.8158151831491,-0.129530785718254,-0.0765867023098512,0.0541881134999339,-0.0485532235066287,0.0966451658989585,0.356914804925452,-0.454277183520423,-0.0522889039220583,0.226602982970039,-0.0366061774808681,0.200689285448167,-0.820107237810139,0.155535737411542,-0.274143070794323,0.0892075206405748,0.205012973658252,-0.593740122847334,-0.0516205454893106,0.0669610013412973,0.237352230624024,-0.149823563258962,-0.387193743385683,-0.10457436458778,0.280360907602777,0.0965339686686434,0.255026115361014,-0.0963248135939507,-0.00930060097948651,0.28306914263202,0.236726662353073,-0.0412864980228135,-0.853022638143525,-0.0243271659140888,0.702358608743873,-0.225510686937527,0.194145157228532,0.0387073103118244,0.168570129027028,0.0746594086498498,0.289900837117797,0.546054673029197,0.624079580040111,-1.07344961598202,-0.22430984971497,-0.620166776633539,-0.0944477070046599,-0.975897944226108,-0.223601503342363,0.0520534899509894,0.316352533595654,-0.081904667025913,0.0682019624150181,0.302751221242384,0.510499968482303,0.316198517646615,0.0315161975726959,0.0353868343578164,-0.0196762914122318,0.103944082348206,0.134182409147947,0.16940281532176,0.189122619934878,-0.177143917296967,0.161974571049808,0.254650545576145,0.286412599767381,-0.310723118543264,0.726074833862663,-0.126362298403373,0.320260102811636,0.18205740146039,0.144186982690546,0.535340298503102,-0.109761818798443,-0.0300337743786465,-0.00152537899704642,0.0792096898654972,0.736571130960471,-0.488004843323807,-0.192891524349794,-1.38829342941704,-1.9311636481468,-0.0373772029760378,0.257968819564775,-0.67730539078148,0.0107009707915567,0.0354251477439475,-0.0501422138933673,-0.0730459009058974,0.190425933400777,-0.575877901266951,0.117033366555643,-0.0581688680020744,0.165972464695709,0.164882245482057,0.0124485526791464,0.0550947061291069,0.0682109686344987,0.304550147934802,0.0120377498331428,0.152370661349367,0.0226534194280191,0.1507865085435,0.248195181070967,0.048543152551961,0.120313419442466,0.167884191603972,0.0676993339803845,0.734107478818898,0.140049793985766,-0.0528227083264487,0.899525204451909,-0.101721092266972,-0.385514739931529,0.00602829139317807,0.0567541509185702,-0.0802967863350615,0.0549094913616132,0.206531297006439,0.119330208256806,-0.930009818900612,-0.042880420572826,-0.197495178210011,-0.0911750393148664,0.107545167871407,-0.0726368477431149,-0.073124584207121,0.263065138767795,-0.0130332100524696,-0.0997898457316844,-0.129569606430066,0.297171190467673,-0.11513029619117,-0.0288524435358873,0.377770371357137,0.00552988169698912,-0.024382227695152,0.0569064285696959,-0.187317270431826,0.225981554419769,-0.0355610315274499,0.0209070958364148,1.6024294753021,0.957292044410579,0.177149775650687,0.35952949630654,0.249190171230935,-0.373953776884202,-0.200772793066463,-0.173995733212778,-1.84571153634549,0.0934317728461958,0.13013917298643,-0.0801918456466815,-0.0508603746687563,0.204738465377056,0.144305257893327,0.0358178885456045,-0.0394047963822144,-0.372791212220356,0.471873783111413,-0.126842954680274,0.0839339033740901,-1.00602234455368,-0.292027148465905,0.254165102777377,0.391773963956271,0.714063251367675,-0.0766893488002545,0.0453438240763447,-0.236398690548699,-0.101221759150608,0.176573707668877,0.360028788411534,-0.237514628301283,0.070232889627493,0.221735717229676,0.228667995333679,0.818127469365662,-0.00350866154029067,0.110651028836883,-0.00956375362777658,-0.114498624897246,0.14638312141925,-0.252016145364445,-0.855539297168888,0.209820650968734,-0.30967000086802,-0.102170549765653,0.307260881536336,-0.256873942188097,0.651934025697884,0.272433734729481,-0.12794463805839,0.163156736203202,-0.080728191319515,-0.269345898170176,0.0873430634714572,0.0932608883194064,0.0406669218058139,-0.180103593326618,0.140617970919603,-0.137737494767871,0.529530345494338,-0.310284069538316,-0.127479232698265,0.485006821710572,-0.172160741091843,-0.27143336320065,-0.403390641490546,-0.0122345510107603,0.0570925874159971,0.258755689055213,0.341197183718391,0.132695957002166,-0.111812497106831,-0.13497202373757,0.168504894970328,-0.129460800200106,0.191770692679425,-0.491455589314333,0.0233935233318403,-0.0604952274783815,-0.0796746667830428,-0.39459645304295,0.107647623252474,0.49275003495405,0.456012332911331,-0.173621939347265,0.309297496942524,0.451610923681161,0.209533123710796,0.0370370292906909,0.0929788970510061,-0.332326301681865,0.0782076445861325,-1.07455887197262,-0.734081118667229,-0.539087612858545,-0.973279242728674,0.907377911491303,0.340951103622585,-0.0549514595523561,-0.116124912674713,-0.108544174423579,-0.0302990119052767,-0.411273564880264,-0.4718834943067,-0.913773835854307,0.216880013663951,0.126148748614049,0.869001730002492,-0.124658171283786,-0.0628507837105989,0.10672067833032,-0.313809978966114,-0.244697004943285,0.0598483206576586,-0.340462112842983,-0.636938946655624,0.183535748477141,0.426108797453812,0.0691114095511198,0.161138164406097,0.058571026070449,-0.367876202638196,-0.363596786214941,-0.322219590122129,0.617987683986705,-0.258092774269634,0.101404392769747,0.322830569214426,0.0843245290821672,-0.111590893617647,0.0311678615393855,-0.0407481563587975,0.0767277671421371,-0.119076662513814,-0.54800822563685,-1.18555528858868,-0.399719158578248,0.175852140324122,0.440428935170082,0.466942224168426,-0.0505409404626699,0.0368076394497675,-0.0295516527077279,0.206130240326251,-0.312640176600766,0.47476309424403,0.149365599798764,-0.250653592542089,-0.0546686185751265,0.329843947572255,-0.935842826089802,-0.00369420864961742,0.0370916533179472,0.290885563924406,-0.107241969308417,-0.189243599829084,0.0647855196344738,-0.560335753835772,0.304750925595517,-0.202500982889409,0.420777676032008,-0.125736347615034,-0.159261768722168,-0.00277061016989878,0.145258104862199,-0.0303054655736308,0.0386925887877978,0.192325606057134,0.395624252908155,-0.350353027837303,-0.239637521491403,0.417542880317007,0.699833090258212,0.393562196048301,0.498314374761267,0.0499034964397292,0.522878346391329,-0.028442622187579,0.083082229046337,0.172191518574633,-1.06056958743655,-0.313461593997682,0.228194238715937,0.446395105583079,-0.157806471404537,-0.411894558117639,0.382498125109202,-0.201596340951451,-0.153687483970376,0.138562464266893,-0.258918024789828,0.0937708650166388,0.0512619166118645,-0.429706105787185,0.100240997121102,-0.0151994052442612,0.129834866420175,0.0768017646061588,-0.734826872724314,-0.107589250581917,-0.34818302656175,-0.0218711712666061,0.0990023725230545,-0.24927686377233,-0.0722310532804506,-0.133817984195989,-0.0239440406030623,-0.119226093180342,-0.0928503196441125,-0.082022187450333,0.0260631394225869,0.013647681913381,-0.00846263600052546,-0.0660420763748764,0.0171782196695476,0.130500456937717,0.207958012660504,-0.417361120942938,0.130831044706781,-0.249196208231011,-0.108090935206747,-0.323800148748628,-0.12232004351912,0.0884777325842526,0.190714995926823,0.0555627885971482,-0.0245632994354908,-0.351464910944754,0.559278959112429,0.0571746116747092,-0.208387047436934,-0.0805469619156781,0.173473633039727,0.0409773174537687,-0.0288236636047694,-0.25346317714649,-0.059009160517767,0.373595307288075,-0.000339577570959948,0.0874404927949499,-0.0409715269001443,0.371145260995966,0.0203522440240304,-0.0502761663422215,-0.0622268801457721,0.0661556077893166,-0.0133629234681574,0.386547851055674,0.0569643377480579,0.143785178377589,-0.28196194652974,0.408097433485711,0.305072462089212,-0.222354040440675,0.129975307873157,0.135949650340313,0.0323335095300557,0.147378185393509,0.122643873447877,0.106136093517884,-0.098402769690817,-0.0455749709909243,0.0503418412148761,0.225919204521803,-0.0161074077219558,-0.167129131366589,-0.170540260112395,-0.410431433807739,-0.166091274143686,0.379646859880824,-0.112111039587117,-0.657443060215108,-0.0979860690650867,0.116018359910709,0.0425627992166425,-0.11752967304393,-0.30721975708921,-0.0924902995360586,0.106428902430171,0.760514525345383,0.0962382733607871,-0.0922417447634517,-0.155790972243555,-0.104084809176173,-0.101898479270597,0.430597995806306,-0.105223808507555,-0.259139584839051,-0.0231212761098019,-0.0479053846238026,0.25175122850267,0.175640106018796,0.113182498433057,0.111181142937042,-0.262180226983374,0.0214180083003583,-0.0785546763171619,-0.00202207142794041,-0.820195422403913,0.108187161939828,0.32477791395729,2.67273865834326,0.170270589294345,-0.25435548127895,0.320530433932808,0.788277597347668,0.715841715611211,0.958053459569309,-0.0794501999332272,0.196731623202392,-0.0429842568760279,0.179071868229157,2.35026299013289,0.236824493432485,0.0359954601894499,0.226196008436027,0.374387967037483,0.0620436094605895,-0.0602050148379311,0.759693972637586,0.230339948202458,0.243893297554529,0.152804679426861,-0.0584265617692194,-0.130084613494275,-0.0634460101510382,0.0165494283522051,-0.266717251525425,-0.10692947672891,0.121726144684102,0.187269659256799,-0.201461797687425,-0.0166215025022122,-0.147732006838105,-0.397848443355046,0.0167465166373292,-0.074364281309306,-0.11821391180244,-0.342174673576733,-0.182606548570246,0.157723439170447,0.0674509340542355,-0.0974857496124824,0.035070598335827,0.201380416139863,1.05379272000818,-0.0166796262298195,-0.271529719346412,0.163379413528398,0.0721731200068969,0.0655432173698959,-0.132064972666572,0.113553933659203,0.0279296703040948,0.00706572919641049,-0.0243245665449474,-0.164564969612332,-0.5701350542097,0.0864158632173289,0.270195120434457,1.07970766424919,0.724552536986724,-0.720405009040152,0.00899880528136649,-0.27195114003557,0.0280671788387224,0.123248395579892,0.204536472493581,0.189149336109775,1.08716571657292,-0.163150976964349,-0.895197893878691,0.194672845859964,-0.419587716072044,-0.172680578087506,0.0847231951724961,-0.252618394658311,-0.182535063514941,-0.0746658400120003,0.0859982298924013,0.600916542378345,0.131853499566899,-1.43576922719137,0.0575633230057793,-0.171275107411969,0.172361044075032,-0.1531782980656,-0.0222545304035641,0.376509130515496,-0.0722553836166638,0.531578384581147,0.196832873898501,0.125529387745652,-0.0224629928597583,-0.642327029810314,0.768988530276846,-0.119853235825086,-0.294301999925674,0.274635681259363,-0.124912463099167,-0.0812541111151429,0.060047354302702,0.297846831124873,-0.679235466221608,-0.0593258675743483,-0.483675375651831,0.334316010892962,-0.0685141208177433,-0.205073831845935,-0.128416351361818,-0.00768550300369192,-0.157321592007214,-0.161825541976497,-0.595919290486033,0.0470990855962238,-0.250840306911194,0.330261464555799,0.070975928566155,0.12516039036112,0.132713905314426,-0.0617248544437229,0.243819688267085,-0.483307560669394,-0.247215519479861,-0.152753026260436,-0.383195640043212,0.861042062636604,0.219484906209967,-0.26522851636092,0.428661906793455,0.0463768763654624,-0.213924564947102,0.220447320542456,-0.17032941779127,0.516399174886437,0.125553848799889,0.468731893780607,0.0751687307712733,-0.174782319262331,-0.761728867684013,-0.207152244427539,-0.794332846386778,0.154224470589618,-0.341923907345175,-0.321579941567396,-0.557938107711348,0.131611259527293,0.0439864391397897,0.24185935253182,0.303972108145332,-0.213541010817055,0.0837241605753126,0.481706071130758,0.00645000534920619,0.08986267950245,0.0288014557303255,0.440791607919877,-1.21475080897049,0.0273543847702003,0.817958581154055,0.0221487054866499,0.0332469382556309,-0.208624408763961,0.0455156047281643,-0.150011191742836,0.210271195410278,-0.216677126119438,0.220171152624463,-0.107982991711907,0.616975869892521,0.0893841764881576,-0.0845004813961127,0.0793686002661452,-0.0620992194320861,-0.278498600055295,0.00728112657032888,0.0476618158058645,0.0581617962715993,0.0856695294619819,0.0217447359660297,-0.489642845127073,0.301164631184232,-0.108810542502858,-0.125685943715632,-0.0298610201272526,0.199515769490537,-0.0782241996126269,-0.019463687533525,0.107426343194235,0.252604093108886,0.942537521417814,0.0212416337197786,0.649383187625371,0.193314837909159,-0.17460629715843,0.163622379560201,-0.308994413206486,0.106480002960364,0.225174128543315,0.685375035336013,-0.247481742084517,-0.526181234196069,-0.871564912734194,0.151693859842917,0.211909348839221,-0.0356184636921708,0.163208627914073,0.770413119731367,0.225087153545939,-0.242477887957973,-0.0411442557529283,-0.137790166958014,-0.0155340784349075,0.257698840466861,0.0997271533753197,-0.0539950052383154,-0.209938785452042,0.193663063812495,-0.254277973972066,0.263552529449096,-0.314823620427918,0.277831446788093,-0.102905291606564,0.162565488284721,0.361154095889332,-0.126582010146759,0.0545678189608219,0.582264811751662,-0.208265083907311,-0.0812664894954641,0.712777318985624,-0.162257141655517,-0.44665096582711,-0.125119845217204,0.293264250977014,-0.0469937954115791,0.0275982130408467,-0.0853504719680303,-0.0376520075319398,-0.244702579853183,0.0882691771778773,0.012860410038364,-0.123674243832801,0.045336779302062,-0.344779239614105,0.447332107166712,-0.0798646034385036,0.0452341033398207,0.00647149922062615,0.107950785875823,0.169849572285707,0.140172998128519,0.453224297501577,-0.114992537043909,-0.119602320667033,0.38219864311509,-0.374466379995166,0.285848729059227,-0.19075734062661,-0.161652244070865,-0.223096540273241,-0.11149870259848,-0.17166879031049,-0.0901963695334319,-0.159771497260169,-0.209376246517432,0.241758827391598,-0.0531771996732742,3.02215155428325,0.127606015261478,0.00152140955839345,-0.363019140837736,-0.163840075808279,-0.655331242617694,-0.117461176861139,0.0215808701243852,-0.0850008731195585,0.167969156174419,0.322317061296836,-0.515744858799334,-0.284443355404564,-0.0224684554909328,0.764572417972552,-0.0214044248231282,0.212580162924474,-0.030646505806454,0.0791283226106925,0.762446193847319,0.119203057169072,0.556169302202637,0.981255187312564,0.541408879450666,0.0482868671078806,0.0191226951950972,0.0157847663937058,-0.30524193952638,0.16880461449727,-0.578478603330099,-0.727524433015552,-0.219398230274242,-0.56675081077505,-0.529405828816042,0.768434286344342,-0.117832805332366,0.652546119331351,-0.158307697505413,0.0498185869342244,-0.174888553844726,0.269924874555624,-0.0743664635674581,-0.0785022795530368,0.7167216697689,-0.260884976334141,-0.060841605632144,0.109244400133616,0.0162460262245599,-0.0641851196182399,0.264682871454111,0.0385472231450026,0.0629437166341987,0.229468605803159,-0.0499354409197618,-0.53603281903899,-0.130825872079977,0.238813880608218,0.196050993787145,-0.391248297885452,0.0719393657553283,-0.297975152701576,0.203799063523849,-0.0868148412744093,-0.429813320867919,0.349419117843891,-0.116844156432934,0.253464688804458,0.0143341014353561,-0.0923664829855784,0.242006117757546,0.150509542010918,0.407454586719822,-0.321897870575499,0.139421242886989,0.126113481550205,0.135188346662656,0.0618394203110942,-0.379928903668265,-0.0114790263889397,-0.187785222710466,0.214182082979145,-0.017123834452114,-0.681531099729804,-0.0435783685059624,-0.201995874115892,-0.169456322778627,-0.00827936395856435,-0.513977984763388,-0.519394683987677,-0.215815953218587,-0.0337474424808233,-0.331879392795158,-0.0594237673339995,-0.0296792596512802,-0.278676581087317,1.07707787656359,-0.311350741103345,-0.037460767195882,0.198744776334311,0.180767947955948,-1.32555415403156,1.00314744476102,0.0216252672132077,1.41805261268649,-0.913945012111089,-0.323574376332565,0.122323101613048,0.340345108998124,0.0374946206401384,0.270259664035368,0.500861567159186,0.250404233300186,-0.208364888959124,-0.0460125530711224,0.0443001140850693,0.458923908847866,0.0383131146337776,-0.0694345339100461,0.0385935404547565,-0.000482811894455388,-0.0557694598526571,0.31554849658308,-0.681363520401143,-0.260372728850317,0.100590397222928,-0.0567448349725162,0.127927278071247,0.108850828837635,-0.0984401692335694,-0.726191235095956,-0.0558769076005533,-0.472369584969717,0.329116160916207,-0.770630709523761,-0.869721109040875,-0.94202302785667,-0.771674636393665,0.2364648530735,1.45493239226885,1.32594251448105,-0.0202202707488175,-0.24025735624517,0.00703984032702553,-0.0804340959557402,0.212176161917482,0.0671561563356526,-0.152328311635618,-0.0272626556638516,0.283414429605379,-0.00521606847976294,-0.223933004633849,-0.210089004591505,0.25436348775035,-0.00935965266103564,-0.0316637968683803,-0.0994868825709558,-0.40614150366983,0.0340379977480098,0.156831448599873,-0.411792251233347,-0.819697580571596,-0.0962284155768223,-0.0238748984904034,0.330485652316057,-0.325620554757691,-0.666047071130396,-0.984239302081503,0.255206284600254,0.209931326300927,0.539274014957513,0.0876014364662564,-0.0713433925136702,0.116574521599821,0.213241670582585,-1.12110137318538,-0.558316349102528,-0.277773183269006,0.126759833519612,0.251367063566368,0.147808628443138,0.195646438600155,0.0187709201690156,0.142638573980499,1.09764764507307,0.43894400025824,-0.0996720669708945,-0.0743997899155545,-0.1651725334765,-0.247776466625146,0.143400334111113,-0.0271354522946199,-0.431145999924173,-0.031706555917251,-0.172601381143188,-0.0743367565452749,0.259746039470439,0.0453074949256271,-0.0235631653905263,0.579924666338235,0.0845980736262247,0.132602552252582,1.08673526262893,-0.130053328050182,-0.293668887055686,-0.247250499876473,-0.129517798241429,-0.0526727517060504,-0.0616804884912014,-2.02434182615829,0.136152973516717,-0.901572614115694,-0.680589621801166,-0.169695444762329,-1.47626571390389,0.371235365926181,0.158338215582583,0.915150966868804,-0.24192901394222,0.183008595380322,0.284319660088649,0.127615129095144,-0.629013699504262,0.286249324840794,0.20869833576537,1.09942131732908,0.193892331721032,-0.00921482570793687,-0.142587339787846,-0.331939521893445,-0.154843273060373,-0.16042044891143,-0.165722292964708,-0.201571170507905,0.0550147645503062,-0.181904002699125,-0.367173745848069,-0.26885941923624,-0.175501721788258,0.0304510350527907,-0.020959185694056,-0.0462839674058355,0.355751397896474,0.895007336584841,-0.24061760275533,-0.204539054892608,0.00278513812399342,0.0122703632010836,0.0218459601226141,-0.194144842928691,0.052287198853948,0.0406648703259022,-0.209666945495666,-0.36697207308222,0.094862356171625,0.357665812149105,1.20545893606623,-0.0910167751280848,-0.0157469612962691,-0.144336612611918,0.168926403132051,0.264289414546618,-0.738246978551082,-0.117893509607074,1.00506526275708,0.046025693678973,-0.30469257031178,0.293991881063577,-0.178959533631386,1.05932957166516,0.107480148009669,0.297818790539957,-0.0651307704053159,0.340774143052092,-0.233416190009242,0.209098631896726,-0.169819552075807,-0.0530015413244046,0.00215685291516252,0.115938032886813,-0.046295536760589,0.460478346063885,0.410085134386028,0.108045543610289,-0.17849579190295,1.75613006924162,0.103597577704633,-0.271479724752762,-0.134656939498042,-0.126233366981991,0.296517199960492,-0.347523245127891,-0.245165253921844,0.0133543520393087,1.90980232199103,1.42437263552898,0.958569680452195,-0.082949452341507,0.019626462506777,-0.147143224014651,0.247334706789689,0.0478720425705742,0.0813371880953372,-0.060572562739498,0.49953098413666,0.403704955940318,-0.18853037393783,0.0664364109099457,0.16138855193148,0.140669894130231,-0.53220592618873,0.0668718596300576,-0.252784183040321,0.113292577461826,0.257581122050709,-0.121373089604004,-0.528780281876194,3.41163844727714,0.116743023577724,-0.0298402365196375,0.219107698455575,0.404236967218078,1.93477442572077,0.512583983352315,-0.0823508788251257,-0.244364566079324,-1.37141572359878,-0.0274790615582787,-0.166940359003898,0.32675109026431,0.257531117389178,0.0265336924649426,-0.287413675788921,0.0745332938168566,-0.205831787376313,-0.0127818053995662,0.0581811217586457,0.343055197581707,0.053069199224018,0.0793904732493013,0.110913764428349,0.193619295101195,-0.0685288931978843,-0.836631598949691,0.150811345614281,-0.07072468570506,-0.259582858742935,0.0478179755214371,-0.124559475128702,0.125816079439052,1.62516788957814,0.136239552625555,0.184088051661087,-0.0855748065472436,0.588395606848917,0.276415837489903,0.197619956182252,-0.131144440471847,-0.330957016607611,0.0983891445345541,0.0741180260469711,-0.144558036245278,0.0550215531465227,-0.488109915251123,-0.714633384453466,-0.072634958945969,-0.311622506591153,0.0774313073519182,-0.122677857143936,0.037352539415388,0.0400481356997753,-0.0279662229924514,0.207476272412423,-0.105405104424073,0.0316200966144415,0.145349908423629,0.0804815253279492,0.105671499278352,0.0474132023730289,-1.16131164949957,0.0988055123099709,-0.109775836281444,0.40456276271946,0.160759345760767,0.165591556139855,-0.17207389001615,0.483703598310888,0.0520733417049238,0.545599863040654,-0.156985311956422,-0.0102843890979214,0.0128957393577977,-0.0612417449652514,-0.294049623653502,1.33109021989943,0.206965081980583,0.0185957268824688,0.229993285238878,-0.0307524367880386,-0.249917265828447,0.04219330863361,0.0918780471110116,0.6771273761423,-0.109885880864755,-0.193385891158298,-0.0969393718866735,-0.145499577581397,0.0191467257743126,0.16273175682589,-0.707122940011464,0.226416428155595,0.485160545062415,0.114649110847201,0.0803683223494722,0.0033410553348896,-0.181362630064625,-0.246524509454077,-0.0222700084185953,0.0537732017980905,-0.00477013807273149,-0.0654666177010613,0.0289824954289353,0.0932716780597203,0.272584893418302,0.0613704497414233,-0.154929058279104,-1.45171019269211,-0.600919226807488,0.552468942851445,0.408316954844582,1.28514441986929,-0.0263402904583521,0.203743945026863,0.411500099524994,0.748622611489032,-0.192084552711501,0.0606852600060676,0.0333195077005737,0.630953270475039,-0.0751733518027893,-0.164231744407069,0.119957046976341,0.0505797034767922,0.381729729787999,0.385094269548291,-0.0501832392670082,0.0149568590024708,-0.10624300484867,0.106051539368001,0.119110972435978,0.321901336880339,-0.0712768791428222,-0.0631502215995302,1.02269763704855,-0.0869167053066651,-0.0739669887614377,0.100996170303173,-0.227346898483294,-0.100401426616253,-0.514145963446484,0.0534285831765181,-0.267001591717244,0.0952482290410452,-0.331833289220317,0.0370652807472569,0.141463301015521,-0.000893187142855046,-0.107641051416411,-0.0254382916121553,0.208765828683882,-0.55596388420367,-0.072880928152917,-0.0242500818764748,0.0481421014288978,-0.188922448483291,0.114882488063274,-0.00738302544131308,0.134868543220549,0.0607534355942123,0.0101907446715462,-1.16585982014655,-0.354082651219833,-0.72221731422435,-0.0961307747554622,1.22689536145635,0.675671392249087,0.0689037865915633,0.297301382240873,0.361454519767154,-0.208927635197772,-0.0758159213304504,-1.14118167074425,-1.02433502124191,0.388317402997318,-0.0585632475676054,0.0551723596658827,-0.551287385352838,-0.195176780626386,0.0998982891847632,0.0614726015455172,0.0484112754109736,0.0839696812861098,-0.0243296935667435,0.121998178263368,-0.0264553261053608,0.267122579004057,-0.157058351532524,0.0510415663390914,0.255100063547158,0.380326202475078,0.560077385706123,-0.0141757254925634,0.104058148474244,-0.228020744455765,-0.282591964574379,-0.583262845214962,-0.99094091220666,0.0175126953339093,-1.30144528011386,0.11679354780176,0.240081053042039,-0.0157686688597832,-0.145447103914452,-0.359395320700287,-0.295867814659531,-0.203570029062029,-0.0155894371961551,-0.029937024040154,0.169258297418238,-0.0229937288342171,-0.0908872222132647,0.508493040140451,-0.134555422020126,0.174120816747221,-0.0902088509427395,0.303530951680499,0.215000752209072,0.325249216435655,-0.0156276603903464,-0.330751432055765,-0.172655608912537,0.313345261159658,0.0836164402364986,0.108348759806536,0.254421284511669,-0.220786960111603,0.153463340394421,-1.50293486845222,0.0440792209625853,-0.0399168376727424,0.0535634923668224,-0.00996489172099722,-0.15097727823992,-0.0182744754437262,0.251483694681153,0.145557448542223,-0.365191926141549,0.0322142528314652,0.8409444250468,-0.0957132650671358,-0.0347639004321804,-0.100894537464,0.17364251580041,0.954763302694869,0.364793746659805,-0.138204739417065,0.24903297993319,0.214591156023361,-0.47510224591461,0.307452202976777,-0.032411534909805,-0.0614072353884676,-0.166098656824824,-0.496930934963988,-0.483897749661162,0.175186055191832,0.180944092355173,-1.92157802750164,0.436444264577235,1.56093747472055,-0.167449146098939,-0.147167979757891,-0.522115700263742,-0.405193373905827,0.305601603387735,-0.541938274845403,-0.0132133528621691,-0.227617293711535,1.05028490744143,0.656558324245528,0.0347952861172117,0.567386465416352,0.176399768285048,0.118652128011716,-0.477909706312743,0.313591992493538,-0.130108759552159,-0.169037455640784,0.177357779529236,-0.159699415045207,0.00483633293322696,0.234398689384932,-0.264583810780978,-0.0188959033354213,-3.64247923614875,0.174649882761075,1.90062575302927,-0.0967273522126341,0.022863121413533,-0.330045879292057,0.182735700362217,0.284810600627665,-0.195014534924163,-0.93850168913609,0.682246013958348,-0.0134250234538584,0.933031031153383,-0.257337336599089,-0.261940642073697,-0.31566727328709,0.0368923138446237,0.0421698702206801,-0.685830854100363,0.00757646563649054,-0.11565380309458,-0.481791810718144,-0.00917165031778993,-0.0188678098549624,0.11168089528468,0.468848140508243,0.092961929836937,-0.148914360604074,0.171271843363072,-0.511795615986054,-0.579803195072543,-0.0940169508642259,0.0766497571728847,0.151112989043605,0.00622309290567932,-0.0627713688715644,0.416083126932586,0.498388013034768,0.26526314481614,-0.216742332694384,-0.0486083024345923,-0.151201890282058,0.188256739249504,-0.186495145745925,0.192219140524002,0.408057633690729,-0.41027693083168,-0.0885130294790012,-0.15220261535722,0.190624344654592,-0.0154392944790864,-0.0826304370647808,-0.681462654789981,0.442458297841848,-0.873569262318243,0.125597818171374,0.228270428555971,-0.148634579096433,0.38728408786755,-0.0577654636031612,-0.284413681749212,0.0495753357314662,-0.0103681364197958,-0.0191843177411289,-0.14560504696924,0.0722244703927473,-2.36049018151575,-0.911990281473757,0.336005262366775,-0.102165237772754,-0.267802517376738,-0.308634819932602,0.0963944194792806,-0.0128163675828481,0.905550963759833,-0.00246206928037617,-0.122518519682266,0.0130709259242191,-0.20787955453552,0.167309119981548,0.772737142078787,-0.144780294025119,-0.0426840175901885,0.0171022473781919,-0.351334224651933,0.0219427105995825,-0.119816629176219,-0.605369754628354,-0.128474039381181,-0.162525119642971,0.0966942778462596,0.0706900206171351,-0.16364129053366,0.141339248080558,0.0735070989085064,-0.241810816719576,1.39069579939,-0.1023742497739,0.0220942649220873,-0.350908552494048,-0.0238882974135906,-0.096052255953239,0.101439775937086,0.535620631361714,-0.23684378735265,-0.173190084933723,-3.51131887563852,0.188918925006171,-0.0727886261238351,-0.0536576705196145,-0.19369317090651,-0.366066641605131,0.0144267946034534,0.205015822803008,-1.06138233411722,0.393514751778852,0.236051274400209,-0.359791237278551,-0.59193439982933,0.0182219128328603,-0.161605188738429,-0.255167297182916,0.719546137477134,0.0212769975492219,0.0957522849650456,0.601881981553419,0.148638512054028,0.421790305438503,-0.231070895120093,0.150596534395547,-0.231973524820205,-1.50052511246977,-0.048560388548901,0.524216465155316,-0.0504423533591224,0.284705365451451,-0.196888195099415,-0.207168168653988,-0.0268972538662641,-0.0763230896181799,0.162978896903026,0.237391786534279,-0.130733187274645,0.0268460793715837,0.227683740556226,0.0183883402872295,0.11002027005604,0.61020235164162,1.63347635929204,-0.369997813558839,0.125107483473058,-0.0910370562426227,-0.524079891101798,-1.92139473718811,-2.60122713850236,0.172612699326068,0.575029920840111,-0.0951957776566703,0.0137315575926501,1.71276187454532,0.0346515919101766,0.299857625276444,0.0564970831996882,0.00200189340754318,-0.149987303699088,-0.115311888981337,-0.192936053271085,0.131099794522537,0.0478566126139668,-0.0336478831374547,0.11497192167475,-0.0445548181079268,-0.0932276053392608,-0.340778174768453,0.119888556433204,0.231268024161081,0.32610095716796,-0.0196079786150561,0.1647072530543,0.0694554571945247,0.122193469893289,0.0513313263373342,0.864111274440568,-0.238271930265566,0.228530234127837,-0.0425413859157892,0.125844469055866,0.0106056719361774,0.100138019943024,0.0663627644605855,-0.210481455689655,0.128664544346679,0.0701599664831234,-0.0948701907697902,-0.121451060984245,-0.35058035171882,-0.00649908690260838,-0.127530617704857,-0.0831900208883511,0.0624825024247042,-0.26346082173715,0.0612788491035357,-0.0701615291498304,0.068723517496365,-0.0714624287735792,0.248382411533617,0.446649160826436,-0.426434087336124,-0.0258575711519547,-0.257575489383251,-0.777619578411105,0.00962754003218302,-0.419395739126383,-0.0362234333608573,0.058105333795681,-0.0555789182873882,-0.0628951322364117,-0.0611347534633865,-0.0491976967634196,0.00717995373471388,-0.0210418404537373,0.149450205997287,0.0880037427740807,0.355008797029199,0.0915264648813637,0.0311917528206681,-0.0218516788338308,-0.0184868497731817,-0.0732811278574172,-0.0628798655053687,-0.0264814364206951,-0.186859262942541,0.324331871235334,1.37832946215822,0.372196766898254,0.0322283707932888,0.0542167595304331,-0.364692153575465,-0.927565388555812,-2.56024231871869,-0.331703736956295,-0.024828411395452,0.0127359258260433,0.259204504746691,0.490722156003709,0.272984397442351,0.0684754834702825,-0.588471373180581,0.121411366581466,0.168656781289882,-0.247291262592913,0.0988839567198271,0.308990528187947,0.0818332768218517,-0.0592296358179684,0.786137220468117,0.284087005125617,-0.0438594484874418,-0.0121832492460417,0.140929393471448,-0.361229963552228,0.0950687392088135,0.454887329010781,0.0169402132508556,-0.265535898553979,0.0610669163269768,-0.171617694090432,-0.198329128801299,0.657278180938872,-0.253217342413625,-0.0495204992693031,0.0610616813983914,-1.10382760915626,0.30930962578279,0.203976488726879,0.565072860539819,0.607450946997432,-0.248817212921638,-0.0713963892260125,0.550518482175928,0.121194015228628,0.430017915446497,0.243274891462338,0.191706391753385,0.275873802749119,-0.418341023342518,-0.0462704195895558,0.220772664773479,0.349257743869011,0.178571168100679,0.688368691113242,-0.0653726001304971,0.236938041110606,1.69120073313943,-1.35492803176932,0.532309117586562,-0.109476452738135,2.97510896989963,-0.241815724543093,0.0410263625416771,-0.0122837178321585,0.437031214357484,-0.373543783516849,-0.0400289975788252,-0.124562287552803,0.0834082887334431,-0.149221354839634,0.131465452788261,0.339373042433747,0.353994700768787,-0.241447502709836,-0.129905338285222,-0.174543046309777,-0.141847403789092,0.0325042907585795,-0.802157378935419,-0.535387079534536,-0.167113189506314,0.347071915485916,-0.206304909478216,-0.269534830517333,0.217769403665359,0.0473027699537161,-0.641652225019918,0.131537443963548,-0.151884811868492,0.135538438104595,-0.261436294107493,-0.051665564666234,-0.125669340614024,-0.138750277615148,-0.0730183498894161,0.0638454454984426,-0.0781109560475366,-1.14940101710322,-0.498738171860245,-0.31248696469556,-0.212620757033186,1.07529194057908,0.998584031306593,-0.0930359066893634,-0.166064565867159,0.196886385441974,-0.0261897493399633,-0.256694479263884,0.41272270998947,-0.131276206594512,-0.786410754735081,-0.102367764813949,0.362017556225947,-0.267244458743935,-0.257066184540822,0.107042040985421,0.199242814317487,-0.196695424041427,-0.436492667604465,-0.595904210275607,-0.0145055285776156,0.0973559574286569,-0.16306760565754,0.158720872455441,-0.0581237738649267,-0.0801686594211392,0.0728409962144844,-0.0425164804140496,-0.0546918791976637,-0.432235857879075,-1.64705042543953,0.296181086422062,-0.399240797324485,-0.280858617836585,-0.0222717923299151,0.126285403239032,-0.106397204934032,0.162730102884074,-0.204462301357261,0.11296174862391,0.087595794512852,0.0480422975583173,-0.0271007055305778,-0.0310596905030026,0.14750577489787,-0.0782937591857081,-0.325401447597811,-0.320361446537245,0.078056575949701,-0.0779562840366989,-0.139028318453713,0.409062835629019,0.202341911979484,0.1839153740211,-0.0415592528111847,-0.391137173104818,-0.143199185093221,-0.0984093425034305,0.153091601755711,-0.952989307060348,0.103250714043111,0.186221545723706,-0.115224656673638,-0.55119660028323,0.352737431734135,0.0812330225723529,0.146540434050567,-0.0781768388309472,-0.0610938385932837,-0.0444039247145282,0.22036623358865,-0.13167023571646,0.0427036422273535,-0.0766539973457497,-0.147100354171586,-0.132981037298726,-0.0941727818347241,-0.093729869503086,0.228806334092504,0.209160161194528,0.0396298392550051,0.225565325329583,-0.0734957120650409,0.304473258477255,-0.0012525746268275,0.118254950089719,0.0921676800980916,0.16226357705576,0.142104090152551,-0.0454327184126835,-0.152224868998218,0.159985084868332,-0.118293926269046,-0.0440361477877621,-0.0380748253596194,-0.0484760563392518,-0.184803733129131,0.0374486141784882,0.182354347267076,-0.0835669234561534,-0.171021109545877,0.0154811400153574,-0.647234110342604,-0.126901625008967,-0.0382826425212466,0.115660519534385,-0.565683249489698,-0.388864591634361,-0.284906342672615,-0.395324168769949,0.727546666138344,-0.460754477519771,-0.248594274648111,0.377936966670935,0.368121626035177,0.14140266290799,0.0168884163744601,0.151643265748705,1.54828337519534,-0.283540338503402,-0.196124877759892,-0.177994343500897,-0.208110475505521,0.0172806605539189,-0.250885092009084,0.0357889521485339,0.590555955347972,1.57155057590765,-0.333345770767879,-0.360464063974891,-0.947538108675235,-0.303451967348163,0.528964784079513,0.46063148963692,-0.0773512888276787,0.215931381906048,0.19292966200614,-0.242654058000639,0.40856385313606,-0.00194056494337777,0.10954975044257,0.18568098155516,-0.153354975498302,-0.0219988027715135,0.387392131429375,0.390095352385427,-0.458872250514753,-0.0595637853561839,0.0769426938161181,0.219039658435331,0.186600652691361,-0.0300196293046518,0.0136099512157612,-0.143966028621838,0.00591621340556483,-0.114749199842705,-0.156149718174173,-0.0767849600893756,0.13346362908981,0.107473432549134,-0.484438347475133,-0.0648681003939945,0.322061623854366,0.125415734768913,-0.150493214057113,-0.692830515000324,-0.412927744694185,-0.231845867958134,-0.0786465739328102,0.242113697774837,0.011948898058774,-0.00252377527108384,0.0643709691099984,0.521412206454541,-0.22046888371019,-0.0761860293833469,-0.0915887493178488,-0.185905670705934,-0.42665925869139,2.30719604061207,-0.0109348512664541,0.177805863336618,-0.153487301998488,0.136356034232177,-0.132236223806552,0.0311644898430849,-0.176597107819145,-0.601960058177132,0.108490405900354,0.0598819103253494,1.58891230341656,-0.170135730814055,-0.136537198394488,-0.0361701287530359,0.410862592265435,0.739402867120629,-0.00353337319396725,-0.113801698809677,0.990245720373863,-0.359470755768732,0.062026037531204,0.327720603283975,-0.315043928955337,-0.140803613393371,-0.109428806350043,0.181698925091364,0.251737095884909,-2.72471647464398,-0.188223333061209,-0.623046125483276,0.195399937652001,-0.863610830385645,-0.06273198434222,0.881791480078235,0.0293159856184819,-0.00138068170465851,0.0741812765934882,0.064981153963811,0.256326690682555,0.101371771663039,-0.108998621036967,-0.291379268191672,0.0788453157758259,0.0828552291991758,0.0539447849829392,-0.0365222347936354,-0.205936570491772,-0.235936452280331,0.0190055053856516,-0.326535038618778,0.102062011984038,0.422827135554392,0.168969467380411,-0.0453397710069858,-0.441430107508599,0.00838400482348013,-0.092951296200675,0.0169017221762875,0.0457265856925996,-0.0171874655132693,-0.0708993261778926,-0.300618995081633,-0.364836353803764,-0.113450891966757,-0.484750924305722,0.0996120871648003,-0.171442002174885,0.162056337660397,0.518587854361255,-0.0405473711282181,0.0349881761307456,0.0749445152118516,-0.322789057146301,0.091924006709629,0.185269365403255,0.144088922548098,-0.777953399754604,-0.111353264885578,-0.164055958344886,0.181665642815743,-0.0466711458236483,-0.191294021572121,0.0338877017775399,0.078221111064954,-0.191859258847863,1.53377512530393,1.36199236486938,0.0944442651569653,0.540889037560355,-0.0920143252105591,0.442844340403642,-0.0852153224826755,-0.0960524285450114,-0.0572270565797088,0.0726301684316568,0.0598176983002113,-0.0310643330897638,0.240993972757313,0.072136851747756,-0.0723963922495447,1.73389041948692,-0.187446490695496,0.268334592201595,0.0736442118141313,0.242401821021214,1.10734468189348,1.7634393828736,0.366483544256914,0.0954387343450518,-0.21851986083851,0.152267800144098,-0.162566908112575,-0.0572040641567567,-0.127192283743271,0.379738660980893,0.225097768190163,-0.0238447424120706,-0.421953352051434,-0.132358874194612,0.0965337268235431,0.203139597281214,0.215193078871297,-0.234017338058504,0.0481495473263613,-0.0213313552895463,-0.0141909618607118,-0.237852472319784,-0.0523001734302649,0.792562293602678,-0.214954386111397,0.00307283850965332,-0.261186479154993,0.532619250583746,0.000530764201381811,0.177863559963177,-0.0962189104738194,-0.131880910005007,-0.139117592649133,0.247993640533227,0.00361710184257457,-0.075287749312297,-0.118937319955054,0.120874530298569,-0.358727361199909,-0.0165496405425119,-0.50370661017748,0.582383195222797,-0.12739269672803,-0.27206846769656,0.217349322999728,-0.74558692265794,-0.0667608210660186,-0.16449736820928,-0.280513133495752,0.187495197979281,-0.00520808907769332,-0.497347680028541,0.0672852464187689,0.307414764631278,0.0113473743187989,-0.729023949746148,-0.0334720117522575,-0.797367019377675,-0.0354790749532993,-0.0570558862533656,-0.0223629244641663,0.027348253551956,-0.232818680208236,0.225208430500032,-0.101686154381621,-0.162335758929649,0.129705567461473,0.137051497003682,0.11256973920441,-0.0809183074868325,0.0114964619855791,-0.916757325964024,-0.308875785056452,-0.301724987523328,0.448168421902414,-0.162602579354751,-0.317916709965879,-0.199269378196911,-0.584240292552204,0.104251313480802,-0.0255408931314999,-0.0585284547998546,0.175063677093414,-0.0151128290860782,-0.868565306724214,0.0544597407516312,-0.0390900158015791,-0.22916942126378,0.174812277386616,-0.225058697683424,-0.11945552628369,0.0396039825340855,-0.311443871676368,-0.615335175946341,0.515392252006472,-0.327449091161309,0.156467082536697,-0.137159738209982,-0.240131660161886,0.399657626952892,-0.0027468953644091,-0.40506732898635,0.0231455893432889,-0.114392235620238,0.14609865355716,-0.073793683052567,0.0431346066461369,-0.344826740748057,-0.209637990389726,-0.352722498751231,0.0572156537192567,0.112196702639995,-0.123805478903002,0.0330886584243179,0.0860877494227774,0.139146433151942,0.354933411350973,-0.122130193843342,0.332126030744084,-0.423371291249454,0.706566627222657,-0.0152511277873055,1.52390375959693,-0.186206282661254,0.315487379195684,0.397401890218178,0.0432108894758987,0.0739896499001574,-0.0165213020384313,0.939771530746114,0.106604443560412,-0.178902879459318,-0.12944174600784,-0.0491086906508777,-0.118996288009688,-0.122788597615077,-0.294713025408672,0.0536727134119931,0.342615766608056,0.330617923887168,-0.217031741916944,0.121918498178016,0.295029734495521,-0.00329118390348709,-0.251621775199162,0.0366610701373696,-0.143453387857082,0.310986438608396,0.0243841563875843,-0.269365232273542,-0.0778331482130375,0.0659481513236101,-0.157574172379908,-0.151129762098015,0.322009708044613,-0.346255385034582,0.317839030392084,0.301475951216126,0.0853003612064703,0.192341355804238,0.0183170731158907,0.238535378056404,-0.978710565382757,-0.0769608506885953,-0.135768798301736,-1.38667292150171,0.0734114704075214,0.190613526241357,0.0112957716385521,0.257655091537073,-0.0404440588822359,0.0123536292006205,0.240443772084743,-0.275203450532813,0.0157519390152394,0.117328335169947,-0.0673454687425865,0.0346949617083854,-1.17099572396684,0.138139014349066,-0.311467101075785,-0.163459870110466,0.100465803965904,-0.0372464767142046,0.392498575693158,0.0350674091931018,-0.0410004127064198,-0.208859883521529,-0.315835435056375,0.0172397386849326,0.187046307806174,0.211583269211731,-0.0138637111778566,-0.253525809268364,0.196437752447224,0.69392254724656,0.025886091056131,0.00100559371095953,-0.00226095496054252,0.143238302926761,0.224627134878427,-0.0969818344260602,-0.0636613599479371,-0.0209699045584507,-0.21241631605718,-0.0275411316886996,-0.128289973753459,0.0508737197099343,0.0195579358291938,-0.118180334659545,0.0372865136831053,-0.0854737817489847,0.141308214141707,0.087672916089057,-0.185086569301286,-0.0973177640884261,-0.447203674817185,0.0513844482937432,0.0777747240197155,-0.0141233944297659,-0.277673377824478,-0.0379547192281817,0.0423138968407988,0.590291287077469,0.0969869647976086,0.36869748530523,0.230395089582297,0.059876810255957,-0.274643054534889,-0.0169985191490369,-0.359417457705317,-0.148766429991021,0.00162828073074412,0.20885638960045,2.14722737882292,0.187958506089042,0.233917921723603,-0.0190052960449304,0.121592276003604,0.196020774989246,-0.103704837601019,0.16523304305822,-0.0277844911633564,0.0178311339800833,-0.197593755286542,0.132622371243312,-1.03287722766772,-0.177379997988016,0.0693112537813683,0.113034196545837,0.03227712554629,-0.0985455256598807,0.270238178928598,-0.854694149929,0.75110163020225,-0.243193094101136,-0.14598712073082,0.045189430773561,-0.1239421417432,-0.0964906305689559,0.297579228055676,0.73858809671668,-0.28574540723033,0.459195951156767,0.0594698960168145,0.124802225576991,0.439221371066802,0.341102923387291,0.197447762172413,-0.083919065391476,-0.106667156814132,-0.663052058813272,-0.147630125167914,0.101470250415982,-0.128711355816645,0.204195322171526,-0.879538639336017,0.104536901967185,-0.218596674150775,0.6234896883061,-0.000228957290016354,-0.00875492048896896,-0.0910890884418623,0.275264052856772,0.0600349461923003,0.293843300956091,-0.0547464069868438,0.0385176464828116,0.0884264449494043,-0.0215143507850409,0.19485280518129,-0.560185113559039,0.0567252984548525,-0.20003013195587,-0.133778155240593,0.061461261624028,-0.223577120249195,0.0274303137359205,0.113980074288211,-0.459171648460263,-0.406524562939208,0.455988029416973,-0.0951514463515022,-0.128224663358014,-0.0528745420347802,-0.0760837505254103,-0.252362991575012,0.0267413100478526,0.414757751392211,0.308853562543907,0.541984016482195,-0.402570738585418,0.274888866629832,-0.236294067470337,-0.0178991934234382,-0.226115588386014,-0.267615159975902,-1.02208155967446,0.347794664610382,-0.140755211442622,-0.0611915904368241,0.481608733093872,0.27622331310304,-0.0441685709108804,-0.131979505358999,-0.198504034094017,-0.152661923655588,-0.136517637065237,0.287899318151698,0.0406765344982614,0.0376506876272388,-0.0347202462146137,-1.7432882070852,-0.61203641485404,-0.195136058651836,0.0499123653240282,0.0382283970225311,-0.081875841559887,-0.24925649656116,0.209429869988264,-0.00138538349505974,0.240874958825708,0.075770502706558,-0.180450923834505,0.127043658779762,0.192567295081365,-0.0185058720299448,0.30375631871791,-0.00997827160307968,-0.39087868900881,1.08756668278766,0.501871980611293,-0.381426412787212,-0.197971186353911,-0.0167108065237308,-0.0074262397278583,0.137710187669726,-0.00762934162226967,0.0419823754580776,0.541420606954144,-0.860020198298659,0.0951342045833826,-0.136437471783477,0.0116981259850943,-0.0939203448029896,0.388253862119517,0.120700058657654,-0.239551855220445,-0.177828622014402,0.0778095783558203,-0.0382132127820587,-0.0512658320654293,0.113684857622796,-0.249023907218844,0.19174493106821,-0.121491520162284,-0.382659188668291,0.100126607030721,0.104229803084319,0.298211173255475,-0.0956615711139462,0.0755605799243294,0.35368080729596,-0.155891807747346,-0.373876461291279,0.22676304230742,-0.0353102121533878,-0.975622115474817,-0.223162625482494,-0.197961718130847,-0.185869976471017,-0.319534008003912,-0.00622704572962966,0.626127072883079,-1.3481424387845,-0.0547785715159559,0.0605982306635396,-0.0337398138938645,-0.0323909374507795,0.244303316525597,0.107125875177187,-0.127948022653905,0.198452746216388,0.0975336523440684,0.114389551146978,0.14831278490118,-0.0489541929708464,0.0230916468372876,0.815449524607603,0.158410851120741,-0.154289351817075,0.0700161576486672,-0.0184396884132425,-0.0908979835674197,0.299570000935599,-0.0462197175330105,-0.165664183929399,0.304528912737212,-0.0844736577241872,0.0853019889364187,0.103601501619114,-0.0735616093158153,-0.258992731772684,-0.0524658399040427,-0.0454842357486654,0.145599235930702,0.0711949940312727,0.0295988123866779,0.537242519484707,0.156978273375422,0.243624334990374,0.125548423645497,0.325845434512467,1.51411893726656,-0.151500854306766,-0.0723694771574229,-0.345489275465165,0.000314936401107221,0.156989283477928,0.0872113273120953,-0.751382387947284,-0.144456529068463,0.030473812072244,-0.252146004137583,-0.29873651089235,-0.175402624568297,0.157766620294334,-0.115160216147927,0.0601203109741362,-0.189608371497276,0.158241355464692,0.322460195504485,-0.177910041319628,-0.0993391771450903,-0.0715746976037622,-0.291752609439518,0.0319175696260626,-0.06914412983539,0.131952776924957,-0.134371917462116,0.0354075420995533,-0.310165510266749,-0.308553971110264,-0.187220784703983,-0.0522588201438675,-0.013913497230101,0.17334233650967,0.123761854475681,-0.15338137097778,0.207485921174825,0.0348872098367617,-0.125516193247266,0.124313095144821,0.252846997673425,0.0570328755383688,0.0497589548448162,-0.404168193966835,0.326097469106585,-0.0581503515166378,0.244750296105687,1.84127065899158,0.101474458292271,-0.109294843056227,-0.218715751551207,0.0608673545475291,-0.135342326868444,-0.223859569290266,-0.0363319233698271,-0.243211811018589,0.0888878427140005,0.267127426734269,0.300380336801487,-0.106215195995307,0.140507332806553,-0.157166631175583,-0.0848062990125612,0.461035400286606,-0.205469198077622,-0.17855443781117,-0.022605171647923,-0.0740178341721114,-0.63335300692681,-2.62656241968574,0.191952555154903,0.206557854305419,-0.0192118927850955,0.177456213747977,-0.133327792978042,-0.194161798253121,0.109745764862441,0.043494128078292,-0.0574850387114181,-0.935819123055576,-0.875431005131,0.530351591888802,0.0605101681031371,0.0731550940337147,-0.12531814650882,0.0946125536040829,-0.275558304411105,-0.0719705416586518,0.0329474056221346,0.304665140088157,-0.0130151088300722,0.00482373839125391,0.364380007680856,0.0175853796571457,0.143862501485626,-0.0293591138385047,-0.127589757021167,-0.0307261351433296,0.45927114314754,-0.152597902150361,-0.025809963843414,3.0106884736439,-0.138006632219857,-0.0066892110175752,0.172901877412013,-0.0482614372974938,0.15011323815838,0.0132113565457044,0.224606185912885,0.540471139250385,-0.0936957323681741,-0.186522986164053,0.00869063912519397,-0.0217045019934834,-0.151444208267574,0.471959937337936,-0.0964570095958129,-0.182421839545634,-0.166610055774634,0.204116252160336,0.0170538462079011,-0.12653391758015,0.0232869517864887,0.116057963941243,0.257781498378874,-0.246937544135843,-0.0489057803619662,-0.104961276331804,0.269659620300662,-0.0383650789527549,-0.415766311443719,-0.123433700841576,0.12786734940156,-0.0948184481146784,0.378507740753971,0.00999132973688257,-0.00268575998757195,0.231028702995285,-0.00170513124264453,-0.277514108512617,0.147549544568788,0.0711096324242849,0.429433611596001,0.190172415007603,0.0634416574271165,0.023013446068207,-0.0434983168241703,-0.605784018901671,-0.12164414823187,-0.213805261454668,0.0748050606854476,-0.111903107537129,0.120378362768989,-0.0538873534205308,0.176484040626994,-0.28559387486131,-0.592485198201432,0.027323222409073,0.34602489092227,0.0535389745513309,-0.557670153838389,0.0378317209679995,0.297166137409083,0.20140422344883,-0.0940131274872373,0.0697535296891511,-0.0883723125432826,-0.0709383914276353,-0.449635646043405,-0.105246910106812,-0.357942693627681,-0.0764400088593539,0.166003344758104,-0.159146679875026,0.0888541897927368,0.896519193389172,0.11964950362324,0.228663064194191,0.0948128071803856,-0.0653570195582069,0.0605134191453684,-0.00426857475007735,-0.100830138610842,-0.346342146408038,-0.0502280838512725,0.565519311082375,-0.176195494636725,-0.239809918241124,0.00766731302497762,-0.450848463591989,-0.0812121249111171,0.310765737672555,0.494667072226789,-0.0455165696378917,0.0689137628202096,-0.229802161563142,0.264171613934321,0.0198112892829035,-0.161427891930827,0.00559859252533612,0.107537006568444,0.0191453860676105,-0.402499967785816,0.048722106475952,0.0553949676960652,-0.209600435721949,-0.254349310504584,-0.454998408841325,0.0351898013761997,0.131375306490721,-0.147299861109,-0.281659646191502,-0.558292304103562,0.311586275251027,-0.00626559490821063,0.0286255149802998,0.0105432244026473,-0.171192653606878,0.699344640286745,0.134561076846763,0.171016264395695,0.140350605855672,0.0139852001176566,-0.321578829477292,-0.0465353882139443,0.262222058848882,0.327907441812799,0.29810978123952,0.567616950788117,-0.331706006494159,0.14880564062235,-0.0583938168950555,-0.316659937225287,0.551150919227269,-0.00711602598525027,-0.0184875790771784,0.104990178647371,-0.0467372201094363,-0.211851773199784,0.0972593763840714,-0.126223764869344,0.164676339844986,-0.016465134691196,0.0947905519314382,0.0577773433111975,-0.13715773887238,0.0796740685023978,-0.484713461048408,0.210230351350914,0.507380801498512,0.282272316902033,-0.210774587864194,-0.104773202173637,-1.08186681435204,-0.145542419807362,0.134019195146505,-0.170554580679812,0.762763004364164,-0.0392235374410544,0.103716393596765,-2.80717642442733,0.19011337839421,-0.0484078924035155,-0.0393473261915034,0.113005253307066,-0.17764559586005,-0.152689024597243,0.126490660445417,-0.0697794362759013,-0.217916117503913,0.0428166038553237,0.00911188058212271,0.210226379167842,0.252395799191018,-0.161900379669401,0.0230827741870211,0.136218835880595,0.764308395014642,1.73767896095619,0.24288586552298,0.770125270762935,-0.849948937825002,-0.523229371466983,-0.180108851785477,0.0228285585723413,0.0677822488225325,0.124705575636268,0.227033032449842,0.241942982182368,-0.195825048661805,-0.222954394611842,-0.64187577479944,-0.0406417295767661,0.0398976933938858,-0.167968209294334,-0.0101132982224388,-0.309604346998423,-0.173346618765301,-0.123509227597822,-0.297475194458476,-0.107748982409711,0.457937271129117,-0.338203772224549,-0.247576186751375,-0.0246719548908048,0.344556080643677,-0.165914806801676,0.315717981086457,0.136390005659366,0.247285762128121,0.160460316286216,-0.141104114525499,-0.0187786842606146,0.76390874942713,0.0494390061265321,0.316271445993678,0.113573887637915,0.280631120382765,0.139037400330206,0.279462210527483,-0.0501345586243587,-0.0404918337687886,-0.0101705687170317,-0.286999138498945,0.0919673267012073,-2.34185518676327,-0.224709674677372,0.0174219358663655,0.3610451908362,-0.125695308392752,0.287993383208363,-0.352954517756866,-0.120544747259596,-0.10711052149549,-0.113737929646657,0.262850298297856,-0.283008487997243,0.0643256482261345,-0.160245099116358,0.0177391376779487,0.0209717699261007,0.113810125187262,-0.065296017582298,-0.213204043235035,0.0560076132953222,-0.199383915026551,0.0304371231025542,-0.17473758149418,-0.0603199954390091,0.130395501878246,0.0647599980901422,1.26099581589493,0.202691475592551,0.0208326326293923,-0.117146237465465,0.757681036249719,-0.0126824196057559,0.130326454834622,-0.312263054756942,0.0612539257018478,0.910616359418808,0.223879649752554,-0.0309666735139622,0.0269277121054475,0.701887231274731,0.19547476066589,-0.341461120773996,0.348163579486676,0.640403299719465,0.394496195573062,0.191086409041681,0.0837474369807213,0.259324734242209,-0.834294958006801,0.291378903050817,-0.137409444307927,0.120191894646502,0.293116410182896,0.109319899235335,-0.00172683252782815,0.314450459361081,-0.0319734481169654,0.231444122732483,-0.200544376199571,-0.426555704469956,0.271042317538432,0.0221087369124192,-0.043514664536726,0.113207797443749,0.0979145120597219,0.0482853276093431,0.906836269006458,-0.17026096951653,0.830330332841355,-0.276262955638346,-0.217923187796166,0.407548051207426,0.347163739784715,0.227121545927593,0.0759471463538996,-1.18690526517673,-0.0313693905328534,0.119139323353041,-0.308004510487204,-0.416032199524512,-0.00999147913981262,0.076359043257686,-0.117589491101397,0.454267633491925,-0.136382720359303,0.135175804950838,0.146039067420301,0.054375022959849,-0.109608032487687,-0.194443138997804,-1.06761804701621,-0.543330406073275,-0.20988059121975,0.209020918070658,-0.132725685452753,-0.0509071728288179,-0.275391081733666,0.0647210974841677,0.00662153657976717,-1.04339493673817,0.0147128924555668,0.171979694240582,0.222347375695108,-0.128004610813483,-0.306570353401814,0.248044031964958,0.0446479143291465,-0.162469002766734,-0.125814424897585,-0.15439674276333,-0.0660426160205112,0.1923335504835,0.220725111768509,0.12356989583793,0.105885164711544,-0.0968214931998256,0.39695671249394,0.0284107402687941,0.0992853245455375,-0.0418091555327396,-0.0962572706955446,-0.109600908466866,0.469381653619057,0.538780843739729,0.270335067589708,-0.101571952689924,0.514154741496106,-0.00415474914509672,0.180707172308647,-0.0305600248148153,-0.00670514085925136,-0.108499063579886,-0.176401383165148,0.0740478189193877,0.573190243519366,0.0168201516470649,-0.863428219774616,-0.56761459623975,0.663563104326704,-0.586394056450484,-0.168067538573185,-0.152809100761493,-0.312498440885589,-0.207479474250465,-0.0951886061290701,0.0195371428753124,0.43635156221787,0.0181704153990103,0.0458386517732809,0.117772208175774,0.0207252967731846,-0.407502336376353,-0.0411939754709591,-0.235548903354849,0.958453542373209,1.70947275341798,-0.288356183494917,-0.253594975229176,1.04994922215363,0.185677228517376,0.0345314761802089,-0.111432993359168,0.118027784498546,-0.539769438361658,-0.0620056825605195,0.297891657017599,-0.0618274108100859,-0.0456876305118767,-0.118193970300539,2.39422293604553,0.0701416259028809,0.0593127284389929,0.729193469385926,-0.134497974488754,0.195393547787341,-0.0812800276643778,0.0811106868804125,-0.100753043362994,-2.8734421734235,0.0252368273888161,0.0101177094430339,-0.453251478821191,-0.361389747362607,0.118202769659833,-0.616236457537129,0.423095666060331,0.125845925971711,-0.650933915112448,1.50664395844272,0.148745975208784,-0.172980145104816,-0.0784011565374243,0.0591218226450231,0.00881568032002965,-0.243894207539529,0.387358401482382,-0.145233639848935,0.304966794029323,-0.40528315875408,-0.294628054053405,0.00923954694423147,0.612569039855494,0.113819088204689,-0.265122408389176,0.017649001701041,0.117145858215337,0.113401006709795,-0.0468702136101104,0.247451637361396,-0.0949681430362943,0.569712783144234,-0.0108214210595473,-0.0758953228603224,0.162826863514571,-0.0347813996088335,0.00285846374402097,0.00833753118607307,-0.0896215270296247,0.134990218650374,-0.0315844700363683,0.361693759460876,-0.0996144259058537,-0.312219092496974,0.213220941107127,0.154559620210552,0.0232494823639181,0.0045655338768938,0.891853616848307,0.149216802301395,-0.140997815090527,0.322322923141473,0.25738237528667,-0.323951623482818,-1.63275335778442,-0.274902417037213,-0.217117896453833,-0.0988271402004577,0.493413391972905,-0.22304143186105,0.208789089663552,0.265081199501782,0.0355820333459077,-0.00921076583090636,0.0412595576860328,-0.0305291342929726,-1.12152275255715,-0.00616907244895422,-0.138503591758927,-0.0367151161018803,-0.0536080611287858,-0.479402214394977,0.572211331412367,0.70922918675357,-0.237777740015877,0.0574565262223376,-0.228700563738954,0.0594292181941454,-0.0872792347028707,-0.0912182383361828,0.579401714932038,-0.0618946389351111,0.74084990371828,0.676478486411126,0.866427144817763,-0.113994782256221,-0.510575747484825,0.328890538027457,0.0525032217078452,-0.132429944992226,0.257041038049155,0.082397595312531,-0.0236645155189089,0.26740451961461,0.118473648286225,-0.114623145408444,0.209901585748779,0.341491008201159,0.675403531728319,0.0594463898631578,-0.112377401122096,0.0218275713567831,0.292351234236173,-0.0829251395383301,-1.43628920149228,0.188450305920637,0.46122424187836,0.0699066747779469,-0.0531233346435537,-0.36240967786319,-0.0248201951892326,0.12684131851989,-0.23112045193363,0.223397812144059,-0.080089800851568,0.0140839411247763,-0.0727456968540576,0.116524867986064,0.0884810151906847,0.675548014982735,0.129757744507224,-1.38120109774195,-0.0116955121755287,-0.0675675572367534,0.0929072320971166,-0.140466015878461,-0.00845895047071206,-0.0719709166529151,-0.314942101882346,-0.0770699711255916,0.0623898020884036,0.0329318212446064,0.713815733296886,1.39548886184263,0.899484783228774,0.0253662638546508,-0.207483337919118,-0.015445140875427,0.070013829260823,-0.696523931019224,1.02766908927976,-0.159249758276247,-0.0400767469527026,-0.00744935838781315,0.0355201678677112,-0.222562363301588,-0.238944523535656,0.770113361715261,0.0627748944588926,0.0461634013930153,-0.468638687180225,0.11433192565556,0.219703413736929,0.0403193549466303,-0.259277511372457,0.317069450724764,0.0168227475445651,0.051964603291671,-0.144129637281459,0.0789407085800635,-0.148883359256084,0.34879432835245,-0.509715818675185,0.107593613013766,0.0556026006807709,-0.699981080139837,-0.0517488524578236,-0.228595809758524,-0.132420766661367,-1.26035976365322,-0.543882696688409,-0.0657296481645431,-0.0382494012605164,-0.267569949340925,-0.118693848622391,-0.40210452323851,1.31864614556258,0.13212454106322,-0.261204877545777,0.0388305399357896,0.0555912082867707,-0.253446110108452,-0.287526334641973,0.040349134018586,-0.123528622867456,-0.782807988844687,-0.233626620595723,-0.481926395315526,-0.148505044572573,-0.191410419506269,-0.244272949742885,-0.0328434034244394,0.0636944256347548,-0.719595067229774,0.0957831283725422,-0.205849263849131,0.267412567053541,-0.1122636934701,-0.352496682355083,-0.342001409111731,-0.0626883286396227,-0.0197317644177693,-0.339709038188016,-0.40231850936376,0.110021240780509,-0.271632320305555,0.0160253620865367,0.437199884043318,0.179978840602598,0.681091545273801,-0.331461156547421,-0.00292978665722703,0.220433120815903,-0.141209287117379,0.0132216684169306,-0.108492043333487,0.143264655710286,-0.0300762299368867,0.110771387285129,-0.0357107984850584,0.0698704337833327,-0.613533719209677,-0.351264104865652,-0.22877669319577,-0.0666593884794285,0.18651665496729,-0.0947964832366685,-0.0414435784042945,0.301561584229552,0.210777834671015,0.16426833677048,-0.0948410905923685,-0.0962519315959038,-0.0543371275460763,-0.102589276074416,-0.436866369312023,0.367447865713881,0.0298351486697592,-0.17697691044464,0.195867435047472,0.793457589268301,0.753935183298846,-0.0241148294855827,0.087363332132627,-0.11846158672335,0.0689879995910034,-0.12744990922316,0.704839658192238,-0.226996285750467,0.47095099355602,-0.14040078334017,0.0316872737569457,0.997965092128956,0.299030426102462,-0.0485523345210999,-2.54101053978943,-0.037891976182713,-0.222982807985557,0.0338201016213442,0.0212972004736041,0.321281611719946,-0.119074277933177,0.0279265065851161,-0.0338185348861317,0.0520050489174281,-0.0445245638026223,0.407402782196779,-0.196636475635159,0.204880350244746,-0.0716038666893376,-1.03730155332439,0.239924157063364,0.0534330967427766,-0.174833958453022,0.244930752133389,0.0325826835454922,-0.18091052553278,0.0830790019531442,-0.683044027970832,0.236994288646068,-0.700129860412145,-0.125438434110583,-0.126734812147302,-0.33558225393783,0.178138335407776,-0.895394416957712,-0.114236941653643,-0.411847462121343,-0.530397780978878,0.184120915170937,0.300905872905693,-0.0360185465162514,0.034872949579227,0.107021134023826,0.0151474306681228,0.446266319464838,-0.18848079703489,-0.0849579287846859,-0.0262369989388895,0.138421292771779,-0.110651874461572,0.431204507084806,0.534993158382433,0.299704743143062,-0.556953858647267,-0.0752038330638341,0.0305394580229124,-0.0336510573160261,-0.349762958039529,-0.173464654765206,0.332413164536371,-0.2449932814572,0.0941369931425443,-1.15984490022729,0.0119716822115351,0.290018961975898,0.0334007084689199,0.0596840491423826,-0.255880710222688,-0.056160299815813,-0.00285227241597812,-0.629196687996239,0.105634363573147,0.00968378848905945,0.799413571529149,0.36531031411476,-0.0885168650322162,0.0679899385289686,0.00459984206042938,0.0277532547514168,0.337272818416711,-0.123916302033227,-0.309959551133513,-0.37417015916791,-0.246301504778339,-0.207731741371906,-0.401096715686248,-1.02084024553826,0.198915089971723,-0.421158600729724,0.0843252999084841,0.255697768367007,1.26404755930797,-1.15887504155535,-0.128050117170886,0.0265105441176004,-0.342590039059548,0.0228969375946467,-0.971122552991352,-0.423200896744459,-0.752221339377896,0.10590414858306,0.118300818380657,-0.0756947941472635,0.172056263612942,-1.06588190609346,0.255459480292101,0.529631220169008,-1.73769599833893,-1.01982278057806,-0.269257413739547,-0.980780482010924,0.0733773720236507,0.599940260715417,-1.30354622257228,0.188788330237799,0.0704409040918819,0.267686091380912,0.948076629980424,-0.0556557289698946,-0.0874460351769713,-0.168947161144802,0.156452260185722,0.0234082787772889,-0.0291559415459759,0.363739183314914,0.152024806544534,-0.0114308291927669,-0.0556149862052928,0.715847158802403,-0.00380250804440745,0.0709220426092016,-0.216979767190424,0.0217339207432965,-0.198590309495901,0.336753100098114,-0.121151386509503,0.11144273238869,-0.218116886879675,0.342871742735598,-0.0914904207587253,-0.528595592070244,0.0760405018121294,-0.258276173603772,-0.211014129229477,-0.0614673840986625,0.266974637853505,-0.105486728583506,0.139809561335391,-0.239254616136651,-0.15947986759865,-0.230106613145703,-0.336968198016405,0.225657556060161,0.172283402938884,0.0967784502935156,2.32037292043521,-0.0498495488688713,0.223527558233794,-0.136194986579458,-0.0356339045579513,-0.194645392376302,-0.0930858015242172,-0.0559206902581914,0.0261876826935884,0.0377080965595544,0.110167087877416,0.260829529388341,-0.0998313580379615,-0.0478870359032703,-0.327611941860777,-0.559745741407801,-0.0113963792166391,-0.221230448093232,0.0685315365918832,0.00344158356029688,0.0264208839645446,0.126827222011843,0.372019119129368,0.100230965647808,0.228390183167388,-0.0715655262767519,-0.0841254968802347,-0.170016706053555,-0.0861812423109381,-0.118658573071918,-0.122217585343385,-0.214119447388419,-0.0425918575624161,-0.298447772139193,-0.0238188910058284,0.278138514875024,-0.742609218575889,-1.05253081530103,0.191459520113741,0.0948512815364414,-0.259253422445452,0.0114769116406759,-0.0749894098121244,0.181672678720206,-0.122995427282906,-0.281936602849869,0.0483422866877883,0.401305015721024,-0.274164827254368,-0.38650772984586,-0.142944891076837,0.086355528878756,-0.420245374265005,0.304528883195386,0.132141924140076,-0.0786234361832772,-0.0129386850378637,-0.0513144646480427,-0.0994161882196703,0.122933258682246,0.447699655616941,-0.289706150423445,-0.476807851774044,0.623060276584742,-0.182885968423141,0.227831398045645,-0.0529951392303012,0.117407458975322,-0.0209347717371446,-0.0481688911842545,-0.0429310198370458,0.170672695512138,0.448241366232633,0.107014405276882,0.0283341225147639,0.127240364946554,-0.397472492457718,-0.653985171436262,0.286143508146183,0.510723683765584,0.199981390890857,-0.162754325538702,-0.134532280381432,0.133745863959674,-0.587790297280761,0.275950480350542,0.604366762536822,0.379973319744578,-0.418001954986492,0.126228110342331,-0.0444595558475332,0.0278249258156447,0.0756699065921787,-0.0682426110362071,0.315614333316493,-0.321411350530099,-1.16037893065674,0.425380957569242,-0.00557189554480536,0.599450476121425,-0.125604003438482,-0.174151420782247,-0.0183315157757176,-0.0132347244450613,-0.0996111688886197,0.0456341293287603,0.0195237946212897,-0.00330979604321241,0.354315068587248,1.12622120611775,0.2846381232496,-0.157530251079879,-0.461824914096053,-0.308863226075622,1.54943628613078,1.32561834837953,0.136891898091364,-0.037205924641941,0.190773069713756,-0.330223500569215,0.0738108190603377,-0.37003610630107,-0.222652473750614,-0.979063294860902,-0.114682756138014,0.151935855935625,-0.259851976347556,0.304403064363704,-0.155499940625054,-0.0393701415745254,-0.247927041568279,0.160521330178407,0.479140852216235,-0.730526727415609,-0.159701784058794,-0.152417919964916,0.153246062867705,0.118568814798979,-0.637271603128127,0.06706199745694,0.820295375319283,0.359550997814542,0.087436038755145,0.308241102041045,0.00544726167469063,-0.0365896222135599,0.0563583229884515,0.217634737455205,-0.222647961532191,-0.0165498332940822,-0.091356809583674,0.265059916225436,-0.0245984128997849,-0.414550129387235,0.216812369806763,-0.00232449279499865,-0.321404826141124,-0.0446238626404933,0.0843431220265942,0.130781549651256,0.421764645931587,1.93795466533813,0.0927998180994034,-0.187326996814396,0.10842033863586,0.110880546415781,0.201423940001666,-0.299074492209373,-0.533933100056679,-0.340529049132041,-0.182685940383863,0.285348083836437,-0.153585445308146,0.0826514277008066,-0.189634817389458,-0.0814299819832889,-0.0527988848788967,0.191628238492149,0.0262819476553186,-0.150169046854646,-0.0771431151055054,-0.0842722602462156,-0.245146637665987,-0.509003123749575,0.117587554831058,-0.0509914845297254,-1.79871265670052,0.164188641632942,0.193278694419404,0.414997383806495,1.12719473710492,0.480506088675497,1.1225110937675,0.291498286336205,-0.391021859967821,0.0166909716663906,0.116197020817869,0.280836343497881,-0.288278863149752,-0.136745488418182,-0.342718798404304,-0.0308068084163991,-0.253630631620254,0.208232915070563,-0.0654063166537113,-0.133940007111099,0.209909574455435,0.247837047702217,0.209342006504224,0.00296412623276311,-0.416553640291767,0.305685844398262,0.339426179787646,0.113530817185413,-0.0429993320043183,0.036907199736959,-0.282904986888159,-0.0189759040183646,-0.11710730967782,0.145096739361046,0.0155403049473278,-0.149222959773597,-0.613302601257014,0.127908343390634,0.00741293708805055,-0.0197503549579798,0.380351791003779,-0.0107065038232414,-0.130237128727886,0.368904556840468,-0.22687749860952,-0.139974642567149,0.279894435784407,0.0228167151321112,-0.26856276420817,0.329883553185603,-0.0666098953409062,0.396908339703411,0.123844039685686,1.02554181428315,0.424169305704106,-0.127272779410527,1.96570850939452,-0.248418002914233,0.18934997570122,0.337114706696656,0.0319502293707838,0.035731070880767,0.106586141383015,-0.467095632076601,1.79478497520745,0.065862371691222,-0.112695926389604,-0.477915651905776,-0.00742290254101624,-0.679718614357352,-0.545599302804388,-0.0406672628071586,-0.404817729291809,-0.969324216381892,-0.877292154788344,-4.16242528149431,-0.19884908042486,-0.32013544797395,-0.0587379552531114,-0.0497599006422875,-0.00921210126360895,0.37670824848936,0.0811745100498408,0.400102414050555,-0.250295500201963,-0.511191979762611,-0.0235778382998805,0.0698665782857689,0.186290192631021,-0.470088626186213,-0.232088648533343,-0.0472486639028098,0.28997531285487,0.07286746514727,-0.253560745264927,0.246573384926808,0.0563756517082012,-0.0768059136556844,-0.13682887147994,0.38681582219448,0.360991075626239,-0.351669199632179,-0.0712617234195864,0.518600403827879,-0.0149812318007385,0.506294162648657,-0.0354463656455263,0.513815360109994,-0.632607557539894,-0.0358559498543443,-0.220258430107706,-0.232731911825805,-0.281438410874864,-0.128936869575326,-0.144693141178324,0.188981521696505,-0.164781617212905,0.176693736028967,0.0283142217760938,0.322915974629215,-0.0722004832017879,-0.632889558841222,-0.124393841209806,-0.0336988925525448,-0.119283061215657,-0.0742564704213304,0.237498093142829,0.461058240826864,0.25675507853188,-0.20476033463038,-0.29026294657831,0.0798791841334965,-0.0798145611606406,-0.140000040691592,-0.0711164606662052,-0.36624297239307,0.0580455233172757,-0.172310102571127,-0.120264852265105,-0.132756323323775,0.208034499537243,-0.000608766126851787,-0.0563862807005913,-0.0462792634834701,-0.051277822597914,0.122541372984037,-0.605195550620843,0.106030168141889,-0.0269968481100996,-0.00992734531025399,-0.189902244199948,0.00160048979822752,-0.195723873860996,-0.046460107971225,0.311378348874316,-0.303169550849459,0.0569891777923029,0.145945381336736,-0.172324818085193,0.17811915876456,0.0445773906585601,0.0691682916793627,-1.57529155384266,-1.78113996133991,-0.492696754892457,0.461290798546435,-0.0654934370759255,-0.121021806021245,-0.0261325923029163,0.182700019726316,-0.129299391862933,0.0445843229463678,0.00520422032117061,-0.0723485730807287,-0.121520690585635,-0.207776490019446,0.129434956554342,-0.0743288549330977,0.263297834015554,-0.100762816354598,0.0516884031901282,0.378255402316036,0.00225465446587268,-0.07813590984222,-0.782108283878096,0.113741865059179,-0.254469064084311,-0.00880139743016303,-0.108921394437478,0.91683507299935,-0.000780456895507152,0.655170265835344,-0.202551454684251,-0.173698125612703,0.160805832433811,0.0382378053183815,0.0826812788326325,-0.559597499767548,0.195505149902648,-0.216913374525412,-0.0441231410724096,-0.200426615701725,-0.219584451947047,0.394666257932228,0.101240212760922,-0.186307054386026,-0.0831087876264782,-0.248354385192542,0.226519403095107,-0.160422807798419,0.142464567652757,-0.00928592609583463,-0.0557222850665111,-0.138208680099851,0.275541121825289,0.0065054823335743,0.145164334263164,-0.40302206593804,-0.111786615066512,0.0223626695594954,-0.030756483417579,0.304185835005389,-0.660511663828705,0.492181300694982,0.0507053645911203,-0.620019645982467,-0.174164837494744,0.722230886707995,-0.0533766486290439,0.0162193370901073,0.407737608782075,0.243622362304426,-0.0211649385848983,-0.454666730050621,-1.03432241660326,0.229723434520234,-1.2249986323238,-0.409205353552711,0.0990763001899413,0.0645250398803786,0.252897863243377,0.0366238011856054,-0.795958104536257,0.0618566749356868,0.431703614770055,0.175183066605625,-0.134879044131023,-0.575467235306264,0.628126636277627,-0.231029012389105,-1.30661256321977,-0.776826757580237,0.0705303947505637,0.331665644008354,0.246978806139527,-0.524220440283771,0.125222870026447,0.370785626482932,0.174735967472046,0.477936866327621,0.312928897334615,-0.107884030190559,0.114350726850328,0.351055703761093,-0.212141781186624,-0.25979356398589,0.413866844943019,-0.518763710589804,-0.292578691386301,0.252175095329362,0.0974776531943972,-0.191774226128894,0.558679589045392,-0.821756827414917,-0.117466255319284,0.274778877213226,0.404560002180763,-0.0763614460740115,0.30607259806144,-0.0789922350049555,0.257831199722403,-0.174841927948806,0.788948875347448,-0.428394628718017,0.148492736089142,-0.964470605387464,-1.98984600919327,-0.137386803135248,0.109049457167523,0.285612265563078,0.0800638975212637,0.232910034869557,0.956685556806857,0.00871319544517685,0.0187121318791652,-0.0548349200921992,0.00366404785133823,-0.00172579794250143,0.299794725790617,-0.016330417557391,0.330704538891868,0.15458589919734,0.0193233301507657,0.215120171654702,0.674266331634048,-0.157699631266501,-0.270847159443498,0.266981108550027,0.0602224437870135,-0.148900149529542,0.338481121961844,0.499832548495197,0.106076788056312,-0.000982809129216512,-0.234124972699046,0.188148463617014,-0.457469699457396,0.214670951720169,-0.684710675124586,0.119622661778281,-0.190892887180414,-0.278835281007922,-0.421484614531511,-0.25113974174563,0.0083843563188717,-0.175461837365481,0.590593300645975,-0.0834644604233809,-0.18298415034606,0.0885552577339987,-0.259633999123348,-0.512023943319552,0.304511959384054,0.0372268924209989,0.00742886548680379,0.335457023265992,0.562088042392329,1.00880546170167,-0.448326056304753,-0.240673786337657,0.423201476686079,0.603407609611906,0.134432589023895,0.150377006388424,0.111057707197398,-0.249962156566659,0.642789714503434,-0.276197795210015,-0.093430315623058,-0.157639675610994,0.0617687785891813,0.1513086388113,-0.136043918935498,-0.188500616271911,-0.91233142861336,1.68257899457512,0.495342723084032,-1.21925786804699,-0.161471304376903,0.0918135887663021,0.247507035079592,-0.296099950036504,-0.178492419613034,-0.0807192060723671,-0.398960112506193,0.0373176520163469,-0.0321941118183007,-0.191404326382822,0.0612510612829844,0.399583920053139,0.258505296235791,0.0250190921177474,-0.120426588091568,-0.255387083009475,0.124334036052616,0.0429341239635025,0.30752994490806,0.165964898404052,-0.35809505296624,0.241995635801418,0.0232499651551685,-0.543518603879205,-0.135514599366315,0.232375949601238,0.120692552152993,0.0201640896098019,-0.0731987231070726,-0.0797476827020021,0.249418675839743,-0.334949990967923,-1.20974049328109,-0.278336364875986,-0.147389562708008,0.0644406818574033,0.0256053375645576,0.789970886823435,0.178353769898701,-0.0499080375614178,0.0647727162209033,-0.0135056593328892,0.249842771259544,-0.640316030998785,0.68868984806942,-0.298341363577073,-0.108468819451205,-0.116729989933283,0.253723567142246,0.580299500169881,0.541793109878602,-0.017511682654257,0.239854981389141,-0.896093518308703,-0.139915404634549,-0.158532403207555,-0.206151054149058,-0.0125877216203138,0.2367765179964,0.193579423323181,-0.0207338347979364,0.0458893438758518,0.132181046237814,-0.128706548609123,-0.0373416784120551,-0.150201973869924,-0.104539721459565,0.0374222936932269,0.0749189955759882,0.281017721850089,0.516472434904018,0.108464444131718,0.0490379039608014,0.436676213588294,0.341347173674642,0.0357566490754751,-0.19049773461296,-0.672261250067883,-0.0190045787167813,0.297167448313067,0.592693146622059,0.0253759388227878,0.305609485829054,-0.0361936411588062,0.421231737533597,-0.107630726895725,2.14763411905486,-0.00198063957593219,0.230246620264445,-0.0331309188087837,-0.554443137987155,0.174808221367685,-0.530353646894922,0.0167123684595866,-0.0653394894359317,-0.117553263210336,-0.498451432022069,0.0745266589684245,-0.0505552200152868,-0.908051150785856,-0.0895716552930532,0.152983991212043,0.0848061613111329,-0.384264240615836,0.214216086344581,0.236633804769179,0.0109400982962841,-0.192035015892722,-0.450500984346731,0.0995446711206092,-0.349001895024897,0.00417881828166456,-0.0679644797420171,0.0528126708895553,0.656438242505811,0.525020262390422,0.853807267489023,-0.597385768846702,-0.306471802664518,0.193316318364645,-0.0126026847797116,-0.0386941107439796,-0.351163654189472,-0.0277465658563352,-0.869401290976109,0.10071025108934,-0.592902261495177,-0.433000400526687,-0.587396002784965,-1.51619827314825,0.112372293116925,-0.147469344225134,-0.216168990058477,0.192045978336528,0.104527534718628,-0.996283945310639,-0.152639941097146,-0.0314982548702688,0.232423859283612,-0.0397022613501386,-0.0932383770937141,-0.951778605788067,-1.09735867244062,-0.0740357216511286,0.0137627745705006,0.0313090565187275,-0.217997886700956,-0.195931996165183,0.051022435691532,0.347042244362208,0.472315097403946,0.0294078888345169,0.519966125727699,0.0493936640095586,-0.783889488352818,-0.741391027737645,0.963818337307799,-0.0268869151024796,0.693876624375512,-0.0967910324640377,-0.0926195097126524,-0.375329644863323,-0.0920006966816503,0.14501983271902,-0.110331626724939,0.000515353967528957,-0.365909460853117,-0.47714168681332,-0.298017183929584,0.159057165414892,0.0285986655522181,1.2165680924373,0.0656821255803183,-0.0708876374824677,-0.396464101684858,0.154051030358456,0.383535073943322,0.222417248094793,-0.119741666921521,-0.00506145773219252,0.0921502188927251,0.4037801272046,0.53652479626893,-0.0135092346180441,0.303016803169706,0.3261244742283,-0.192745470859755,0.0606308664102396,-0.143407022288647,0.808848191921116,-0.164665618491029,0.0732224012086112,-0.15932412927675,0.0550120021316503,-0.571000071665556,0.106812555737368,0.116748063865063,2.61816022850635,0.0259291726911157,0.128933712682767,0.0477952038042966,-0.181345268346604,-0.435179778475983,-0.554490823515073,-0.214930711868135,-0.584271246300962,0.222236688936663,-0.087161389620011,0.234796411590985,0.207418728472947,-0.0430201386024772,0.31673002257682,0.253444011925528,0.203776852677423,-0.544145507688669,-0.624376857051608,0.236918792612118,0.248691644416983,0.352021274855938,-0.945041896179835,0.132068611158384,-0.0572933261066383,-0.0447091903946428,0.854312318313111,-0.68602744428045,0.21251616650789,-0.0102662394240478,0.213668240645381,-0.179231202512414,-0.039624133819509,0.115346793792263,-0.958822825321021,-0.315015313641147,2.32790732819798,-0.680606271739918,-0.218421188642069,0.0218026366078952,0.0260854165862104,-0.0164307142662282,0.698376545141597,-0.444005703247051,0.509917273683998,0.0450540489486805,0.0917933531589287,1.25308633934443,-0.224149958316306,0.315667222108962,0.0719004807407383,0.0107771242620662,-0.359192129688196,-0.108574061178219,-0.315889775360797,-0.0867203705313371,0.0815221523132751,-0.0108748245530693,0.306319098661698,1.70449719475573,0.0989715512546338,0.184924166765072,0.310668834522738,0.801209673150601,-0.0308381195792693,-0.120188553751849,0.324209089938026,-0.349590968557988,0.191317755996346,0.0781724549714392,-0.0195432500057897,0.0830981671318487,0.31794468139097,0.388685194464276,0.11280022353868,-0.227355119593702,0.135471463876023,-0.62927380346612,0.0602047596278838,0.408715576915284,-0.0520442807494158,0.219380406566875,0.270345089945748,-0.187571613001976,0.354929285190143,-0.0821018809259226,0.339257632546156,-0.89911622172994,-0.90486477602515,-0.0429077392024745,-0.110306797187869,-0.141966821272671,0.518935573865188,-0.000666307024727125,0.556324203225256,0.633943040790018,0.918439766466052,-0.121577828017312,-0.252689923779959,0.153976350379611,-0.369438679289213,0.645023356150488,-0.0717907797834435,0.324182745200766,0.140145054208238,0.0781579082909592,-0.760537434885099,-0.732525218835667,0.357455418351697,-0.661435629191023,0.107186385393373,0.00168611512665471,-0.0877015547219679,0.344155207449188,0.163057549718656,0.508724945523598,1.2399776671346,-0.37276245567791,0.467471357536971,1.10336280468746,0.542066899699474,-0.162104813575821,-0.230804092909993,0.161623305987976,-0.821775381746465,0.0651360298628493,0.0978612254521844,-0.00532212920022323,0.0692977352330715,-0.062995200331952,-0.51093933655979,0.52709499249709,-0.235513538417871,-0.262343376936986,0.361162535693716,0.056226132286109,0.000642084204685803,1.8106824778966,0.120838358546431,-0.185634490910316,0.439924027326849,0.128911903777069,0.947628401018571,0.226955720868823,0.625107423067426,0.148133675089588,-0.549865734140908,-0.0869130481756086,-0.0232911976653623,-0.0988236021019761,-0.416229895989762,0.145837342809077,-0.277670761377481,2.0152960766571,0.14424696479908,0.370290546586928,1.68688318050229,0.127176636596094,0.138830503161902,0.202537792301879,-0.762818866812482,-0.0426189340245722,-0.213483052166117,-0.274108549141254,-0.0169130941746479,0.114202074900666,-0.116099281355147,-0.0678246817772522,0.0952622329792602,0.120003476941071,-0.0698255245427633,-0.1087470874316,-0.123588436329351,-0.01568393106575,-0.065357882555771,-0.0927094584466848,-0.102187350518145,-0.0959360750384125,-0.249082826038101,-0.24969436872157,0.47075610051692,-0.238167031880518,-1.03280098444528,-0.134329528667276,0.625746063325712,0.0783365852592243,-0.0180294897291021,-0.976969187505996,-0.0368974669999099,0.336725628841374,0.0123146478170286,0.0617988657463058,0.206349226609562,0.693074344758627,0.0916538727354734,0.134645704757952,1.0030303766868,-0.377012382054042,0.675123879769174,-0.45137646092,0.402272552957665,0.275436575582372,0.415004563624987,-0.0603850211787772,-0.111003474188445,0.01772594760841,-0.322274744623828,-0.175194695110289,0.166746646337319,0.393248576776397,-0.165009322741293,0.0954465573554497,-0.556299160801545,-0.166508527155389,-0.0666790805162546,-0.025630259091627,0.27944062822664,-0.270733187646271,-0.0221303511912659,-0.0935293025866151,-0.137304762143211,-0.233927595555747,-0.327759530657276,-0.569068127016839,-0.231919365172739,0.0769580594124853,0.550960496615307,0.284922195017232,1.22062936083794,0.158320456475547,0.148591636611968,0.0453753236192085,-0.0808819979054562,-0.0969597676427468,0.0912567221818694,-0.435772185473523,0.197448578819242,0.415408734468105,-0.913530918034363,-0.234217761493883,-0.0668318176943346,-0.150099944895787,-0.102429435315415,-0.367113333681001,0.453655465943486,-0.0459994469941231,-0.00880246210391813,0.0126878274946817,0.151110195399374,0.0523727505333931,-0.0451894465981641,-0.177479917327476,-0.0172906095784696,-0.0757604746925168,0.0341779425521929,0.237027341366858,-0.259471714307502,-0.208165438446476,0.216725715087212,-0.399024796389269,-0.374626727648698,1.30897303610474,0.00672085076571472,0.19547258986027,0.251106093881494,0.154388597404358,0.00337369934728408,-0.5586423236942,-0.0139736065176969,-0.208866482149419,-0.285789084907006,-0.108678653693551,-0.0836426197366585,1.29635051486651,-0.0183018597082731,-0.438376841686273,0.0594009331531672,0.414147019929323,-0.274602519228342,-0.0588200201614028,0.309130164862591,-0.221471751685739,0.0499016329541015,-0.00500562657474956,0.322034758694144,-0.0615511596949858,0.0597580504992764,0.0125663010774296,-0.422526427241995,0.0897600529104989,-0.17715682342533,0.257275633024927,-0.30763786659362,0.340334206379598,-0.193835710776101,0.471032329086831,-0.0859549057188531,0.113416817905161,-0.552815630268482,-0.575332108003325,-1.77715403360062,-0.343783949785014,0.111633247780912,-0.159281084889901,-0.07790554184525,-0.194877441738371,0.439607506315889,0.0800885554473286,0.635996245827682,0.12097508978131,-0.226387872741709,0.349432517012454,0.0362967574533609,0.465685002813361,0.082438458764005,0.0623601119825257,-0.192464653343102,0.601179707821358,-2.20287080419761,-0.11971147957724,-0.663999975882873,-0.221949513064578,-0.273818926349423,-0.0293677963040161,-0.290880061931866,0.459536963518478,-0.0242522541090889,0.1306513462752,0.0433185433392625,0.0865814709459378,0.33670693296976,0.33388893996885,-0.096678464966945,0.0273589562632795,0.610666969090325,-0.297845322958958,-0.0161286922472397,0.657955336087718,-0.0726897865144319,-0.0790831577628803,-0.0615256192533048,0.0716837721278985,-0.114455933504698,-0.429449263072082,0.21017120966862,-0.604634240823347,0.131280161057151,-0.111559135268755,-0.928493287877096,0.165384235856288,-0.36240837256392,0.16168543137702,0.292230734602314,0.234791702706976,-0.0699654954285357,-0.0755591695333708,0.185001222725027,0.422979386933326,0.00270569502944897,0.155754717776511,0.101946370022826,-0.0386666832791938,0.228946804675681,-0.438675108691177,0.0663361435646257,-0.267994056678294,-0.122153196131522,-0.0699165649875551,-1.10276750703219,0.262891816952363,1.60350988908146,-0.165432430016524,-0.0536319781303828,0.12161944323771,-0.256268735295678,-0.00601077927604308,0.356817101955617,0.324102271888865,0.0419697173218899,0.0941513650354263,-0.125832491382776,0.593809640692198,-0.0168791981553413,-0.182704465250506,0.0627321407854184,0.324958534638275,-0.260165315060572,-0.207218029956411,0.00632930582063053,0.208857889652721,1.26995615903244,-0.998832985341003,-0.151226767277285,-0.348467979171617,-0.560860711841152,0.118700169982607,1.31561147297955,0.120176056193753,-0.179949068317458,0.173690644986495,-0.245075519464381,0.135849772168883,-0.752766650290504,-0.456983461692598,0.116126050534454,-0.0523871885883559,0.0996576583316957,0.165272577884116,-0.0663807712141407,-0.0921526362615131,-0.248474945518009,0.375293209654349,0.0581506414676375,0.388946549226568,0.473368642576223,-0.0124823510556194,-0.737156265777722,-0.573615845135305,0.279080012807232,0.0624692552459231,-0.619564358027302,1.57530516639429,0.159103592008457,0.224751359337224,-0.498486308914613,-1.95991298138778,-0.0158766904700004,0.282106404550197,-0.196461634377447,0.829264444995362,0.246330788704947,-0.781365635319527,-0.142613376736957,0.084088980287276,0.0327478152587237,-0.456080731563868,0.143893458971848,-0.200488781982404,-0.0969646452637923,-0.0225228695923673,-0.220275277700047,0.137487619264304,0.233527909434932,-0.239606339337593,0.542166359805578,-0.0716516682746574,0.414101179848596,-0.0258497005945144,0.823587588586051,-0.335303369320525,-0.0981409482615584,0.043936421949371,0.0785423340436512,-0.104904523829663,-0.00638250756489248,0.695803413572401,0.0390826492536423,1.18728854140807,0.460820219049867,-0.0204384626158699,0.313665111202578,0.0171223319937391,0.242521832815504,0.0997059807323699,0.196398870928117,0.475227488377115,0.253863117079153,-0.181726063954496,-0.238297869640481,0.135235448850595,-0.191028135493301,-0.0861522470754626,-0.520713736092221,0.0195879263961847,0.517226435808315,0.365614403629725,0.258092708518215,-0.0830063269956055,-0.150882930207998,-0.153436198717974,-0.0435008199684576,-0.0315038819782935,-0.0362508110665262,0.330838685616736,-0.358271012570267,-0.107632801225208,-0.248891598946773,-0.0176469874503367,-0.016379424564745,-0.134373468503002,0.66885297913377,0.0777202641481768,0.0450352699415632,-0.179532669890486,0.095528768664554,-1.45247104143666,0.327096516479975,-0.188065361420133,-0.157847214847291,-0.499789712759359,-0.397788550249886,1.69636602580579,-0.41663487955024,0.278238106540336,-0.108606938920965,-0.479227112498644,1.60045976545638,-0.229609503981321,0.164960259479235,-0.0170251387604988,-5.93456097202361,-0.457050192836856,-0.160297105011139,0.144274925114128,0.909887637123972,0.563249602000235,0.0279840591746675,0.486671299205812,-0.00190152644249944,-0.779550119871036,0.270312669241203,-0.0449333172481909,-0.147512659391048,-0.0261896948761551,-0.100413045588671,0.123730978210281,-0.086039538741141,-0.497681976134539,-0.034025805976143,0.706575307016807,1.03415078449619,0.522262227415111,-0.0633255398072598,0.64991717163709,1.0250142929026,0.953133948037124,1.50849442383854,0.461900421684339,0.653039278762273,0.140551069843935,-0.329542365920698,-0.240775787411448,0.0235898522146763,0.335275933072231,-1.31687194095139,0.355470619814695,0.0228250808774733,0.617501763161462,-0.130744852971492,-0.198591781508848,-0.69858151934831,-0.458623630948625,0.325819558525826,0.261172770901066,-0.393166416847499,0.0967576429672001,-0.01145863431967,0.878642668828819,-0.260884604686855,-0.241268422335826,-0.0265095657372964,-0.154235442543754,-0.208582033743425,0.56625275354683,-0.176237198859677,0.0958414486380109,0.235148439984364,-0.893080265067575,-1.01750079337727,0.618954001987365,0.221187813698227,-0.119156965305572,-0.358510350571346,0.143605904903977,-0.196700309691061,0.0421713286372784,0.624753803017376,0.0592710742921204,-0.278754375453929,-0.208519857212673,0.139561975561861,0.147709851137076,0.228851302172399,0.167074800971459,-0.0284909483661652,-0.407731113477115,-0.00491680021157027,-0.208070343691305,0.0676439864647888,-0.177098708471349,1.26485909758699,0.475262550283668,-1.68289834387711,-0.235657845130387,-0.177135332323315,0.408720892238801,0.796246870211989,0.864242670796593,0.320980014987317,0.275809247168484,0.0569281979626032,-1.37073930290241,-1.3093252867826,2.79936793599592,-0.0183318893879795,0.00267765614630442,-0.872920000892001,0.0774921591432275,-0.0893884717548477,-0.366534632102915,0.139281547315989,-0.270737146354353,-0.806192609697023,-0.0031428052318099,-0.182752766490763,0.445981792947603,0.666586533892686,-0.0761393563405484,0.817119513059907,-0.206068001357601,0.150377251120804,0.380092155251616,0.203737066754983,-0.171528393900278,-0.238491603574067,0.482845429471246,0.113131760284296,-0.135075160910247,0.28216915956942,-0.349243111527139,0.580602333027116,-0.198187443138103,0.111272818889154,0.0862727195920118,0.0741424938591655,0.0248742867180078,0.0432887736129504,-0.60404598085599,-0.622608446439467,-0.609175161140653,0.165197855240748,0.203054649649483,0.206142147725512,0.19472316037437,0.0751285693104722,0.156656772244491,0.174357975355189,-0.22417836195127,0.776224774199278,0.0968880855819919,-0.455853836453894,-0.180091720872363,0.155753664171088,-0.0301819112862071,0.0364087822345135,-0.216955186868956,0.248059653142923,0.188016040437318,-0.808221244262232,-0.0071344418023707,0.112593652519513,0.679585523303916,0.558962529957459,0.247130974137969,0.188844048535917,0.817603249841477,-0.0523117268845849,0.0663103183486587,-0.237633616147682,-0.23239710974256,-0.240221233941248,-0.0950158976305421,0.129785078372428,-0.243898177965886,0.267004911206497,-0.410705700878824,-0.274828173522872,0.229102201688417,0.215780080215742,0.233385362618261,-0.380874042875623,0.196947529648882,0.17043873432431,-0.610776670683005,0.0943870800700477,0.397560658049332,0.0347050133133323,1.21173025789049,-0.0783791574465348,0.160256311158965,-0.660980545644192,-0.439484556280424,-0.241761512338083,-0.0914656036145317,-1.29809507295334,-0.411500917548734,0.218968298960279,0.0911292118952088,-0.360742448359684,-0.27255746373122,0.518925522007437,0.00615906619631553,0.77687281442216,0.0762180814421668,-4.91772330402325,-0.150590709497013,-0.000313212885196101,0.0815558567744112,-0.239616906648866,-0.707571880551609,-0.200958316589091,-0.0143157484813208,-1.04301544538651,0.0785734472881476,0.262949257794392,0.463318090638652,-0.516702726494338,-0.461411397393283,0.101576982536097,-0.58296350238411,0.926528058294919,0.0510276146212256,0.94461840340722,0.074104709687071,0.325407891253066,-0.228243408094088,-0.0787000237575087,0.114583443240809,0.3734478437495,0.341291511800153,0.181813271095212,-0.23801465717439,0.707481848664823,0.0243807000789911,1.15898268636166,-0.0818747144008451,-0.0352140717312188,-0.0209892528258343,-0.440002857865247,0.179376912553988,-0.166017797259465,-0.0505014328581864,-0.868725399949454,1.01355791763758,-0.225831989676099,0.0400803356683778,-0.0899864788542417,0.253025422407702,0.58107229541505,-0.423253084467503,-0.10954260538706,0.199542355246267,0.411694654409869,0.498408716040319,0.485981395293515,-0.0629015812797597,-0.0824021851177177,0.378817126734057,0.1259868529631,-0.835442266699665,0.00368923555230329,-0.682534247083223,-0.728263241987074,0.696393659575178,0.208024826332786,-0.0291431505827377,-0.629444603136935,-0.464599811793582,-0.0853695275422953,0.0821398000571434,-0.197248207845329,0.127492401434168,0.30074023175158,-0.0532953222885543,0.363353898798761,-0.0255132086778862,-0.0790895150722994,-0.201096786564235,0.021682289160542,0.226929774962312,0.731660216058518,0.0926936068329703,0.367677539733631,-0.433867785315881,0.1540340707141,-0.00762209736027913,-0.137506463367107,-0.235105870528915,-0.535207863135939,1.361933785715,-0.185683775720028,-0.00129204325610184,-0.0187860021856796,-0.272216729773315,-0.0375747031588297,-0.0838648647043041,0.00393628260580499,-0.149602465538348,0.00825727264146555,0.19202358856486,0.190229348811548,0.161387832943802,0.279851752340157,-0.503979989661588,-0.203460116002453,-0.0277504786886774,0.0117587514688059,0.330371357650312,0.0659239632286663,-0.0436198955079442,-0.265103203803886,-0.136620258338473,0.00142353434072699,-0.243453023446958,0.0625670346361186,-0.651344711661052,-1.04655302352758,0.390477553736908,-0.317757565150003,0.133065980675113,-0.180640352980459,-0.529753852230963,-0.102149182794064,-0.0860571750167065,0.00692850131591811,-0.313306116811501,-0.195522706032904,-0.0187679877165175,-0.207238326410693,0.164705038762601,-0.00888830936857198,-0.253457566111853,0.0865594351944634,0.0583822700334635,0.22644068306445,0.780872276734904,-0.20005041720598,-0.0897487063081769,-0.529776155326206,-0.152610379712852,-0.0362215625931216,-0.0382752376793764,-0.417221275797755,-0.397033954674149,0.059150721618011,-0.638583268965843,0.00728867942261952,-0.39877606795381,-0.539292709985234,-0.0971748726451219,0.0563677264207551,0.0154816418643773,-0.00671560511359103,0.0669486354677875,-0.207271263243282,0.242860540505326,0.141174550767167,0.194246218562889,-0.0918820565578454,-0.0217550351565355,0.767083988457628,0.0301008625234558,0.067777469206621,1.80744668699422,0.201458705480898,-0.223036023827823,-0.162852993311743,1.12345473410214,-0.137412407317363,-0.132588683795486,0.0684863858179394,-0.18962812178238,0.0489028848550372,0.189637892678368,-0.0577786658507291,-0.442180309763595,0.163452068550998,0.0235387183927119,-0.131524283476273,-0.143164936168505,-0.287705376589718,0.252130953987193,-0.306102776680683,-0.0387255231720471,-0.0893180533880433,0.0498425649678043,0.895850411880863,0.431793264478852,-0.123094253176813,0.303232159409522,0.205757689004109,0.00195144723056819,-0.205398471060673,0.11811722559954,-0.508652597049487,-0.4793880663812,-0.0110070345964776,0.0805094638176281,-0.0661441632195253,0.254022616816244,1.23259929584633,-0.298510926086419,0.428696381921111,0.0131662049393951,0.158261592606339,-0.143179759842772,0.0744482741668632,-0.209665543659146,0.844204393564887,-0.00175975226793896,-0.336698280443494,-0.682045459570431,-0.0428191978527414,0.0694340436205133,0.504320107884441,1.88246859578451,0.578900984241061,-0.161101299453921,0.204202112020174,-0.165686679652618,-0.257354693265524,0.506552045886239,-0.513872158341731,0.639160343029902,0.242689768598582,-0.250087894853938,-0.0308310488653852,1.40318914558096,0.106006838771927,-0.354600061448698,-0.74802490101602,-0.696039531737154,1.73190819688184,-0.0174257115007142,0.0662901875094122,-0.212746362938753,0.371639437627157,-0.528954397616025,-0.0624214429747893,-0.00144624609694034,0.259221200333078,0.288595652041498,-0.184821938680763,0.161049526678981,-0.115789561644521,-0.255318044017737,-1.59838077172425,-0.719876188797865,-0.959294262206712,0.155724649474757,0.307064662919543,-0.108221080787079,-0.201087537152157,0.043975568409799,0.198704660599866,0.380280588217245,0.0636252080791861,-0.0436658657170507,1.13432545537808,-0.054063177853672,-0.859159625287347,0.0231388398933551,-0.246343490557392,-0.0841887727289471,-0.0933866156277781,-2.9064152500583,0.147555132221898,0.128637779973438,-0.577263159247882,-0.172132645178434,0.93532177462904,-0.0678124925018811,-0.17849421697657,-0.164342260060558,0.0947362270729306,-0.271477731051069,-0.0727260070752581,-0.453931565268443,-0.425662344106041,0.701979395124242,-0.019289872954968,-0.764983770623072,-0.186623326736628,-0.254440582918831,-0.136547645537765,-0.268080818822022,0.194868009788858,-0.560941692700387,-1.02924559722044,-1.78444073415104,-0.869471801756824,0.203396222240582,0.686216775513023,-0.111169353031684,-0.374809957466401,0.56918677345654,0.929513804299221,1.30611399049728,0.0109236042525614,0.0409846308627475,-0.141467919150254,0.00833587145277125,-0.632469389438587,0.0267048635727435,-0.128744518252184,0.185132931110752,0.126596536227645,-0.176474080474325,-0.088932708509536,-0.098476150772811,-0.249314952504781,-0.257525963345439,-0.00474678319263001,-0.284759847352101,0.0702703096580255,-0.106422673373256,0.0604426850253751,0.457837378040757,0.403867136282918,1.21843845085396,-0.158790730781359,0.0348930789861113,-0.29991970648954,0.349580912896363,-0.0510075884974031,-0.206447252170341,0.0957536306301416,0.266563415683691,-0.0674502674256688,0.00276793305590743,-1.03506875276743,-0.049757971542372,0.0895670769184988,-0.0863248555949672,0.0186104829321528,-0.508255571581812,0.224363259129467,0.132859116442517,2.38840035857711,-0.118889421236041,0.23506540451569,0.532586022730579,0.17043472715096,0.0657166744010344,-0.184783231185898,-0.11624375415704,-0.146162800972445,0.077197281617795,0.345461746888539,-0.144960063483337,-1.38136667032864,0.68524458688053,0.021970759204616,-0.328425710827306,-0.189171314610394,-0.0826691217119267,0.052286622084348,0.46622349256182,-0.213534985830983,-0.0973041488849796,0.246856814977237,-0.00323246347411957,0.452998788072306,0.0198214145834333,0.481924379032289,-0.170781415480218,0.32444526374498,-0.526818041755621,0.617179185170328,-0.628999301372856,-0.31158000914055,0.0243150074920119,0.0394290829646534,0.19042382137769,-0.101632663245122,0.0217629419909938,-0.0482191331624779,-0.216766853148751,-0.116236108783554,-0.355769230941103,0.0641634992536202,-0.815012049776829,-0.174335807559166,0.0214533335470241,-0.0942323851182585,-0.144287171508069,0.166667574433801,-0.000836115929188439,-0.15739042976031,0.202470542371668,-0.558644497468428,0.418972942012055,-0.142633219272744,-0.211234314081272,-0.239862848350972,0.240618007363904,0.859264231398373,0.0695624941646761,-0.304823954792091,-0.149971892772064,0.00746720855700826,-0.287923759663895,0.185579074484774,0.588723311290163,-0.129791500098608,0.288012034629304,0.220427776572314,0.14086342359327,-0.241246924647982,0.106874849612036,0.825941049205969,-0.224157967897589,0.076626565471001,0.248089381441031,0.0824113934055076,0.0388366065830018,0.116460735218078,-0.21544801389396,0.120768649917805,0.0108395605979407,0.0924939624197819,-0.154289043528324,-0.485319600191434,0.259774325220448,0.293957575316269,0.614219802834763,-0.250937627407058,-0.0775059267537184,-0.321553983409934,-0.0717644072461548,-0.179478371670989,0.10639763749599,0.197223899983736,-0.173690314552925,0.155046563476941,0.156165610443311,-0.180593834292376,-0.528053433912621,-0.208727616652906,-0.458962553592492,-0.103375650724495,1.28088011066073,-0.364403901922632,-0.937099897260118,-0.0123194450220159,-0.124107836050194,-0.0058994144769705,0.00761955996922548,-0.211724423391836,0.0332222544622099,-0.485405119836637,1.07273724651542,0.153437677010696,0.194151475777907,-1.14314421643357,-0.0942499633181315,0.0699172190459008,-0.225887882005143,-0.100181582311636,0.590596127440515,-0.198195852394841,0.045379945456569,-0.373723005285701,0.154029914383968,-0.10914673978496,-0.339719052330832,-0.00831369917876037,-0.334619239697486,-0.0188311264053203,0.59229794457818,-0.0111071943436581,0.223277201131998,0.15705900902891,-0.111115060667633,0.780084606120329,-0.120643999238684,-0.0960790350123708,0.0725814164255122,0.231147791781055,-0.254266726613448,0.581632906660543,-0.279165144390868,0.228777297918117,-0.362664967897306,0.247181208325906,-0.141573044621975,-0.0934728771956496,0.310426505167026,-0.192665633754051,-0.171163849414485,0.125204601115794,0.904466952843323,0.501111333901865,0.0349543608058271,-0.236945396305011,2.31082866953372,-0.434346516618999,-0.19030704581088,-0.0906383650516303,-0.224782817772446,0.975514956383802,1.51967362982559,-0.121246194597299,-0.267380850476648,-0.0686563357747785,-0.220857070902429,0.0969543587534909,-0.00220574366324722,1.14184650618312,0.190199953374448,0.186754827978039,0.0318198369334541,-0.0556731203612968,0.509906194038195,-1.11818301548124,-0.0761881165093637,0.0510703753924694,0.0880909830637749,0.690940265764469,-0.258360372372842,-0.502141132255208,0.164133662603519,0.104314844890634,-0.0140241615098137,-0.202101299096037,-0.196432059059031,-0.0792331214082978,0.0605190364889699,-0.168638723228976,-0.0285623565857712,-0.0655511860483942,-0.161001357136016,-0.551466679224778,-1.82844264078026,-0.566987952320647,0.185329825032148,0.125682564667382,-0.328127557479344,0.0876675157543455,-1.89611983922565,-0.299325018875565,-0.0972283692231682,0.653135551723841,0.244719138803728,-0.887176396531615,0.611077880064411,-0.107712269817059,0.693627684689252,-0.0877384732619338,-0.296697593467423,0.295532889722098,-0.958822964597638,0.0127194420456129,0.0153924162130335,0.163570514198072],"y":[0.00121304115851171,0.540604931494252,0.0825818392824401,0.0252575824864749,0.0684394952173052,0.128279232702469,0.777942310072269,4.72772490939141,0.142524262563551,0.0168094838384131,0.243704417215347,0.152409597855197,0.139551597426331,0.716089564961085,0.142631983206131,0.0158634862926648,0.360883821688659,1.38034688015477,45.1579851842688,0.0316361073828435,1.06843273894035,1.56056152171697,0.00419084931358526,0.114353957333331,0.332011330904214,0.142524262563551,0.00411683845347271,0.214799001636142,0.0614688361424385,0.11257503223522,0.0570257778453027,9.11123328603494,0.255243341098463,0.115597204501953,0.00947518536003681,2.23489639217426,0.051052548967674,0.123825470520158,4.45048097418933,1.51950378412725,0.555324324293309,0.132680099583533,0.0468791300281877,0.0971362738478571,0.0021523107667342,0.0147363309021572,0.232287047020016,0.140441827618829,0.393502134068128,0.0114484087671866,0.016936726755383,0.128542528765901,0.345502048510554,0.107354964735127,0.069814215314912,0.0234933792595118,0.00459848528213147,0.0804205837138711,1.47818889763321,0.0603748403667264,0.25356564723164,0.130751309049599,0.0549043056750132,2.09965740820589,0.0375060843610187,0.011249800747522,0.0208809999112162,0.227280550920699,0.109193370418435,0.0344255500471701,0.134113164488797,0.777191668087034,0.694186818860114,0.146764443869745,2.92147457523071,0.458049022222307,4.28269097748201,0.0278937932733824,0.0202307610424076,0.00460997542369454,0.0614688361424385,0.490413406181966,0.0166923597787139,0.175124664174564,0.141829045667774,0.00170783886867989,0.181872908667897,0.00407996707515448,1.45871734002972,0.197078049408265,0.0358044923638088,0.108421809761933,0.0726013805644724,0.0898788814722309,0.115713176180213,0.00947518536003681,3.26200375102729,0.315056045811215,0.520922167029769,0.0692641240983059,3.53697669189693,10.5731482586756,0.122456569618017,0.676361667979867,0.00385631494084208,0.861903347868282,4.30025102618811,0.0725951439134972,0.240087043690842,3.58345248955439,0.0649545376936062,0.00411683845347271,0.135638733190319,0.0738240472895363,0.0762960487173458,0.0158634862926648,0.513941207411948,0.0293766753549939,0.0711581260335338,3.22573031154754,0.0149236544925937,0.0818233581757009,0.0614688361424385,1.2847863275775,1.26901476924371,0.0451549731464553,1.54044540424671,0.0234933792595118,0.0688140767070753,0.0159185958779729,0.168161584727465,0.127492827213836,1.14494857581857,0.419462067012117,0.165744288344279,0.0107767401614758,0.201628194149819,3.05288422140308,0.154590008104649,10.353122514854,0.132715346044724,0.483020961590955,1.38499031999341,2.14776359951949,0.239509517939684,0.16581544056764,0.111536947553299,0.512169988268594,0.0197576796784808,0.0375060843610187,0.362896457419757,0.154699480913639,0.0204109004922536,1.29191950588707,1.05496587971326,0.276387003336763,0.154699480913639,0.239755279819947,0.0780563463060434,0.251049302928911,0.0370373374431246,0.147497384945501,0.0965000651465204,0.0672674435219996,0.00827862905112156,0.0124299458379158,1.45202085356399,0.0889080927701457,2.1807081088157,0.0368401375339735,0.739914160767582,0.00298562862325312,0.00329111046875399,0.0149236544925937,0.0435811396153694,0.399071280990619,0.127112452673322,0.00141021433832185,0.0880904578378599,0.0644195404711431,0.689134005066903,3.6269488642233,1.02168977484812,0.615703275462965,0.114353957333331,0.0979520601416119,0.237433192934264,3.24877425716094,2.45355289210793,0.0104526353142326,6.79282118159986,0.199841863296602,0.950889178596605,0.03690087050643,0.0454863584200561,11.9814819010727,0.120891445029521,0.018213394299291,0.0966819321127051,0.0672674435219996,0.159955258527847,0.0353007315049884,0.028873429718069,0.539410118298742,0.0209226495669109,0.0691723142136732,0.039951814594055,0.520881715694177,2.87450259171683,0.0637183101492261,0.0565739586011443,0.0036929116085014,3.56623657629027,0.320219386917268,0.00434189191494265,0.200444144814504,0.0324334366906665,0.116546719657008,21.7795948020172,15.9897325421615,17.3006593161248,0.290384458127712,0.0563675539190095,0.363215370771923,0.179903226159086,0.462983435856105,0.328432089316972,0.00782002579161583,0.0166923597787139,2.9462995959976,11.6400122607782,0.06717159959534,0.0953324795338304,0.0436931412956842,0.377944688328723,0.130751309049599,0.0104526353142326,1.99123182285677,0.218315004292681,0.166581454304626,0.322399741108452,0.27668572828989,0.111267690768199,0.0362734805403617,0.154590008104649,4.8821549028696,0.180958083293105,0.0234933792595118,0.349360784294961,0.086473061367089,0.154874628505491,0.160490047961719,0.641761703806222,0.527449010052268,0.376245366238701,0.0195276971424756,0.080040595786052,1.87434685734114,0.182613452305892,0.026346937631785,3.14395004815886,0.0103230553331435,0.398264454570971,0.608160843623456,0.328596331332857,0.143543163200891,0.0368401375339735,0.0375060843610187,0.0326795945322925,0.185567600030243,0.0316361073828435,0.342432287531116,0.0672674435219996,0.143753060707225,0.218315004292681,0.239688994827829,0.00481868046262364,0.0280354687584875,0.0715473711912393,0.455622881495625,0.0197048238177622,0.0721113079456428,0.118230199542716,0.0972884686457311,0.0370373374431246,0.0971362738478571,0.0672674435219996,0.0166923597787139,0.0435811396153694,0.086992843136258,0.0036929116085014,11.7425685861047,0.00883507889890769,0.0780274377942296,0.0217902084804504,1.5345094205911,0.0197048238177622,0.123639312193052,0.026346937631785,2.50131969521873,0.00454888400368057,0.041642515680023,0.216704140792974,0.06151247459663,0.0118003362744535,0.140139117126687,0.084929341663334,3.59678618443687,0.0209171002807837,0.110184216159282,0.366321704727898,0.48658612801172,0.305429414165508,0.0702572251429715,0.0672674435219996,0.136595277896716,0.106774017517753,0.565288584887757,0.0455106100474715,0.00947518536003681,0.114066567488366,0.139580531528287,0.112375649207978,0.0711581260335338,1.84235365134298,0.00230106010368019,0.450569072659819,4.1723800884589,0.567594322777414,0.0147363309021572,0.0428138877325034,0.17073973588619,0.269749005971493,11.052596711747,0.497063280896497,0.396067018720274,0.250923667922217,1.9697125324063,0.00349599865640438,0.0256352826026638,0.0450129006423602,0.562661050419632,0.127112452673322,0.0372477760158091,0.445635278711868,0.32026188269458,0.127112452673322,0.050102442800017,0.171946257404244,0.0197048238177622,0.419618305475357,1.64764046798962,0.125051568964389,0.0195276971424756,0.270872524317462,0.0563537107044377,0.0739045990177743,0.0168094838384131,0.00790968930391987,0.0218767093611828,0.0414001377668117,0.0818233581757009,0.225523022083862,0.00659116107410934,0.0672674435219996,2.55727407077523,0.0579469719086449,0.00686155703517166,0.197279060457972,1.64058512760293,0.0277073537847336,0.0590238363256675,0.0232712094423213,0.011094478772892,6.03056340047747,9.47499441509725,0.114766438533409,0.0570257778453027,0.0905351945635842,0.0751862524179104,0.339053376642566,0.0256352826026638,1.23950818188024,0.0248045111492735,0.927615016941145,0.0702572251429715,17.0722145717345,0.00593828021807046,0.236894849615149,0.0602428265616625,0.282258689478725,0.06811620739093,0.41993529271671,0.161882422645581,0.800643092853569,0.170530270372194,0.485263613777942,0.0682695801490116,0.547033021235577,0.139712324894668,0.313769971330188,0.262316767177885,0.0795496161702012,0.013766600510077,0.285741913890512,0.276518464457158,0.0484235414086104,0.569228794705177,0.365769080306612,0.179903226159086,0.407977547678173,0.412590154340158,0.0440351249654628,0.0536593860418449,0.0157949646038045,0.0370373374431246,0.0217902084804504,0.0896599668422461,0.194296877830236,0.192660491908561,0.00626713213883022,0.257913903198268,0.906464415110111,0.0561139370689358,5.25214319179113,0.0535159485215416,0.509845605508263,1.79797190564614,0.0672674435219996,0.323981847619161,0.0221622331632297,0.0382919160356111,0.270872524317462,2.2905303282683,0.126144350068723,0.00593828021807046,0.00606966185826972,1.64270546850986,0.220247495760268,0.408099377049679,0.621512719226079,0.838546822655075,0.00454888400368057,0.103798330234893,0.0234933792595118,0.00329111046875399,0.0338393027746236,0.00141021433832185,0.0918343913949572,6.78969789096756,0.0511957384793949,0.0591683754868001,0.00606966185826972,0.0372751825812681,0.0158634862926648,0.132715346044724,0.134533924565777,0.00315561947602539,0.0647295615861959,0.0528618138018548,0.0597508523350764,0.0352016040845777,29.3873734420999,9.59654062457913,2.19940974718876,0.0747212588098682,0.0166923597787139,0.0319768447348491,0.193794080389875,0.00141021433832185,0.0217902084804504,4.86283993633202,0.268564980493208,0.0834034906019596,2.31564430316161,0.144969953327665,1.03639419034985,0.00407996707515448,0.0715473711912393,0.0275149914177536,0.143543163200891,0.442124535537161,0.325881386097665,0.697216633115848,0.154590008104649,0.0492985567306844,0.231941279301644,0.424374462822048,0.0234201704074267,0.0838811819084171,0.0450129006423602,0.280869524116628,0.00407996707515448,0.0211611751863212,0.086992843136258,0.0213313778665293,0.534986947383463,0.0903550141797481,0.114766438533409,0.0195276971424756,0.691756235500866,0.018127883182096,19.9757259332548,0.086473061367089,1.35696224327875,0.193794080389875,0.957965799019688,0.11017481866874,1.31552247493151,0.404631359511178,2.01113059840299,1.01132996467137,1.67591097406106,1.01414116656921,2.0173805925885,0.619699920957624,0.00031765305898337,0.0514832796374831,0.104187503304971,0.0736322880563328,0.0468791300281877,0.543185937676671,0.213647439158468,0.0999374459332967,0.018213394299291,0.00947518536003681,0.0653364468487771,0.199841863296602,1.5101862522636,0.164279067288117,0.0760852880259302,1.17987754094913,0.00906073420687269,0.113938020039937,0.0978062116951104,0.028873429718069,0.171181934420027,0.892624655861539,0.0166923597787139,0.0234933792595118,0.222451888340823,0.0102545440838995,0.0464017913773774,5.11818075280334,0.00407996707515448,0.0288714684638814,1.13003004351555,0.0104526353142326,0.150491008464522,0.27782027496032,0.00593828021807046,0.026346937631785,0.00459848528213147,2.14776359951949,0.423108585142048,0.366755076786113,0.0197704517689435,1.01392071243063,0.0278937932733824,0.0894111904965788,0.0865962984195445,0.753079662094615,0.431237708638608,0.403847558861173,7.64974728147222,0.0134742627195686,0.0217902084804504,0.0281881093361584,0.0158634862926648,0.254077374455244,0.0611957404177207,0.0663326584292509,1.8579995720501,1.08262837411284,0.06811620739093,0.325134097131939,5.93484997882308,0.404884252332191,0.0876009793385781,0.473133873353317,0.115597204501953,0.0195276971424756,0.285040791283006,0.159955258527847,0.102791677567528,0.0205929766871262,0.102446216999783,0.47403262163327,0.167227212152352,0.209189098478238,0.276207910406349,1.28452153699458,0.165744288344279,0.0218767093611828,0.0362734805403617,6.01937325512428,0.0168094838384131,0.0166923597787139,0.0788591939294461,1.10320638020395,0.59406639971811,0.270872524317462,0.132715346044724,0.127492827213836,0.00407996707515448,0.0030054913774704,1.80213743435645,0.022275509239016,0.80140000993453,1.73323112742379,0.143352684829643,0.0467529612935936,0.590132424814733,0.119453263291254,0.00460997542369454,0.00593828021807046,0.0217902084804504,0.103863396275375,0.80140000993453,0.133272819079903,0.00298562862325312,0.450440003775203,0.791861018325946,0.159941977547238,0.0158634862926648,2.75474235377323,2.76601177079024,0.00407996707515448,0.1817063743229,0.0798668339486119,0.495537435738104,0.988930126355185,0.0978939252014066,1.04922910789765,0.0218015032544538,0.100626175904096,0.0281881093361584,0.122378034288297,0.323981847619161,0.0890324487110214,0.521404031228414,0.162487444684875,0.0215769217855509,0.0653269443253174,0.637351112409298,0.162487444684875,0.0656883926956565,0.416812246361136,0.125051568964389,1.60891035160113,0.00329111046875399,0.428780032292995,1.95672146768887,1.13905509447008,0.108491538733203,0.048975141042381,0.16197689883158,0.388231262967331,0.0221622331632297,0.067654278117622,0.0796592061137124,8.15525234678446,0.131555993431154,0.0686147573237967,0.00459848528213147,0.0509089323214273,0.0380218048667962,0.287210597864711,0.205794750492552,0.119453263291254,0.417297484771028,0.213342996857309,0.154874628505491,0.0402056440408849,0.0158634862926648,0.106254328567417,0.107869078610365,0.0104526353142326,0.0234933792595118,0.320219386917268,0.38265234050099,0.506493728927312,0.132715346044724,0.0467529612935936,0.658337133533676,0.0850238552782123,0.0233293789963836,0.086473061367089,0.0527229242072276,0.0451549731464553,0.92643607827133,0.0218767093611828,2.35526620489374,0.0275149914177536,0.0888144635109516,0.0248072680145259,0.0458301838355025,0.0154821369438163,0.546552494594594,0.0847795768993234,0.173624221284198,0.0473263681284882,0.0442108253457593,0.123864742290287,0.0905351945635842,0.239589405634418,1.11042916928748,6.38017082427968,0.0614688361424385,0.00298562862325312,0.301921895040769,0.315450838822319,4.91684575537689,0.0853543878004371,0.0933654710249695,0.00459848528213147,0.0865962984195445,0.0971362738478571,0.00947518536003681,1.56454193373808,0.0647065170226978,0.305680844304382,0.31696833193685,0.0672674435219996,0.0056492655075572,2.47981563928621,0.37923540049615,0.0442108253457593,0.0211611751863212,0.152047798370628,0.186842172927274,0.591302195996823,0.225445845543885,0.126105807845551,0.00543147291271282,1.02012024442237,0.0257987873052996,0.0946349501008994,4.15492639295878,0.332118778649055,0.0394506369694468,0.00659116107410934,0.0595649184046842,1.44894596074541,0.0023534462217772,0.139551597426331,0.00298562862325312,0.0168094838384131,0.137672640302506,1.79258375591057,6.21369035802396,5.09137317388042,0.239791708013277,1.61125822190671,0.312176782074709,1.10858801441335,0.0280354687584875,0.00476218922256842,1.20328742216874,0.211308039781596,0.125376737316837,0.0218767093611828,0.0739806170536602,0.099541494890991,0.198680521213169,0.274860270431155,0.748284793171897,0.0362734805403617,0.0710495876331265,0.474941362421679,1.99304693638553,0.00459848528213147,0.211308039781596,0.088374348777414,1.71131718437698,0.0168094838384131,2.89891363703658,0.3124717320817,0.25489053748679,0.0308553045328201,0.181190396534799,0.180958083293105,0.0684394952173052,0.0356391500831134,0.0248072680145259,0.0450129006423602,0.133279956295073,0.00459848528213147,0.0394506369694468,0.0290341322999814,0.714388659776437,0.03269631705356,0.0234933792595118,3.72089499794428,0.408404278262663,0.00329111046875399,0.0897302419562838,0.11257503223522,2.41831381274057,0.0702572251429715,0.00738336360458375,0.159955258527847,0.258856019391655,0.0536593860418449,0.0655408566197993,0.044900200029293,0.0461219326844996,0.0699056385767817,0.0818233581757009,0.0979520601416119,0.0356103344810315,0.00298562862325312,0.0805441812205181,0.0451549731464553,0.135838918788729,0.127082372867785,0.0206385049319028,0.538008664404916,5.21713518743386,0.634920015229307,1.05294135411851,0.236808015071122,0.347667433121975,0.663275368136992,0.0353007315049884,0.544511330098354,0.719921518027277,0.246654502095659,0.331980241196796,2.87022285213575,1.27734335767019,0.0511205834228772,0.74795930528417,0.0435811396153694,7.57882300144156,0.0747212588098682,0.135469474501536,0.0833471713831507,0.0715473711912393,0.0921973812992743,0.357186996858633,0.100626175904096,0.363051347649499,0.352437525279642,0.015338392640947,0.0036929116085014,0.0293641058665195,0.190701316890876,0.619699920957624,0.127566320797387,0.0164067793647608,0.00750441389946724,2.17474019570145,0.460683894167464,0.516576311902631,0.11257503223522,2.25914824396248,0.06811620739093,0.1359132967779,0.693527802688114,0.166581454304626,0.0580026618595194,0.523252359969084,2.58752712613323,0.208322218241635,0.745926335929779,0.270872524317462,2.75474235377323,0.108491538733203,0.0369925985146603,0.387817876582288,0.4749052974546,2.37776241788612,0.0589486888360361,0.0293641058665195,0.0195276971424756,0.11657208517791,0.296895070260511,0.1612491608445,0.0197048238177622,0.304259910596341,0.349434387278569,0.567594322777414,0.343057184152043,3.45383965045143,10.7827167635182,0.234815621699189,5.50871488041112,0.00401548166356793,0.121781990663643,0.0451549731464553,0.141656394990252,0.0702572251429715,0.355052324419712,0.559169273866907,0.0890324487110214,0.038616932751444,0.00593828021807046,0.445635278711868,1.33144247141756,0.00121304115851171,0.329441263272636,0.037325723533133,0.00454631076683781,0.0488140139804341,0.0672674435219996,0.00459848528213147,0.143753060707225,0.0145213193944826,0.179903226159086,0.315540152260485,0.152409597855197,0.208322218241635,0.0890601374366632,0.166648331846914,0.240087043690842,0.0648511641583757,0.789873896841488,0.922815444616418,1.93451285412164,0.167097952926445,0.055086724641985,3.60506187230955,0.185200807430027,0.00411683845347271,0.00606966185826972,0.0372752573181502,0.122378034288297,0.0168094838384131,0.277512516244089,1.27211278777793,0.0648209704704071,0.123894859606519,0.0274649116480148,0.360023334896927,4.40045796088191,1.23270901739913,0.638617713594059,0.0672674435219996,0.357458104081827,0.112947893184963,0.0408362763240822,0.476742611646738,0.169134060409173,4.33392914242096,0.0589486888360361,3.95048770869428,0.0511957384793949,0.0724069996852267,0.171714971769673,0.493261463737139,0.0509113179148502,0.179829445327717,0.358547473586084,0.8399859749391,0.0908576759166715,0.0486810306255043,0.0262356666186366,1.86043107022829,0.0702572251429715,0.206621115785932,0.0818233581757009,0.0104526353142326,0.883660414881033,0.147849009907857,0.569228794705177,0.95694263120013,0.000876057544547143,0.181872908667897,0.0450129006423602,0.0166923597787139,0.0711581260335338,0.569228794705177,0.0313154358062983,3.67490387468606,0.0784701086209545,0.112506122298816,0.00883507889890769,0.559325465633358,0.0446081133619268,0.0494464413743751,1.36320178764824,0.277512516244089,0.272268327877101,0.0890324487110214,0.0145213193944826,0.0536593860418449,0.0760852880259302,0.324468724874127,0.00116380427287773,0.0197048238177622,4.72531405015067,0.0289181750853702,0.670044754476321,0.00404081638338247,0.31696833193685,0.147497384945501,1.01795574727841,0.320219386917268,0.139551597426331,0.0195276971424756,0.0693703585998147,0.00401548166356793,1.29607282708929,0.0589486888360361,5.41577183542422,0.0972301811406793,0.0218015032544538,0.35956619017736,1.71169344210693,0.0408362763240822,0.489641467673164,0.734370019130202,0.0509113179148502,0.0880904578378599,0.128166969165267,0.0865962984195445,0.00459848528213147,0.0137588701180091,0.122732679224458,3.09600772170997,0.0435811396153694,4.30025102618811,0.646229070631471,1.22384987395097,0.138785729274634,0.749977018183376,0.0812983042046511,0.0796105337802797,1.02418596516548,4.91684575537689,0.0339799027559752,0.261381856204261,0.4208552272897,0.283666048183297,0.38180321101221,0.0248072680145259,0.264065902578283,0.16581544056764,0.166581454304626,0.0358044923638088,0.0458301838355025,0.529319732003221,0.0479261967394173,1.59022795926608,0.00315561947602539,0.0789084234280331,0.411791309651475,0.0653269443253174,0.0451549731464553,1.14094367719775,0.339639092055912,0.268708811554021,0.274226743757498,0.0791692510235405,0.00411683845347271,0.125051568964389,0.0677190760133282,0.0702572251429715,0.06301345805018,1.3941002572221,0.368404356264804,0.227330068184355,1.57233231684146,0.050779317312943,0.0467563303602578,0.105208262580619,0.958895324528815,0.214011976307974,0.0645938043462783,0.690958756438785,1.54314881056974,0.138996795478563,0.161592712272654,0.1026859616258,0.00315561947602539,0.397814565576244,0.747682786275648,0.00141021433832185,0.0353007315049884,0.0394506369694468,0.16197689883158,0.319606200885697,0.0671524936478646,0.0168094838384131,1.72954978567044,0.0525459415451182,0.00315561947602539,0.00460997542369454,1.48421485790977,0.317961178061264,0.0702572251429715,0.0702572251429715,0.0979520601416119,0.0218767093611828,3.74091704027218,0.0197048238177622,0.0375060843610187,0.345502048510554,3.41769100593248,0.0880904578378599,0.62885710337542,0.178647090301883,0.0794764611025657,0.572535710311603,0.00329111046875399,0.0481498192638804,0.0036929116085014,0.206504473070241,0.0408362763240822,0.0614688361424385,0.00407996707515448,0.665456447376267,0.100626175904096,0.213342996857309,0.0771667172825306,0.0212149250965154,0.0979520601416119,0.0220942953367839,1.03747889450713,0.267268122237729,0.0294599736810421,0.451103048780114,0.00593828021807046,0.407545606258617,0.132143350990551,0.108077065988984,0.0116773790911124,0.00407996707515448,0.0708418517605244,0.0604456738943895,0.716911352120118,0.870348892879163,0.448603207825026,0.217537835690649,0.0415099352417681,0.0188941321505588,0.0381084933975253,0.00659116107410934,0.118282737570121,0.124496247530311,0.00121304115851171,0.0711581260335338,0.0364774306440582,0.0750295576116174,1.23002970756969,8.39467787977262,0.0197048238177622,0.041642515680023,1.23800049126337,0.354078023438058,0.0315847795870568,0.14140666996898,0.323981847619161,0.270492530181306,0.0149236544925937,0.845352377607299,0.547176633397108,0.00606966185826972,0.857720045990041,0.187133153861838,3.26200375102729,0.112947893184963,6.65112160690437,0.297828514755134,0.00473174806234623,0.0688140767070753,0.0297255884908239,0.110572284773651,0.242449105026232,1.02012226293847,0.0253805703473298,0.0403277505967767,8.42274844515373,0.741253124384253,0.050779317312943,0.139580531528287,1.08656559510203,4.12976163708193,0.497063280896497,0.332905181108188,0.0915836791139002,0.748507442622767,0.147497384945501,0.223863520223173,0.315056045811215,0.0614688361424385,0.307622786160835,1.15311348474348,0.00185273569939584,0.08264702096919,0.419462067012117,0.00459848528213147,0.32794839832181,0.402766964581626,0.768001120315757,0.325881386097665,0.267268122237729,0.507860687004746,0.439797539101734,0.0375060843610187,0.0356391500831134,0.0195276971424756,0.347364126697632,0.240087043690842,0.0896599668422461,0.677447449227901,0.200783076682715,3.95363360063235,0.723409143501444,0.0168094838384131,0.00626713213883022,0.597088856420344,0.10393050620414,0.407662897684194,0.0461219326844996,2.4280871741365,0.0622351421916546,0.411791309651475,0.0195276971424756,2.82295085960226,0.273785007619018,0.184572302455561,0.601703072225002,0.0944351713915657,0.100942193736438,0.120165856383946,0.0854771605495596,0.0652882672422695,0.171714971769673,0.0450129006423602,0.128770266148453,0.242449105026232,1.89083541505142,0.0222139917248024,0.137672640302506,0.200783076682715,0.166971424957768,0.793267084368073,0.161982867175784,0.534986947383463,0.00883507889890769,1.26901476924371,0.058423750528285,0.049139366916818,0.981237217245642,0.114066567488366,0.00459848528213147,0.499429590820014,2.24191063514568,0.0833775144840563,0.00947518536003681,1.18251232256637,0.429520002461895,0.0618751050272665,0.0538566549074897,0.0400753596802403,0.529319732003221,0.00454888400368057,0.0108307155591846,0.122256335448388,0.488302390662843,0.0056492655075572,0.0660054216157546,0.0293641058665195,0.29721610124596,0.491822904754046,1.10320638020395,0.0375060843610187,0.5564255045703,0.225523022083862,0.0672674435219996,0.140682253480382,19.06830690457,0.231179028699573,0.859972156609328,0.0276319973730302,0.0645938043462783,0.821391313562383,0.014661865093209,0.220582915864296,0.357458104081827,0.243704417215347,1.84501013344656,0.0168094838384131,0.0402056440408849,0.2558106850751,0.0339799027559752,0.1359132967779,0.223863520223173,0.0604456738943895,0.0257029326308624,0.0979520601416119,0.727058690357005,0.23941547143042,0.00593828021807046,0.0056492655075572,0.123058776142951,0.34923111096301,0.435180342308126,0.18637485166302,0.000812948016292426,0.387189026044898,0.425559465026273,1.85629579663446,0.0215521295131734,0.0949805696796929,0.0589486888360361,0.264065902578283,0.87971380439442,0.139712324894668,0.0664206568967991,0.0672674435219996,0.0536593860418449,0.108187611798949,0.155242598661533,0.0826797667647909,0.0187543436867197,0.0195276971424756,0.00411683845347271,0.29721610124596,0.15225344922641,0.0006626888108606,0.290999007158141,0.0674214193994059,0.0372800057604592,0.102446216999783,0.000876057544547143,0.404118915224736,0.00905388074928039,0.558025382523306,0.0760852880259302,0.510889114260664,0.984739538095503,0.0614688361424385,0.00606966185826972,0.116504743482068,0.0104526353142326,0.11017481866874,0.366910343794684,0.552216831869672,0.0218015032544538,0.0158634862926648,0.220582915864296,0.11257503223522,6.02937340487434,0.0702572251429715,8.3352879512136,1.37587198889175,0.146764443869745,0.823848864570809,7.51485704668457,0.0788591939294461,0.0217902084804504,1.06720851534183,0.00659116107410934,0.0702572251429715,0.0865962984195445,0.436055652833192,0.669014265845929,0.00407996707515448,0.0158634862926648,0.18915195371068,0.329296981973122,0.818782819980794,0.00759819506386321,0.183926389770086,0.00473174806234623,0.236808015071122,0.239907307577072,0.062321021378901,3.36784755612013,0.0795905483797196,0.4749052974546,0.729798808830232,2.18587132355549,0.0441918045103556,0.028873429718069,3.20580095839808,0.0653641229937024,0.114066567488366,0.0145213193944826,2.05980725030094,0.378944767854216,1.38893670358892,0.0678720674317296,0.140139117126687,0.139580531528287,3.86156608007048,0.772971386813549,0.0971362738478571,0.166971424957768,0.0206385049319028,0.0787542265628141,0.00407996707515448,0.253295754378762,0.0671524936478646,0.18637485166302,0.775529691213294,0.429520002461895,0.0343414645022923,0.25569487652148,0.039951814594055,0.0479261967394173,1.63085802398507,0.00485280918750653,0.508521431323551,0.0702572251429715,0.549018115318202,1.14494857581857,1.11411429569313,0.220148126842577,0.125051568964389,1.28477646877316,0.430021972204687,0.0281881093361584,0.0197048238177622,0.197797192545651,0.11017481866874,0.0213176508508624,0.0218536705005053,0.0141000991332968,0.0492667681812746,0.0380208847945878,0.0168094838384131,2.19380003247443,0.213342996857309,0.0217902084804504,0.127112452673322,0.00947518536003681,0.0708418517605244,0.0479261967394173,0.450556003029832,0.0690082726024044,0.0234933792595118,0.176647343233519,0.189204587236502,0.00790968930391987,0.120748776837183,0.151186263156311,0.203508139916298,0.0104526353142326,0.00407996707515448,0.313542894887032,0.123551685773283,0.00459848528213147,0.277512516244089,0.102791677567528,0.214011976307974,0.0971362738478571,0.651007070684547,0.811119783314346,0.0195276971424756,0.00883507889890769,2.61274485777867,0.403171500401637,0.435319409721699,0.0168094838384131,0.0166923597787139,0.117778310517897,0.00686155703517166,0.277512516244089,4.07438078118181,0.0166923597787139,0.00148341115078733,0.505609480331729,0.861235805115969,0.493261463737139,0.0380208847945878,3.26200375102729,0.00329111046875399,1.56106387970433,0.0377191980411576,0.37011790758766,0.0889080927701457,0.00329111046875399,0.06151247459663,0.507569066951375,0.328432089316972,0.349434387278569,0.116546719657008,0.534986947383463,0.0978993601540305,0.268416910135073,0.00121304115851171,0.640350771275166,0.0451549731464553,0.108077065988984,0.109496667515653,0.0818233581757009,0.0104526353142326,0.220009673422181,0.0370373374431246,0.541962357619476,0.0426005121118614,0.0840103977456048,0.0217902084804504,0.0589486888360361,0.0410340343834169,0.0257029326308624,0.0851303181753314,1.4159585467273,0.0234933792595118,0.0159185958779729,0.038616932751444,0.193141125880745,1.83400932399523,0.00405427139052615,0.00454888400368057,0.174284449075951,0.62885710337542,0.184572302455561,0.303529940392609,0.0339799027559752,0.240723501842226,0.553746445241948,0.0672674435219996,0.769961797936431,1.13905509447008,0.067654278117622,0.169982046311221,0.160222861825975,0.433497750173421,0.067938277501381,0.0743282447144101,0.2558106850751,0.0577938378524624,0.0394506369694468,0.388593183689036,0.00398389888372882,0.199725602217908,0.106774017517753,0.0393561990639805,1.99123182285677,24.6218996401847,0.161685592327123,0.56489545332861,0.0645938043462783,0.0854172969512036,0.00626713213883022,0.0971362738478571,0.32470819810687,0.00459848528213147,0.0257987873052996,0.254599496072089,0.0451549731464553,0.476301263135945,0.15261163394772,0.0481498192638804,0.566621491615729,0.351365526453055,0.342432287531116,0.0157949646038045,0.526474171796221,0.0716689639488025,0.086992843136258,0.338299794303057,0.0136027177129889,0.143178543270171,0.166971424957768,0.130751309049599,0.848362613973338,0.0168094838384131,1.01132996467137,0.0147363309021572,0.127082372867785,0.0944351713915657,1.05763641282567,10.0853471586745,3.05093498112702,0.822499082425479,0.0316879936483847,0.114066567488366,0.0922029316571254,0.0206588798271408,0.0796592061137124,0.0168094838384131,0.268564980493208,0.00106898696268826,0.197797192545651,0.234745643751415,0.29721610124596,0.901587534234326,0.00756414078815883,0.575157303698369,3.42704296577283,0.0757102268223784,0.800730106286925,1.03639419034985,0.00116380427287773,19.8343759903105,0.673087645580452,0.591143186619663,0.050830217403903,19.6085209724405,0.555324324293309,0.0812675906611075,0.0211611751863212,0.0979520601416119,0.127741389958166,0.123639312193052,0.0273364578475598,0.0979520601416119,0.0206588798271408,0.0168094838384131,0.0464017913773774,0.00265924793670972,0.028091858780483,0.861235805115969,0.413979749182254,4.50359173345734,0.082735060584521,0.377944688328723,0.0525736740085782,0.080040595786052,0.0439723049111307,0.0168094838384131,0.198040870403191,0.473558225506477,0.127112452673322,0.198680521213169,0.0987422917741626,0.141829045667774,0.0702572251429715,0.0328741096585519,0.314908449115843,0.0328741096585519,0.0303089288227088,0.0195276971424756,0.0706672089775578,0.178294305315247,1.64801681582293,1.75801595243402,0.135838918788729,0.0479261967394173,0.0648209704704071,1.52981892709688,0.0402122274036898,0.152670270522971,0.1359132967779,0.448793643609886,0.086992843136258,0.992149269797961,0.00407996707515448,0.0672674435219996,0.0984052560382996,0.00606966185826972,0.0672674435219996,0.0277073537847336,0.370269147846954,0.0788591939294461,1.56109605617001,0.220148126842577,0.0372752573181502,11.4580097423227,0.127082372867785,0.212028338619143,0.0281881093361584,0.13351706753188,0.613994555623594,0.171181934420027,0.194800051088586,0.183685342799604,0.0742420183969878,0.00606966185826972,0.0365387457853382,0.144190642300856,0.214799001636142,0.467452328013943,0.00298562862325312,0.135838918788729,0.105275190435028,0.00329111046875399,0.399071280990619,0.276449007674386,0.866973463794472,0.0164067793647608,0.011249800747522,0.0343640697906325,1.27706127335596,0.0166923597787139,0.0166923597787139,1.08996903852807,0.0652882672422695,0.151565801348704,0.745442919850882,0.0164067793647608,0.105208262580619,0.00593828021807046,0.11771716526715,0.577894460444501,1.05851198278994,0.0277073537847336,0.0193198465505753,0.361102243748291,1.5218902172666,0.0699032425336779,0.0104526353142326,1.4403967851018,0.0804205837138711,0.0672674435219996,0.294272218359504,0.0118003362744535,0.0281881093361584,0.280916943494393,0.139551597426331,0.0164067793647608,3.16485430554629,0.0256352826026638,0.230374177552424,0.181872908667897,0.155242598661533,0.0307572237431331,0.122378034288297,0.0830883297600061,0.00947518536003681,0.723409143501444,0.0762960487173458,0.0221141481581544,0.770439372836797,0.0946349501008994,0.102791677567528,0.0760852880259302,0.0249964838592474,5.34213254138141,2.44156490086778,1.32484622088363,2.24963958561621,0.0684394952173052,1.28476028701103,0.197797192545651,1.03639419034985,0.313542894887032,0.5854517543877,0.709326288799336,0.677226289148063,0.708628129091459,0.00606966185826972,0.00141021433832185,0.138504957656793,0.00434189191494265,0.0402122274036898,0.0739806170536602,1.35983927556736,0.988130972310288,0.0056492655075572,0.0739806170536602,0.0195272682659921,0.0838811819084171,0.141487854541546,0.0825818392824401,0.264065902578283,0.104257587805067,0.0204109004922536,0.0232272978244537,0.0920057935468886,0.234745643751415,0.260836285894463,0.0148655927726722,0.156442988488283,0.98437277681776,0.119453263291254,0.0495809894680694,0.10827065906627,3.00978676031731,0.00569817735165309,0.42314768381533,0.0402122274036898,1.56805563777701,0.0138895788396268,0.0690082726024044,0.0818863834804944,0.723409143501444,0.0281414757580683,0.127112452673322,0.0463351197509933,0.0536593860418449,0.0865962984195445,0.193863255710462,0.0564530643128249,0.0614688361424385,0.459215185078108,1.06523068194658,0.0106031621346388,0.0979588081185319,0.609295899690258,0.00883507889890769,0.491822904754046,1.03098487497761,0.127082372867785,0.0739806170536602,0.161685592327123,0.332905181108188,2.66965751990843,0.18637485166302,0.0217902084804504,0.257974418335539,0.178816622135615,0.0370373374431246,2.77738477335047,0.0614688361424385,2.31553719004652,0.10965467486682,0.0793391320060809,0.0023534462217772,0.0589486888360361,0.0603748403667264,0.0716689639488025,0.0796105337802797,0.00756414078815883,0.0212220249015296,0.00415831872242659,0.112947893184963,0.297465307354734,0.0711581260335338,0.0614688361424385,0.0660960450124907,0.0672674435219996,0.258856019391655,0.00526552289067343,0.00358117086348743,0.256063481414924,0.00827862905112156,0.114066567488366,0.0615350227233221,1.41618029810107,0.606675266200296,0.00659116107410934,0.00459848528213147,0.379045229285591,0.675642380712841,0.125148618462469,0.0200925978977868,0.538008664404916,0.0833775144840563,0.125813024092088,0.0256352826026638,5.55661208135797,0.426678398059656,0.0394506369694468,1.43740695454777,0.174822974370871,0.0819983600707709,0.0166923597787139,0.0851746660495717,0.0319768447348491,0.609099442533923,3.29881292577662,0.420321241912227,0.5355160017489,0.0614688361424385,0.0164067793647608,0.821208686845829,0.0965000651465204,0.0596106501921475,0.594297656479166,1.4159585467273,0.132680099583533,0.119265082205604,0.0702572251429715,22.3126820127702,0.301921895040769,0.334755553665429,0.00298562862325312,0.0854771605495596,0.0370373374431246,1.8638828777159,0.0531595544706274,0.794759573215123,5.31568300034284,0.626165782074011,0.010755650529379,0.0205929766871262,1.1309514490229,0.167227212152352,0.501460561874647,0.13991393500292,0.00947518536003681,0.00659116107410934,0.127425489299559,0.0218767093611828,0.198397576263175,0.00659116107410934,0.0195276971424756,0.0190271316556984,0.179903226159086,2.48056808553314,0.14872925706803,0.142524262563551,0.00593828021807046,0.112947893184963,0.00543147291271282,0.0222139917248024,0.239589405634418,0.0464017913773774,1.46084124721252,0.615425154432416,0.0521577164702833,0.791861018325946,0.13276848348873,0.349434387278569,0.639584467538902,0.28194505290561,0.176647343233519,0.00431164095251631,0.00606966185826972,0.00407996707515448,0.534986947383463,0.0379033786577836,0.0375060843610187,0.00272520147235411,0.0137578450631309,0.43933034515539,0.222255573559327,0.0854771605495596,0.00411683845347271,0.00817736899861541,0.0420257274415236,0.0103230553331435,0.0453725391854269,0.55665809275023,0.114066567488366,0.428780032292995,0.0106031621346388,7.04284185839144,0.018213394299291,0.0861908744219117,0.429102059964555,0.0211611751863212,0.1359132967779,0.611707783203397,0.793267084368073,0.223863520223173,32.9169749040973,0.0400753596802403,0.332905181108188,0.0166923597787139,0.132715346044724,0.0704652852712834,0.0346731617383663,0.623261090232879,0.00459848528213147,0.115597204501953,0.793267084368073,0.0197704517689435,5.56360510557687,0.0767799682327379,0.0370373374431246,0.0990147880697231,0.815398197503196,0.0603748403667264,0.00793295388501706,0.328047559396122,0.161685592327123,0.116504743482068,0.0838811819084171,0.0435811396153694,0.00407996707515448,0.0979520601416119,0.4749052974546,0.0941027396022316,0.0589543514677482,0.00947518536003681,0.0317714935858366,0.04017965282136,1.05496587971326,0.10393050620414,0.00606966185826972,0.0959766526649428,0.015338392640947,0.333906630325922,0.550072274270647,10.3194322558024,0.357458104081827,0.0663963811889072,0.178294305315247,1.56805563777701,0.428515700451037,0.866674160511146,0.171714971769673,0.11257503223522,0.0652882672422695,0.0838811819084171,0.0624392742538957,0.124579879373261,0.00385631494084208,0.0179041882630937,0.161685592327123,0.450556003029832,0.670044754476321,1.81587107925518,0.166581454304626,0.0341535584114983,0.0760852880259302,0.0854771605495596,0.0972884686457311,0.0979520601416119,1.50763367048553,0.018213394299291,0.166581454304626,0.0653269443253174,0.0189188080507413,0.0168094838384131,0.443796329322799,0.809906321775336,0.828048045597703,0.0164067793647608,0.179829445327717,0.0159185958779729,4.15190363507648,0.838546822655075,5.41413153633167,0.0566067255934518,0.235241132500741,0.124496247530311,0.731830118142868,0.354879201704377,0.37011790758766,3.52595727615643,0.493261463737139,2.44343515670071,5.05579250022273,5.56360510557687,0.831812394620213,0.086992843136258,0.968409336058904,0.0030574054071175,0.125051568964389,1.02658399721003,0.0739806170536602,0.815435634496934,0.133101625794071,0.811007947826258,0.0770238392399662,0.0356103344810315,0.0234933792595118,0.0448748525927228,0.00454888400368057,1.16553235552538,1.11042916928748,0.0614688361424385,0.0672674435219996,1.96912663245712,0.0838811819084171,0.0926056828503804,0.00542347084586741,0.176647343233519,0.0954495555153234,0.155242598661533,0.113612820733828,0.0749585254543165,0.0168518802392531,0.00419084931358526,0.0653269443253174,0.0760852880259302,3.03694689367258,0.297828514755134,0.4749052974546,3.12074546632558,0.239089394132639,1.340976565671,0.122944688288505,0.0954495555153234,0.197279060457972,0.010798078778355,0.76329275061048,0.739914160767582,0.0511857792419379,0.381068134290834,1.04922910789765,0.18667665825874,1.12760799793578,0.181872908667897,1.20850538345629,0.566070256013332,7.85021419236835,0.193794080389875,0.180787544745946,0.0271044776741718,0.0356609883149435,0.90916523727296,6.43722225822277,1.30961739484927,0.075707697959015,0.094283855123758,0.163839447445812,0.139580531528287,0.822499082425479,0.183685342799604,2.35819920204489,0.32794839832181,0.00298562862325312,0.117547265407849,0.0197048238177622,0.85346882073596,0.0645938043462783,2.33221410191016,2.6117426607229,0.411791309651475,0.421249556142679,0.0615350227233221,0.0217902084804504,0.0118003362744535,0.175124664174564,0.181626929543518,0.202441690777921,0.0168094838384131,0.872104041613047,0.0421820176991698,0.182308749323054,0.845352377607299,0.351365526453055,0.300780644440393,0.893064480944466,0.0722995605337732,0.0494943416052741,7.28140751800237,3.34368194448457,0.00923612899724914,0.286999379376735,0.0221622331632297,0.0602428265616625,0.00569817735165309,2.58752712613323,0.0380218048667962,0.0840103977456048,0.0106031621346388,0.37011790758766,0.163419205600022,0.412575407643809,0.00947518536003681,0.584823994574904,0.524248041543289,1.31309577678004,1.51541071432245,0.231179028699573,0.11257503223522,0.243590516752951,0.59406639971811,0.0168094838384131,0.0017348219626119,0.276449007674386,0.186842172927274,0.00431164095251631,6.41692831814497,0.158834134323555,0.141829045667774,0.114066567488366,0.31696833193685,0.0197048238177622,0.0248072680145259,0.0739806170536602,2.54553501215664,0.00139918589257872,3.69415650138168,1.60606096119451,0.159941977547238,0.00459848528213147,0.0104526353142326,0.069814215314912,0.0840103977456048,5.39219716591267,0.154590008104649,0.734705265716782,0.134500410888056,0.00782002579161583,0.0649545376936062,0.120748776837183,1.2540095381688,0.103863396275375,0.0164067793647608,0.101952406658243,0.00460997542369454,0.0197704517689435,0.330474801833405,0.00569817735165309,0.534986947383463,0.470975881187938,0.0958039046797313,0.0200995742484223,1.06695125253939,0.0692641240983059,0.418149131751107,0.127741389958166,0.0695971501675616,0.0538566549074897,0.894973056481956,0.558434412804878,0.549036014309833,0.180561623540819,0.940218077256494,0.18272273423756,0.139772387784669,0.122378034288297,0.594373947087004,0.0684394952173052,0.0672674435219996,0.0380208847945878,0.821391313562383,0.0281881093361584,0.0615350227233221,0.0168094838384131,0.535493229091547,0.0248072680145259,0.811007947826258,0.0442108253457593,0.105208262580619,0.0369651588949079,0.11257503223522,0.146519796962383,0.0467563303602578,0.214611042307936,0.0450129006423602,0.0158634862926648,0.341889577893,0.476301263135945,0.0454863584200561,0.133367645199529,0.858892766908458,0.139551597426331,0.0740110016637447,0.0702572251429715,0.0451549731464553,0.0672674435219996,0.743685431979854,3.59467203689914,0.791902597756607,0.493261463737139,0.110693361465161,0.216783058690278,1.07424752453573,1.14839576775598,0.287210597864711,0.147497384945501,0.0278937932733824,0.0693807584889081,0.00196710152911757,0.662797455558691,0.114766438533409,0.398965103665105,0.132252086222209,0.0473263681284882,0.0818863834804944,0.0831044442602011,0.00459848528213147,0.770439372836797,0.222749765123598,4.53554331089413,0.0576344832890614,0.103443184645079,0.0589486888360361,0.0990147880697231,8.14692206018099,1.18392664634476,0.15511231908647,0.0158634862926648,0.0702572251429715,3.96239604397732,2.07742810029321,0.0689009961671803,0.180383540022676,1.79258375591057,0.239688994827829,1.98869253517305,0.00554465977811824,0.805150588636702,0.222749765123598,0.129179319472705,0.571041744945995,0.0130013793455995,0.39607208234861,0.0293641058665195,1.98245722691963,0.0833775144840563,0.0164067793647608,0.91306541018145,0.0165094669358971,0.0197048238177622,0.0375060843610187,0.122944688288505,0.0672674435219996,0.0511857792419379,0.132742374352844,1.13905509447008,1.05453236764813,0.0164067793647608,0.0195276971424756,0.0290341322999814,0.399071280990619,0.906464415110111,0.354845354781611,0.127102735562851,0.36466617530845,0.0136027177129889,0.790419902293761,0.221565510702022,0.0168094838384131,0.162729743376864,0.0702572251429715,0.0444282422240755,0.00139918589257872,0.236844831363697,0.995166008445084,0.537583946769745,2.51876381022282,0.00947518536003681,0.0356103344810315,0.0357231972671091,0.0450129006423602,1.12092865502688,0.00161138485580163,0.0538566549074897,0.159898986010081,0.1026859616258,3.17218998284575,0.0941027396022316,0.0398944169870788,0.11257503223522,0.0365387457853382,0.0865962984195445,0.294462991629555,0.734836329499352,0.0544495745810078,0.0206588798271408,0.203508139916298,0.0168094838384131,1.05496587971326,0.151723465649993,0.00883507889890769,0.398264454570971,0.0543596495378718,0.032161006716023,0.00434189191494265,0.0494943416052741,1.19226499014867,0.0198428994896227,0.086992843136258,0.00385631494084208,0.14140666996898,0.0614364470365818,0.0678720674317296,0.00614931567930242,0.0890324487110214,0.0377855186980461,0.118158133474985,0.0281881093361584,0.0668861547243075,0.90057371872741,0.0653972489406986,0.0018835297482147,0.0769895762651347,0.15371317096278,0.786107353931988,0.216582287941036,0.328432089316972,0.0660586253442284,0.0182955290652585,0.0017348219626119,0.0672674435219996,0.0645938043462783,1.16492886160862,0.0734827074428003,0.534986947383463,8.45249056846172,0.0441918045103556,0.0364774306440582,0.0159185958779729,0.000485568677475681,0.0277073537847336,0.0596504757710883,0.0189773911010558,0.476742611646738,0.0295579699424944,0.0481498192638804,1.04922910789765,0.0951798824116844,0.900429269706737,0.310482337696921,0.00329111046875399,0.0217902084804504,0.0276319973730302,0.236844831363697,0.0168094838384131,1.20963538018143,0.40942691722469,0.438536443342242,0.00315561947602539,0.232526994078927,0.800654509811821,0.0164067793647608,0.104257587805067,0.0563614323245143,1.7775282296573,0.0363592437800154,0.277512516244089,0.0838811819084171,0.975097500151541,0.023194764608665,0.0830883297600061,0.0343640697906325,0.151565801348704,0.0402122274036898,0.0653364468487771,2.05936104721347,0.0166923597787139,0.234815621699189,0.00407996707515448,0.0084654132368613,0.0168094838384131,0.414558010144188,0.38180321101221,0.845352377607299,0.172841599929998,1.04922910789765,54.3098224688406,0.0865962984195445,0.0200558260488684,0.0365387457853382,0.102791677567528,0.0604456738943895,0.329025534407141,0.0788591939294461,0.117547265407849,0.181872908667897,0.0159185958779729,1.15338168724045,0.225329499496512,0.120165856383946,0.1026859616258,0.0468622799937739,0.408160607911106,0.0402056440408849,0.0528618138018548,0.0799541881680441,0.166160540189493,0.0197704517689435,0.0372751825812681,0.800730106286925,0.357579329660536,0.108462620284522,0.198680521213169,0.1140983740879,0.0298543410974583,0.225903580352166,0.375663799783372,0.412575407643809,2.17123374454828,0.062377621797737,0.109294298749448,1.33005371348986,1.00022216874956,0.146764443869745,0.0825818392824401,0.164896948085131,0.572535710311603,0.0139374711613066,0.184572302455561,0.106567183910213,0.00839375177350195,0.635572861141521,0.0796105337802797,0.47419934597336,0.258064927875471,0.614303994416613,0.015338392640947,0.0880904578378599,0.00407996707515448,0.0498555705517042,0.0492310720780484,0.0234933792595118,0.412590154340158,0.32026188269458,0.10393050620414,0.00516427843486195,0.115597204501953,0.174222473063539,1.01392071243063,0.0979520601416119,0.138224812062143,0.0442108253457593,0.213647439158468,0.185885540938701,9.90692164010896,0.226050787439536,0.155394148119172,0.90621722768677,13.6017377017191,0.0647065170226978,0.10312114630122,1.88986071541169,0.815081153223971,0.774329935269415,0.734836329499352,0.239581551894305,0.687139558805104,1.00330919146326,0.0375060843610187,0.183685342799604,0.197279060457972,0.0036929116085014,0.398264454570971,0.406989169586797,0.0933900764726269,0.207214877406833,0.315450838822319,0.0252325255677302,0.580346654694217,0.088374348777414,0.00516427843486195,0.0706672089775578,0.0648511641583757,0.0416258592695452,0.00116380427287773,1.99304693638553,0.0441554547999769,0.0413083051753416,0.00431164095251631,0.0444282422240755,0.0693807584889081,0.077508169240904,0.193854242218128,0.034366002325725,0.31681450305845,0.468779932642312,0.0672674435219996,0.0189188080507413,0.143543163200891,0.0985927871700709,0.100036647315322,0.523062964869626,0.0473263681284882,3.81816056890429,0.445279553102564,1.04849019663374,0.238855027868265,0.0784701086209545,0.324044844567747,0.0220048422299199,0.0166923597787139,0.840132691051013,0.100626175904096,0.682085555395832,0.0370373374431246,0.00329111046875399,0.086992843136258,0.0437169091307368,0.0176786975345918,0.0394506369694468,0.106774017517753,0.329025534407141,0.0125400876763405,13.3842921411041,0.132715346044724,0.415869189054016,0.811119783314346,0.0747212588098682,0.371358254486124,0.743478252512291,0.437441042685696,0.0614953759361274,0.366199086548791,0.0540790211492507,0.815935980778152,0.154874628505491,0.0168094838384131,0.178647090301883,0.0612159065878276,0.114766438533409,0.220879106936892,0.109735593357188,0.0168094838384131,0.162279905666064,0.0767741931631283,0.00281822804816019,0.220582915864296,0.0168518802392531,0.0248045111492735,0.00577346048977,0.10017495195107,0.623360642102921,0.0450129006423602,2.31232073124486,0.0614688361424385,0.151186263156311,0.135838918788729,0.0256352826026638,0.0277435643331768,0.0965000651465204,0.011249800747522,2.38992472959447,0.0380208847945878,0.277724574501198,0.0164067793647608,1.49303113178213,0.00329111046875399,4.77783118301919,0.0672674435219996,0.0234933792595118,0.0536593860418449,0.0416258592695452,0.0166923597787139,0.00141021433832185,0.00407996707515448,0.0589486888360361,0.0356609883149435,0.357458104081827,0.0747212588098682,1.71169344210693,0.698491771233296,0.132715346044724,0.0672674435219996,0.492507260379483,0.248182336631564,5.94643782202376,0.123825470520158,0.399071280990619,0.0158634862926648,0.267254374669945,0.0356391500831134,0.245978580977023,0.126519839533059,2.04350054669933,1.64133039724428,1.1249257922934,0.0234933792595118,0.00196710152911757,0.0654008455209546,0.0294497582946361,0.11257503223522,0.0104526353142326,0.357458104081827,0.139461583975561,0.406450576929125,0.0228147992712077,0.0017348219626119,0.329437495127202,0.00460997542369454,0.0521577164702833,0.115597204501953,0.139551597426331,1.54882747439373,0.0200760204770733,0.00454888400368057,0.225893350019311,0.0509089323214273,0.0402056440408849,0.114066567488366,0.132644947994632,0.685561764428103,0.158872345450678,0.0309123152511477,0.139215044694534,0.149019021844506,0.323284011831274,6.28112707240507,1.02479164580398,0.0416070136836051,0.0166923597787139,0.127112452673322,0.299333869074544,1.20804606383215,0.86870110871256,0.0691723142136732,0.215895834297239,0.355105007728052,0.212475464448455,0.509287949512919,0.0903550141797481,0.102791677567528,1.36617186431296,0.00569817735165309,1.63601070256898,0.0890324487110214,0.0437169091307368,0.127082372867785,0.491822904754046,0.748507442622767,0.132715346044724,0.0833775144840563,0.59442798830849,0.119741500989086,0.29721610124596,0.0762960487173458,0.193794080389875,0.0724069996852267,0.00407996707515448,0.244479246784708,0.00473174806234623,0.220582915864296,0.194214674956053,0.00116380427287773,0.0197048238177622,0.075707697959015,0.181872908667897,6.5340160613562,0.0317644788478658,0.187133153861838,1.03747889450713,0.0444282422240755,0.362896457419757,0.0159185958779729,0.0365387457853382,0.014661865093209,0.115713176180213,0.0784701086209545,0.0511957384793949,0.359253881461244,0.251049302928911,2.51835325877788,0.0270303960634119,0.00141021433832185,0.48664559628603,0.00659116107410934,0.0804205837138711,1.54343928962303,0.545375773596892,0.0328741096585519,0.133187348953427,0.0142690548867,0.144969953327665,0.00315561947602539,7.90522769659088,0.0056492655075572,0.0168094838384131,0.086992843136258,0.30979490215918,0.217029851601869,0.00385631494084208,0.0990147880697231,1.38590864943896,0.00459848528213147,0.0648511641583757,0.226500084155321,0.0536593860418449,0.028873429718069,0.00230091183741603,0.0536593860418449,0.670044754476321,0.0941027396022316,0.227280550920699,2.22354253540234,0.239589405634418,0.304821017657104,0.0234933792595118,0.147497384945501,0.00407996707515448,0.527524776597855,0.0653364468487771,0.0319768447348491,0.609295899690258,0.276518464457158,0.147624330348209,0.0138895788396268,0.118282737570121,0.0256352826026638,0.534986947383463,0.327855572596143,1.6636134122983,0.0848389022301814,0.0838811819084171,0.481778351373681,0.276449007674386,0.0166923597787139,0.0252833486296207,0.520577164032037,0.0460067961984981,1.48253912003761,0.0221622331632297,0.00626713213883022,0.0965000651465204,0.0672674435219996,0.0364203501376111,0.18667665825874,0.0614688361424385,0.136127634391891,1.18223141746241,0.159018166067558,0.00419084931358526,0.0804943338538985,0.0158634862926648,0.06811620739093,0.0277840733794765,0.0880904578378599,0.0147363309021572,0.480621462365362,0.00593828021807046,0.284280723734994,0.0739806170536602,0.00121304115851171,0.160675170990038,0.218315004292681,0.328432089316972,0.605951007733236,0.0327887007582918,0.140441827618829,0.0104526353142326,0.355105007728052,0.00419084931358526,0.125051568964389,0.605690917511909,0.0158634862926648,0.0779553694527797,0.0528123240589885,0.160297931024884,0.469931299720132,0.119453263291254,0.258051887051628,0.0106031621346388,0.0564663031912013,0.0416949645516445,0.00606966185826972,0.275547622392463,0.0450129006423602,0.239589405634418,0.10232134426748,0.222749765123598,0.00298562862325312,1.95978053692986,0.0614688361424385,0.00460997542369454,0.0460067961984981,0.213342996857309,0.0365387457853382,0.00905388074928039,0.0898788814722309,0.106943988523261,0.032758357455683,1.01887496135198,0.270487754708997,0.0825818392824401,0.0833775144840563,0.0825818392824401,0.0166923597787139,0.107869078610365,0.142011492662573,0.0205929766871262,0.0435811396153694,0.0197048238177622,0.774329935269415,0.290140878765139,1.20850538345629,0.189615505253664,0.0611791659231674,0.0372751825812681,0.0479261967394173,7.57062176168748,0.178817840983991,0.0890324487110214,0.132028184977637,0.0615350227233221,0.152047798370628,0.102791677567528,0.37011790758766,0.0708418517605244,0.0140097434011857,0.0118003362744535,0.0576344832890614,0.179724070469211,1.31552247493151,0.00141021433832185,0.0205929766871262,0.0158634862926648,0.0855239831172985,0.0645938043462783,0.0189773911010558,0.333822954939191,0.0217902084804504,0.0726013805644724,0.114066567488366,0.990016718748193,0.552216831869672,0.0160902603051206,0.762957391202675,0.127082372867785,0.0840969254626935,0.0260332064508296,0.0252833486296207,0.127112452673322,0.00459848528213147,0.0257987873052996,1.97946307738828,0.0840103977456048,0.0804943338538985,0.2334263708634,0.332457453097218,0.0971362738478571,1.00330919146326,0.0865962984195445,0.503876948254088,0.18637485166302,0.0132481317498369,0.717606033123193,0.0122447960612203,0.167014211007613,0.0141000991332968,0.0788591939294461,0.0356103344810315,1.00022216874956,0.29721610124596,0.050779317312943,0.086992843136258,0.0278937932733824,0.123058776142951,1.98331862538068,0.0277435643331768,0.140912714373964,0.197279060457972,0.160513639763373,1.62070137613404,0.224996968048284,0.0317714935858366,2.46678264861281,0.158396691486826,0.0375060843610187,0.0449747937540372,0.0458664718188478,0.29721610124596,0.329025534407141,0.00569817735165309,0.199447989245459,0.220083516426305,0.0799541881680441,0.547033021235577,0.00121304115851171,0.0176786975345918,0.1359132967779,0.147497384945501,0.0874973214262932,0.807868506383144,0.123058776142951,0.0158634862926648,0.245554099643693,0.108421809761933,3.62202810028447,0.890121716554107,1.65029382909868,0.0204109004922536,0.467707216570234,0.0440351249654628,0.116546719657008,0.0561299131065681,0.0234933792595118,0.0570257778453027,0.0755580722852818,0.286999379376735,0.299607075307626,0.178202622471464,0.20856170318365,0.143388112419094,0.148333758266021,0.264855801651674,0.0660586253442284,0.354478280090437,0.399071280990619,1.4179552591366,1.10077327591728,1.32484622088363,0.00569817735165309,0.0488140139804341,0.10393050620414,0.154590008104649,0.146727455715937,1.97946307738828,0.0925366296544678,0.0418648385834954,0.220879106936892,0.00401548166356793,0.0022229228535356,0.276518464457158,0.0256352826026638,0.759237674965495,0.0234933792595118,1.63601070256898,0.057492475836355,0.0316361073828435,0.0450129006423602,1.04214903532347,0.115597204501953,0.0521577164702833,0.0166923597787139,0.179829445327717,0.0614688361424385,0.0854771605495596,0.0399382181621259,0.0690082726024044,0.359208306430935,1.23250300359772,0.049966353668149,0.102791677567528,0.0444282422240755,0.139215044694534,0.233857033877066,0.171714971769673,0.0615350227233221,6.49616477070659,0.0715473711912393,0.443481342205309,2.55752601514943,1.05315992379101,0.0278130368520151,0.199756583886579,1.02168977484812,0.0702572251429715,0.0812983042046511,0.271317295866564,1.8777116305973,0.270872524317462,10.4179362836412,0.00141021433832185,0.640638791204819,0.530361228027758,0.0672674435219996,0.0056492655075572,1.99123182285677,0.0649545376936062,0.0197048238177622,0.997846803129985,0.699036145182037,0.236281298062111,0.0796105337802797,0.038616932751444,0.0253508299531895,0.363051347649499,0.109462044918209,0.0672674435219996,0.105275190435028,0.0715740008786597,0.0684394952173052,0.00528645926860618,0.0319768447348491,0.0536593860418449,0.00659116107410934,0.0855239831172985,0.535761521197531,0.0649545376936062,0.0776538523214883,0.0343672820985998,0.077508169240904,0.109535946913381,0.114066567488366,0.096341028752406,0.00731661718127994,0.0552608323582036,0.00407996707515448,3.12407895239735,0.712699555640077,0.127112452673322,0.133743058944992,0.0180448691320751,0.150491008464522,0.605232904985226,0.0618070204388145,0.154699480913639,5.44885176160076,0.0416949645516445,1.60891035160113,1.73286238489288,0.147497384945501,0.0324334366906665,0.155242598661533,0.419522114036753,3.18465227380168,0.00593828021807046,0.144969953327665,0.123551685773283,0.0684394952173052,0.0779430474348763,3.20078288302151,0.0257108343001713,0.0394506369694468,0.0402056440408849,0.0987762894302276,0.086781536241353,0.0249621032426942,0.0437169091307368,0.356654192452932,0.445635278711868,3.68300445206497,0.00407996707515448,0.00329111046875399,0.11056174765258,0.0179759732781862,3.43844051068992,0.00947518536003681,0.0604456738943895,0.123825470520158,0.0623843738112641,0.096617683909033,0.0277073537847336,0.154590008104649,10.6560715732285,9.59565343320493,0.181872908667897,0.171181934420027,0.0277435643331768,0.010798078778355,5.3362470699171,0.145760744369984,0.0182955290652585,0.29721610124596,8.19717871572631,0.12351449600632,0.0458301838355025,0.0206588798271408,0.0435811396153694,0.0671524936478646,0.351365526453055,0.596374522983821,0.0580026618595194,0.211308039781596,0.0281493562394956,0.491822904754046,0.641897046430971,0.0854771605495596,0.941112131078339,0.105907780768471,0.0861908744219117,1.08554159635502,0.0615350227233221,0.0275149914177536,0.0784818378065301,0.104257587805067,0.176647343233519,0.0854771605495596,0.571755287880514,0.0248072680145259,0.181872908667897,0.0107767401614758,0.00298562862325312,0.115620724680052,0.357458104081827,0.0343640697906325,13.6050834263614,0.0139802896287417,0.351365526453055,0.179903226159086,0.220879106936892,0.0255282110855254,0.226258928432434,0.0528618138018548,0.8399859749391,0.549163491021169,4.18024650469246,1.48012353767732,0.127112452673322,0.44192626341535,0.110693361465161,0.176647343233519,0.445279553102564,0.0370523248401019,1.10320638020395,0.06151247459663,0.039774259322378,0.445279553102564,3.2299082800264,0.678642854433977,0.0209679217005323,0.0312093817019672,0.015338392640947,0.0256352826026638,0.133495203649,0.0402056440408849,0.233486812635519,0.0999374459332967,0.0993393147468768,0.419727678150197,0.0451549731464553,0.0563675539190095,5.8169264822789,0.0130013793455995,0.0804943338538985,0.0702572251429715,0.186842172927274,0.200783076682715,0.151565801348704,0.0140097434011857,0.00136212981132986,0.0702572251429715,2.46764344799579,1.58645124915753,0.497063280896497,6.53148759356459,6.79417623332282,0.0197704517689435,0.696075744578418,0.655409432124841,0.00315561947602539,0.0147363309021572,0.0234933792595118,0.0614688361424385,0.0277073537847336,0.0925248104359321,0.213342996857309,0.0545541220167345,0.0819983600707709,0.303529940392609,0.00459848528213147,0.0428138877325034,0.0257987873052996,0.32470819810687,0.00754325657883153,0.169982046311221,0.010693866983192,0.115713176180213,0.301921895040769,0.0168094838384131,0.0785688473429434,0.0240426889317082,0.0283021768645347,0.239589405634418,0.0865962984195445,0.0274985885485839,0.222749765123598,0.0708418517605244,2.06637628659801,0.00385631494084208,0.028873429718069,0.0614688361424385,0.0392177290751502,0.216704140792974,0.114766438533409,0.179260381790001,0.0234933792595118,0.132715346044724,0.0622997709104631,0.11056174765258,0.0421820176991698,0.0645304689742235,0.479611118555517,0.00593828021807046,0.0878067492419905,0.1359132967779,0.625172194134489,0.129371360167829,0.0221622331632297,0.201628194149819,0.00309271121898999,0.00459848528213147,0.0368401375339735,0.472928553329943,0.234815621699189,0.0197048238177622,0.0104526353142326,3.34615562013576,7.07800653917507,0.0218767093611828,0.534986947383463,0.050109550356437,0.0826470000185111,0.599116152359974,0.169606571700878,9.03251853494307,0.00738336360458375,0.127112452673322,0.0458301838355025,0.0450129006423602,0.179903226159086,0.0536593860418449,0.00407996707515448,0.028873429718069,0.0672674435219996,2.42273722173672,0.0878067492419905,0.0538566549074897,0.349434387278569,1.34536551347366,0.0672674435219996,0.0618751050272665,0.62885710337542,0.0536593860418449,0.0182955290652585,0.301921895040769,0.0789789838719478,0.17792457015105,2.50298338848375,0.618858347244875,0.026346937631785,0.23807282911153,0.183685342799604,0.277512516244089,0.0023534462217772,0.0535678493924506,0.00407996707515448,0.0693807584889081,0.120891445029521,0.665456447376267,5.44207773835475,0.129082199597598,0.939627372063465,0.0760852880259302,0.681366448651368,0.619551087512229,1.81987223206743,0.324468724874127,0.120891445029521,0.125376737316837,0.0450129006423602,0.0356103344810315,0.050797908136273,0.015338392640947,0.0211611751863212,0.0767741931631283,0.251259069774278,0.381298125208919,0.600453152665349,0.954396692555429,0.086992843136258,0.114066567488366,0.221244292832211,0.0653364468487771,0.11099722976912,0.00883507889890769,0.00569817735165309,0.794759573215123,1.67062097254876,0.0771667172825306,0.0494943416052741,0.124139693551738,0.131669335655648,0.0556014841243996,0.613329212561002,0.143178543270171,0.00883507889890769,0.0402056440408849,0.0650789942206767,1.09284615344724,0.0818233581757009,0.0854771605495596,4.15492639295878,0.222749765123598,0.47419934597336,0.59406639971811,0.506104119381677,0.0277073537847336,0.0833775144840563,0.0536593860418449,0.0599451873645366,11.4116480165989,4.46161038080762,1.63644302241281,0.530361228027758,0.357458104081827,0.321457306653641,0.0428138877325034,0.0481498192638804,0.12178671210635,0.0160902603051206,1.7328830656416,0.0946349501008994,1.13359579347678,0.0276319973730302,0.110693361465161,2.6304389383173,0.166971424957768,0.00947518536003681,0.0672674435219996,0.614004067188387,0.323420803151992,0.0188249865621577,0.111121490592266,0.096825409027854,0.193334566978367,0.224450026435123,0.0398757891342887,0.220582915864296,0.0649545376936062,0.822499082425479,2.18587132355549,0.867651621132305,0.821208686845829,0.743478252512291,0.140879977786088,1.17194532885516,0.0479261967394173,0.0734827074428003,0.0104526353142326,0.0218767093611828,0.0440351249654628,0.0444282422240755,0.166648331846914,10.7742141820818,3.12530021195244,0.419628126194369,0.277823633722508,0.915265249077549,0.0440351249654628,0.0343640697906325,0.0168094838384131,0.111121490592266,1.00788201091662,0.480621462365362,0.181872908667897,0.497063280896497,0.0191334351438423,1.67880103051023,7.67928489010006,0.00275377972964126,0.0197704517689435,0.392830367587236,0.0536593860418449,0.109735593357188,0.041642515680023,5.82764575426201,0.840914278716581,0.172718602450644,0.405639699701347,0.166581454304626,0.270492530181306,0.00195802312323055,0.105208262580619,0.0168094838384131,0.0197048238177622,0.0221622331632297,2.86451780979694,0.496533114983416,0.0388902511214588,0.915265249077549,0.454719075434713,0.141829045667774,0.105208262580619,0.0253359751394339,3.12407895239735,0.00930235994024686,0.0771667172825306,0.114066567488366,0.318820057256344,0.87971380439442,0.186944063020789,0.512731373389147,0.187133153861838,0.552216831869672,1.40821916403864,0.0316231041130952,0.149019021844506,0.142011492662573,0.122944688288505,0.0188542634863658,0.0281881093361584,3.22573031154754,0.116848928150799,0.00815315331791984,0.047451881650676,0.0309123152511477,0.239688994827829,0.0672674435219996,0.856355920514581,0.0104526353142326,0.0697474360284256,0.43344367334862,0.0343414645022923,0.127112452673322,0.00454888400368057,0.0645938043462783,0.0719642168944171,0.0205567367472687,0.00543147291271282,0.00645589028828067,0.00407996707515448,0.0168094838384131,0.00407996707515448,0.10232134426748,0.13276848348873,0.626165782074011,0.0838811819084171,0.117790907998257,0.163839447445812,0.271317295866564,0.282321217662704,0.0467529612935936,0.0890324487110214,0.0528680716964271,0.00827862905112156,0.347364126697632,0.144190642300856,0.014661865093209,0.4749052974546,0.034366002325725,0.114766438533409,0.0182955290652585,0.0168094838384131,0.0458301838355025,0.0281881093361584,0.117778310517897,0.00033083665495648,0.0182955290652585,0.018820896512589,0.885398449427781,0.0136027177129889,0.0237393202984525,0.028873429718069,0.0536593860418449,0.0036929116085014,0.977482571549716,0.0218767093611828,0.0954495555153234,0.228757676424797,0.700583579130216,0.457942077623825,0.132715346044724,0.187462599728554,0.0797121272029794,0.0147363309021572,0.0316361073828435,0.123894859606519,0.084929341663334,0.0946349501008994,0.0205929766871262,0.0311561231972035,0.321457306653641,0.00506405161994418,0.175124664174564,0.177183261384205,0.0896257407819922,0.206018846923082,0.751453608297794,0.0794764611025657,2.04937063295103,0.0979520601416119,0.201852724440237,0.0336641489431937,0.048975141042381,0.0449747937540372,0.0428465666071815,0.124964619845553,0.376503541901391,0.0672674435219996,0.0379269966281466,0.144525564225123,0.133187348953427,0.0375060843610187,0.220582915864296,0.143400818508631,0.441302076553921,0.0158634862926648,0.0316361073828435,0.506094009080211,0.159941977547238,0.0971362738478571,0.0691723142136732,0.750600773534233,0.0157949646038045,0.0827362053912822,0.00117552329551341,1.95531157172771,0.0375975226228227,0.187133153861838,25.4790230225055,0.304821017657104,0.457942077623825,1.50643177907499,3.42327364549672,2.2424411716599,1.36694249792289,0.015338392640947,0.193794080389875,0.0324334366906665,0.0402056440408849,3.80787511657816,0.357458104081827,0.0158634862926648,0.0979520601416119,0.220582915864296,0.0157949646038045,0.0352016040845777,4.18024650469246,0.0944351713915657,0.836780833514187,0.399640159717293,0.0402056440408849,0.266622874173771,0.0430624262349775,0.0118003362744535,0.375322060857722,0.0195276971424756,0.0166923597787139,0.238855027868265,0.791861018325946,0.0106031621346388,0.18416689123457,0.31681450305845,0.0158634862926648,0.0779553694527797,0.127112452673322,2.2845790130404,0.0294599736810421,0.211103404764852,0.0353007315049884,0.0515391942593044,0.021169488761108,0.159941977547238,2.41883446633665,0.00845139272583984,0.0273364578475598,0.311670221349399,0.076383150017142,0.0580026618595194,0.185414279342433,0.175789107303533,0.0139625771057593,0.00407996707515448,0.015338392640947,0.106051643000397,0.811119783314346,0.100574872396968,0.473133873353317,0.339639092055912,1.51212378797432,0.201628194149819,0.00606966185826972,0.584039998231175,0.00434189191494265,0.0788591939294461,0.19446031938516,0.328432089316972,11.6249832353055,0.165355818532667,2.34184720337722,0.028873429718069,1.12885723374175,0.0890324487110214,0.0402056440408849,0.626165782074011,0.442618938478636,0.0971362738478571,0.0400753596802403,5.67890990169227,0.0442108253457593,7.02059507735066,0.0604456738943895,0.139551597426331,0.314585288599412,0.10520214126983,0.0164067793647608,0.122930855374187,0.050779317312943,0.1359132967779,0.0784557389321442,0.0706672089775578,0.0195276971424756,0.0771667427470897,1.204758826085,0.0788591939294461,0.0375060843610187,0.0568806219311672,0.142011492662573,0.0742176463993055,0.0593635254365486,0.748480129858454,0.581764819781412,0.0767741931631283,1.99123182285677,1.39886477682163,0.0422126723104046,0.456498655829432,0.135375822880032,0.00358117086348743,0.144478046881196,0.194062562086564,0.11257503223522,0.0158634862926648,0.320219386917268,0.107509225564609,0.0356609883149435,0.158872345450678,0.0788591939294461,0.0285185404524636,0.0994520499286496,0.222749765123598,0.442124535537161,0.0615350227233221,1.22148580491772,0.182638250549365,0.353678509895338,0.233368135317465,0.451061140101351,0.0197048238177622,0.640638791204819,0.143543163200891,0.0604456738943895,0.833577528815967,0.0716689639488025,1.01118172968909,0.0580026618595194,0.0804205837138711,4.9787539834116,0.310163489465506,0.628842586006387,0.127741389958166,0.769961797936431,0.230300048234622,2.02405842990956,0.0889080927701457,0.0212220249015296,0.139551597426331,0.595671592281534,0.472649120930349,0.0195276971424756,0.600292755078881,0.00411683845347271,0.0536593860418449,0.0145213193944826,0.386731369915065,1.56433978344595,0.00343377486905861,1.34418584718094,0.0106031621346388,0.015338392640947,0.0678013515876239,0.0296564643223139,0.133187348953427,0.151236535247784,0.549018115318202,0.399071280990619,0.0870977162421182,4.71487148451713,0.110693361465161,0.0245112648384883,0.0450129006423602,0.0135316751001583,0.236445377101403,0.00401548166356793,0.0182955290652585,0.0380208847945878,0.0686831843255189,0.0129608917901793,2.93826540140312,0.320219386917268,0.116546719657008,0.051052548967674,0.0205929766871262,0.185567600030243,0.105208262580619,0.00901402907892245,0.102224479852055,0.2475424605632,0.736883502169741,0.01349860600608,3.04886267579968,0.329437495127202,0.366412490636833,0.299607075307626,0.866674160511146,0.0979520601416119,0.107903145087254,0.168773105492031,0.124560809529937,1.84062783312445,1.00330919146326,0.100942193736438,0.484265529001832,0.0249891481114933,0.123825470520158,0.808593412748645,0.0372752573181502,0.801231010876257,0.0204109004922536,0.197519404037069,0.00431164095251631,0.207462448041871,0.143400818508631,0.0428138877325034,0.342347811273132,0.412590154340158,0.347364126697632,0.424043591073531,1.1989454106603,0.673153034015182,0.177183261384205,0.0702572251429715,0.164896948085131,0.0671705243414448,0.026346937631785,1.9067223617084,0.509845605508263,0.0650449372464769,0.120748776837183,0.0946349501008994,0.437367035459403,0.0463137376004234,0.065285360412143,0.00816402245135151,0.0217902084804504,0.111121490592266,0.00606966185826972,0.793267084368073,0.104257587805067,0.00459848528213147,0.0994520499286496,0.0141569450874245,0.201628194149819,0.100626175904096,0.0118003362744535,0.00606966185826972,0.00329111046875399,0.0205567367472687,0.0458301838355025,0.0672674435219996,0.160490047961719,0.096825409027854,0.150211855860997,0.502773087378392,2.46758896075418,1.04031903540814,0.185885540938701,0.0854771605495596,0.777942310072269,0.151186263156311,0.327855572596143,0.086473061367089,0.154699480913639,0.169328388263735,0.050797908136273,0.0402056440408849,10.3065496563699,0.0168094838384131,0.00121304115851171,0.0702572251429715,0.373904691191459,4.4965598747005,0.176642004796691,0.0122447960612203,0.0719642168944171,0.116546719657008,0.559325465633358,0.113811058420824,0.199841863296602,0.00459848528213147,13.4412848084886,0.00606966185826972,0.28204827172935,0.0104526353142326,0.0855239831172985,0.32470819810687,0.239755279819947,0.31696833193685,3.72786107280382,1.68519614798723,0.0277840733794765,0.0124463904386346,0.00606966185826972,0.0511957384793949,0.225523022083862,2.64749839090723,0.128001469671201,0.0826470000185111,0.14688205927712,1.0415037487867,0.292501644949302,0.0159185958779729,2.56529341968983,0.184572302455561,0.0307572237431331,0.287210597864711,0.495754133928346,0.0663326584292509,0.0979520601416119,0.772971386813549,0.418149131751107,0.0408362763240822,0.0979520601416119,0.00883507889890769,0.0363592437800154,0.651403317998296,0.0256352826026638,0.0488140139804341,0.155242598661533,0.0311398607999489,4.45048097418933,0.175313891249716,0.106774017517753,0.264065902578283,0.762405188973884,0.0721113079456428,0.267479743248493,0.381027913487168,0.0589486888360361,1.90069213517115,1.96912663245712,0.116504743482068,0.362177050086721,0.00720418064600042,0.0896599668422461,0.91306541018145,0.0200558260488684,0.115713176180213,1.1235325467146,0.144525564225123,0.160490047961719,0.152047798370628,0.026346937631785,0.117323832052037,0.00473174806234623,0.280323531233836,0.155242598661533,0.0158634862926648,0.240459825405001,0.0217902084804504,0.42759207853168,0.268311092509908,0.00136212981132986,0.132715346044724,0.320219386917268,0.509391874698079,0.0204109004922536,0.270322222221566,0.0653364468487771,0.00815315331791984,1.10309684748288,3.14681675804586,0.95184407633718,0.0281881093361584,0.130341961103517,0.0740608825039602,0.658225528205876,0.227280550920699,0.00298562862325312,1.44363652139823,2.31207517479951,1.8291024271631,0.0979520601416119,0.197797192545651,0.0210884701616462,0.459189144717777,0.177183261384205,0.516323738211185,0.161685592327123,0.0370373374431246,0.0373942370556637,0.10746395828625,0.00434189191494265,0.0394506369694468,0.0369651588949079,0.000345389407912768,0.032758357455683,0.867651621132305,0.180787544745946,0.0420257274415236,0.0672674435219996,0.0544495745810078,0.14107584819544,0.0999839347150616,0.154699480913639,0.670044754476321,0.0312263573907013,1.16650159776701,0.687139558805104,3.35835914242572,2.96647781177287,1.87505118517377,2.08345703368798,0.100133745881125,9.07271893073245,4.24274295986432,0.00407996707515448,0.34923111096301,0.00315561947602539,0.0614688361424385,0.762957391202675,0.0672674435219996,0.305680844304382,0.0168094838384131,0.356654192452932,0.00272520147235411,0.213342996857309,0.203664562357125,0.0614688361424385,0.0056492655075572,0.0158634862926648,0.11056174765258,0.216889984338719,0.0195276971424756,0.177037993178653,2.9127133028853,0.191116154022653,0.0104526353142326,0.0164067793647608,0.802975375101497,0.125376737316837,0.466141521873863,9.19402905610474,0.294230617208167,0.354343494370438,0.774329935269415,0.0464017913773774,0.06811620739093,0.0449747937540372,0.16197689883158,4.0272633012066,2.9329101978798,0.239791708013277,0.0366580940062409,0.354195460995922,0.232744669221038,0.311547848305952,0.00947518536003681,0.11330065730121,1.88698270371573,0.435738231721279,0.168068209561292,0.0796413838387279,0.325134097131939,0.622558710984682,0.0168518802392531,0.0168094838384131,0.220582915864296,0.0168094838384131,0.259136253830165,0.0581039990395819,0.300102086319107,0.0388902511214588,0.00947518536003681,0.800654509811821,0.0498587736724115,0.0735536342280046,0.450556003029832,0.171714971769673,2.43206438092877,0.473133873353317,0.11099722976912,0.0220278793288452,0.0253805703473298,5.12738403007637,0.0145213193944826,2.07456299626198,3.80558478946996,0.177183261384205,0.815935980778152,0.0589486888360361,0.0767741931631283,0.872104041613047,0.497063280896497,0.10232134426748,0.377445717304472,0.131001225702531,1.13572653117798,1.13572653117798,0.282365305567232,1.4609233604252,0.111267690768199,0.00385631494084208,0.0672674435219996,0.0476555687723794,0.0660586253442284,0.123551685773283,0.169606571700878,0.0695971501675616,0.00593828021807046,0.164640997621989,0.853894811568249,0.815081153223971,0.220582915864296,0.0166923597787139,0.0122447960612203,0.0339799027559752,0.0509089323214273,0.246121977966571,0.880059336139283,0.390468749379621,0.00121304115851171,0.00659116107410934,0.0103237965146232,0.438632804903988,0.00659116107410934,0.0166923597787139,0.29721610124596,1.20704501089265,0.0372752573181502,0.0949749328194669,5.51570601313714,0.0838811819084171,0.00606966185826972,0.135838918788729,0.0107632805068111,0.15225344922641,2.7607230996184,0.0979520601416119,0.318820057256344,0.0298172710384958,0.47419934597336,0.977482571549716,0.197279060457972,0.119453263291254,0.143388112419094,0.864183333314225,0.0450129006423602,0.589144595636144,0.362814867265735,0.0645938043462783,0.0804943338538985,0.0529027543094091,0.00121304115851171,0.0402056440408849,0.026916738884102,0.171714971769673,0.1359132967779,0.132715346044724,0.270492530181306,0.984739538095503,0.0211611751863212,0.154590008104649,0.147555826816082,0.231941279301644,0.532907280770234,0.853303135249357,0.607781693296782,0.00569817735165309,1.16968648651883,8.65546642642186,0.8399859749391,0.0626706940572397,0.0104526353142326,0.232526994078927,0.931822759707084,0.0293641058665195,0.0481498192638804,0.0645938043462783,0.0573567651190744,0.107407435725463,0.118585026119806,0.0440351249654628,0.278210289803958,0.129638694146595,0.180787544745946,0.0400753596802403,0.224996968048284,0.0834034906019596,0.473133873353317,0.126962208743481,0.682656671026117,27.6527536813374,0.111586988706968,0.0158634862926648,0.0479261967394173,0.552216831869672,8.86383561159093,3.43145610722599,0.0281881093361584,0.357458104081827,6.32798090131161,0.0168094838384131,0.221359895688013,0.501460561874647,0.412518179658821,0.00516427843486195,0.0614688361424385,0.0379269966281466,0.410765223052957,0.00431164095251631,0.0197048238177622,0.668798361776908,0.0283021768645347,0.0671524936478646,0.0611957404177207,0.267479743248493,0.05455075009054,0.213342996857309,0.062377621797737,0.0368401375339735,1.52134285767056,0.0442108253457593,0.147555826816082,0.147497384945501,1.42226210843601,0.20254293633253,0.0372477760158091,0.0248072680145259,0.146308514790656,0.187133153861838,0.116287187971872,0.133101625794071,0.17073973588619,0.100771933470826,0.0492310720780484,0.114152776790755,0.039951814594055,0.11257503223522,0.198807000493054,0.101955433341905,0.764744878158135,0.0458301838355025,0.018213394299291,0.0359937230721944,0.0232712094423213,0.0104526353142326,0.390468749379621,0.110085635989634,0.0166923597787139,0.133367645199529,0.06151247459663,0.0645938043462783,0.027281126693858,24.7932169818438,0.133495203649,0.144969953327665,0.197767579465517,0.536579815349066,0.14107584819544,0.0479261967394173,1.83400932399523,0.0328741096585519,2.31523491868349,0.218608536188943,0.00606966185826972,0.00790968930391987,0.0446081133619268,0.0570570307457038,0.47419934597336,0.0695971501675616,0.00606966185826972,0.0653269443253174,0.0168094838384131,0.0647065170226978,0.0252635968956367,0.0580026618595194,0.132715346044724,0.109496667515653,0.294209144505384,0.0428138877325034,0.114066567488366,0.00329111046875399,0.116504743482068,0.175359729526217,0.377162630879585,0.193794080389875,0.084468165308589,0.0750295576116174,0.00161138485580163,0.0568806219311672,0.2069623033854,0.00569817735165309,0.0253805703473298,0.00407996707515448,0.0197576796784808,0.0107632805068111,0.0760852880259302,0.292765205599443,0.018213394299291,0.297828514755134,21.9085145847806,0.674995379843054,0.230993003364834,2.58920889473892,0.77137868202989,0.019707338020414,0.0978993601540305,1.13359579347678,0.320219386917268,0.136806143731623,0.0237393202984525,0.0218015032544538,0.791632215579513,0.0825818392824401,0.25569487652148,0.125907081203253,0.0568806219311672,0.0715473711912393,0.596949718959995,0.0215769217855509,0.0056492655075572,0.165394713354411,0.0776891094684477,0.0614364470365818,0.742797376161623,0.0370373374431246,0.0672674435219996,5.25214319179113,0.130073992263818,0.0702572251429715,0.169089560165579,0.203902181662322,0.0600873322836296,0.0750295576116174,0.00593828021807046,0.716089564961085,0.0645938043462783,0.0511957384793949,0.0158634862926648,0.226258928432434,0.00116380427287773,0.132742374352844,0.0158634862926648,0.130073992263818,0.872191860349633,0.0139374711613066,0.011249800747522,0.0256352826026638,0.243216778813986,0.0702572251429715,0.00407996707515448,0.216704140792974,0.0492310720780484,0.00407996707515448,1.05496587971326,0.073496173982879,0.424507584859063,0.0946349501008994,2.31232073124486,0.293276041857867,0.0375060843610187,1.33144247141756,0.0564530643128249,0.403171500401637,0.0692641240983059,0.85346882073596,1.23892702474705,0.0854771605495596,0.0159185958779729,0.0363592437800154,0.475376475932767,0.445279553102564,0.0450129006423602,0.0623256070064101,0.0451549731464553,0.125051568964389,0.00759819506386321,0.210602160552019,0.0166923597787139,0.151236535247784,0.0979520601416119,0.0328741096585519,0.313542894887032,0.362814867265735,0.6125478457417,0.00593828021807046,0.0672674435219996,0.364269810653563,0.171580840180864,1.600310787697,2.06971596885056,0.0104526353142326,3.23788071428665,0.170065911221715,0.160222861825975,0.00516427843486195,0.49761963332858,0.236808015071122,0.191758364845341,0.286999379376735,0.00407996707515448,0.0168094838384131,0.0333750806784521,0.010798078778355,0.0529027543094091,2.52017939767646,0.138821785004941,0.0898788814722309,0.0951798824116844,0.655409432124841,0.396902225637102,0.0691375828781023,0.00718858733121906,0.0663963811889072,0.410171541217819,0.330982440116378,0.0769574550728935,0.208322218241635,0.0538566549074897,0.0281881093361584,0.123639312193052,0.774329935269415,0.0176392161306243,0.0234933792595118,0.0435566654250721,0.00434189191494265,0.140912714373964,0.0104526353142326,0.609251354013497,0.216704140792974,0.366513571223223,0.0281881093361584,0.455798784652487,0.0103237965146232,0.0221445022163917,0.0854771605495596,0.177830892049282,4.85893781293891,0.0818233581757009,0.0614688361424385,0.187341164750132,0.3618099840324,2.17123374454828,0.438664596210051,0.0197048238177622,0.0578808951494907,0.147497384945501,1.76502774808168,2.14776359951949,0.146727455715937,0.188201581504034,11.1366368695726,1.28770879220816,7.17465306436012,0.29590174707171,0.0788591939294461,0.287210597864711,2.79339711470453,0.421183283024042,5.38709718198797,0.00883507889890769,0.181872908667897,2.63801249393823,10.3194322558024,0.00419084931358526,1.38034688015477,0.306724912706086,0.0777800747953343,1.16553235552538,1.42753894759009,0.178454855544629,0.177037993178653,0.26385861396632,0.107217505294622,0.00298562862325312,0.323981847619161,1.38893670358892,0.00935623776165214,35.5214252073372,0.436055652833192,11.6994031512953,0.0309123152511477,0.00526552289067343,1.41454585401049,0.297409547950422,0.144969953327665,0.43344367334862,3.34222383255597,0.230331082855626,0.00541847726975198,0.351365526453055,0.995166008445084,0.371358254486124,0.621218883663659,0.0166923597787139,0.03537453040137,0.268564980493208,0.00411683845347271,0.0553998456362821,2.08345703368798,0.00419084931358526,0.00947518536003681,0.0865962984195445,1.60772464263292,0.0689510588577876,0.0672674435219996,0.1117341894595,0.954396692555429,0.401588013109418,0.0854771605495596,0.0316906421371129,0.143543163200891,0.00407996707515448,0.0529027543094091,1.67155592483846,0.117547265407849,0.721734284275401,0.236808015071122,0.0444282422240755,0.148340745378453,0.323981847619161,0.514180995584335,0.0672674435219996,0.639358191145854,1.27407274917396,0.0738240472895363,0.176647343233519,0.0247487854387873,0.00883507889890769,0.0563675539190095,0.198040870403191,1.54882747439373,0.269681893107705,0.1117341894595,0.0468552681873348,0.0898788814722309,0.793122984323734,0.0614688361424385,0.704224719143292,0.0188941321505588,0.0056492655075572,0.0050838323988797,0.1359132967779,0.0249964838592474,2.51632902396987,11.7127344965216,0.0897302419562838,0.156442988488283,0.840914278716581,0.856355920514581,0.0622997709104631,0.00606966185826972,0.497063280896497,0.00139918589257872,0.127112452673322,0.00315561947602539,0.0755580722852818,0.176647343233519,0.198665166604565,0.286932727875795,0.0106031621346388,0.00298562862325312,0.95184407633718,0.0139802896287417,0.145140499901302,3.67838445299179,0.186509463249524,0.0979520601416119,0.0702572251429715,0.0333750806784521,0.0416258592695452,0.15631330397492,0.0855239831172985,0.214799001636142,6.42590241281108,0.11257503223522,0.00385631494084208,0.238874958571622,0.0141000991332968,0.0716689639488025,0.0380218048667962,0.419618305475357,0.407977547678173,0.313542894887032,56.2815830525894,0.0614688361424385,0.0536593860418449,0.026346937631785,0.287732335612117,0.257974418335539,0.00754325657883153,0.329286985835253,15.8857340760672,1.00022216874956,0.115092020456585,0.181872908667897,0.126962208743481,0.0104526353142326,0.244286836927974,0.594373947087004,0.110085635989634,0.00973201219475736,0.0962088048460175,3.05288422140308,0.0874710816595967,0.371069390674875,0.126962208743481,0.145167079757024,0.393552810544579,3.17218998284575,0.0248072680145259,0.450556003029832,0.0281881093361584,0.214011976307974,0.0195276971424756,0.56484233893272,0.0215521295131734,0.0221445022163917,0.158834134323555,0.362814867265735,0.192660491908561,0.0154821369438163,0.789487688579368,0.00983063083816985,0.0925248104359321,0.38180321101221,10.3306600315819,1.48253912003761,0.0380208847945878,0.0671524936478646,2.62301126207904,30.1979841900734,1.40275042202578,0.161685592327123,0.157154293888498,0.0834034906019596,0.00459848528213147,0.856105571133433,0.0248072680145259,1.21063464722964,0.0364684556180969,0.00136212981132986,0.0762960487173458,0.0907743006388371,0.34923111096301,0.129714149929555,0.00569817735165309,0.0168518802392531,0.132680099583533,0.00523541645349658,0.0106031621346388,0.509287949512919,0.119453263291254,0.067654278117622,0.644000803262263,0.0118623681966753,0.176642004796691,0.0293641058665195,0.110693361465161,0.0394506369694468,2.77104605492002,0.327855572596143,0.0916409699409624,0.0218015032544538,0.132715346044724,0.00454888400368057,0.110085635989634,0.024829219808277,0.612125007022305,0.0394506369694468,0.022275509239016,0.0692641240983059,0.116546719657008,1.94753011065569,0.00459848528213147,0.154590008104649,0.0623256070064101,0.0450129006423602,0.282365305567232,0.0467529612935936,0.0767741931631283,0.0400753596802403,0.0898788814722309,0.912220806832437,0.0855239831172985,3.56623657629027,0.0150014926423921,0.57660759070772,3.59104805482037,0.00459848528213147,0.677262995863995,0.0294599736810421,0.0408362763240822,0.0468622799937739,0.0538566549074897,0.039951814594055,0.0316361073828435,0.0036929116085014,0.0112475600748757,0.161364628742817,0.0779553694527797,0.381027913487168,0.0788591939294461,0.0168094838384131,0.0104526353142326,0.00815315331791984,0.0846507772101082,0.0382919160356111,0.0166923597787139,0.202878729124266,1.2612860675305,2.51835325877788,0.06811620739093,0.00405427139052615,0.0252575824864749,1.50090365543082,3.32233242737225,73.0882837834004,0.127492827213836,0.01349860600608,0.00569817735165309,0.0380218048667962,2.24102292545732,0.0451549731464553,0.0538566549074897,0.169982046311221,0.187341164750132,0.332905181108188,0.547033021235577,0.0890324487110214,0.432978330630874,0.0450129006423602,0.0614688361424385,2.9085809991153,0.493261463737139,0.0536593860418449,0.00473174806234623,0.0375060843610187,1.27407274917396,0.0941211497349001,0.0795496161702012,0.0104526353142326,1.03747889450713,0.0234933792595118,0.239755279819947,0.349434387278569,0.176647343233519,0.306740880560016,0.051281384783662,0.0546949760379639,8.51377339368512,0.677262995863995,0.62885710337542,6.27476395909038,0.881562402130479,0.277512516244089,0.0645938043462783,1.37529342555745,0.0702572251429715,0.149916664463383,1.00788201091662,0.135838918788729,0.205452151162825,0.257974418335539,0.0248045111492735,0.38969903595669,2.41883446633665,0.223863520223173,1.2469093400222,0.0116453971485285,0.357458104081827,1.08622932044637,0.357458104081827,3.623814095536,0.0702572251429715,28.6922933464611,0.242449105026232,0.0277435643331768,0.00883507889890769,1.27706127335596,1.20733813533795,0.0464017913773774,0.122944688288505,0.125051568964389,0.0451549731464553,0.103033186151681,0.963107436336445,0.441302076553921,0.518153628195256,0.0221622331632297,0.332116174962393,0.172718602450644,0.0234933792595118,1.58940898350912,5.18747643938621,0.16581544056764,0.122217162201907,0.371358254486124,0.681366448651368,0.226258928432434,0.0375060843610187,0.674189376329781,0.108421809761933,0.167725377066011,0.0458301838355025,0.774329935269415,0.0160902603051206,0.11257503223522,0.181872908667897,0.0570135994457705,0.0243483376776383,0.0437169091307368,0.435319409721699,1.19233645285579,0.0979520601416119,0.0653269443253174,6.72974009531579,1.26416075610886,0.100626175904096,0.276449007674386,0.10393050620414,0.0145213193944826,0.4749052974546,2.35526620489374,0.0402056440408849,1.53508494984416,0.0776538523214883,1.5676447448723,0.759237674965495,0.329025534407141,0.113612820733828,0.0538566549074897,0.157274872151295,0.175124664174564,2.49624913013258,0.00298562862325312,0.0880904578378599,0.143753060707225,0.154590008104649,0.0479261967394173,0.0610643916579056,0.0364519868070057,0.0249964838592474,0.0217902084804504,0.99094937289997,9.58796017852587,1.26901476924371,1.94924895303249,0.558434412804878,0.0056492655075572,0.0484235414086104,0.0228074254169459,0.0458301838355025,0.0702572251429715,0.0205929766871262,0.00883507889890769,0.049139366916818,0.015338392640947,0.0200995742484223,0.154874628505491,0.0740110016637447,0.47419934597336,0.0356103344810315,0.0104526353142326,0.0929411513749569,0.32470819810687,2.55932921991562,0.111536947553299,0.140879977786088,0.0218536705005053,0.0638985257481875,0.233107723866566,0.0818233581757009,0.238165550613691,6.9356390722939,0.0702572251429715,0.287210597864711,0.0494943416052741,0.178289051516615,0.687139558805104,0.0880904578378599,0.0684394952173052,0.0284753346801675,0.0740608825039602,0.0283021768645347,0.43283187345558,0.133187348953427,0.0364519868070057,0.0479261967394173,0.167493733810359,0.123551685773283,0.0400753596802403,0.105208262580619,0.824321111567357,0.152047798370628,0.0218015032544538,0.246662721564788,0.0664206568967991,0.15225344922641,0.00116380427287773,0.11257503223522,0.0366119015957425,0.117613604446854,0.205887746445083,0.0212573040441202,0.0986985088851143,0.225523022083862,0.178294305315247,0.0166923597787139,0.00569817735165309,0.0218015032544538,0.123551685773283,0.0182955290652585,0.128489677275006,0.0166923597787139,0.226050787439536,0.010755650529379,2.16737773186825,0.0894006012358975,0.0343640697906325,0.0915009219984114,4.12976163708193,0.0890324487110214,0.569228794705177,0.351365526453055,0.266628846630413,0.115713176180213,0.311670221349399,1.21924314078731,0.549163491021169,0.122888408401505,0.0104526353142326,0.108462620284522,3.0515541272889,0.574311767879177,0.619699920957624,0.138224812062143,0.218315004292681,0.00606966185826972,1.11025433434739,0.0106031621346388,0.359253881461244,1.79258375591057,0.674951283811639,0.551712660434867,1.99123182285677,0.881562402130479,0.132715346044724,0.108491538733203,0.0684394952173052,0.0306933962709174,0.134442108338223,0.386256955027076,1.02168977484812,0.00139918589257872,0.0623256070064101,0.181872908667897,0.213647439158468,0.00967030279187589,1.77797313978614,0.808646275704249,0.114066567488366,0.0375060843610187,0.05455075009054,0.0946349501008994,0.133101625794071,0.00606966185826972,0.00459848528213147,0.276137607518688,0.00407996707515448,0.0727957118453141,0.114820046300988,0.0461414541080635,0.0634079629219563,0.0663370287615567,2.02551975145362,0.0204109004922536,0.13276848348873,0.0962088048460175,0.0755580722852818,0.362177050086721,0.534986947383463,0.492507260379483,0.0890324487110214,0.301935355358765,0.00593828021807046,0.00117552329551341,0.0370373374431246,0.190302838809674,0.0946349501008994,0.0402056440408849,0.0788591939294461,0.493261463737139,2.25317354740266,3.49736215527021,0.00459848528213147,0.236281298062111,0.195906981519117,0.062377621797737,0.228683342823997,0.00901402907892245,0.0994520499286496,0.356654192452932,0.0780274377942296,0.0702572251429715,8.59206150162136,0.0672674435219996,0.114152776790755,0.0168094838384131,0.266628846630413,8.30623651838889,0.00298562862325312,0.202441690777921,10.5808174922068,2.18738455117968,0.0402056440408849,0.0618751050272665,1.56621924518524,0.127741389958166,0.125376737316837,0.199841863296602,0.0323758120927884,1.87505118517377,0.499260599776372,0.43933034515539,0.185885540938701,0.25569487652148,0.0614688361424385,5.74860160678749,0.0195276971424756,0.00116380427287773,0.0402056440408849,0.0206588798271408,0.529319732003221,0.0990147880697231,0.0647065170226978,0.138224812062143,0.0771667172825306,0.0408362763240822,0.0166923597787139,0.0157949646038045,0.0833775144840563,0.460357155308212,0.0119486213331832,0.803444198274217,0.12178671210635,0.0837339461499816,0.161882422645581,0.0233293789963836,0.124084143327283,0.00121304115851171,0.0684394952173052,0.00973201219475736,0.01349860600608,0.00947518536003681,0.0570257778453027,0.284052060200538,0.692997948692464,0.114152776790755,3.85834129981127,0.0536593860418449,0.114066567488366,0.155394148119172,0.912220806832437,0.0247487854387873,0.0220942953367839,0.0400753596802403,0.0825818392824401,0.0678720674317296,0.220208947754637,0.0615350227233221,2.64749839090723,0.0818233581757009,0.239509517939684,0.0898788814722309,0.0498587736724115,0.266628846630413,0.0139374711613066,0.0672674435219996,0.29721610124596,2.80295393810061,0.95184407633718,0.0227483494152608,4.58695626690652,0.0796105337802797,0.0971362738478571,0.0499380213358411,0.0971362738478571,0.0572979678024011,0.0733973846470418,0.00606966185826972,0.00459848528213147,0.522842987159693,0.067938277501381,0.078948126661234,1.72759879041324,0.181872908667897,0.108462620284522,0.0540898568145164,0.132715346044724,12.9095937203426,1.6500516131931,0.601703072225002,0.0804205837138711,0.179234316904833,0.273785007619018,0.0597792089934429,0.0458301838355025,0.114066567488366,0.105208262580619,0.683841925708274,0.0168094838384131,0.109910230318687,0.123639312193052,0.0890324487110214,0.2401726704794,0.111584481498461,0.501460561874647,0.0158634862926648,0.00593828021807046,0.00593828021807046,0.479840176417155,0.0278937932733824,6.38578462714156,0.122240182048584,0.00116380427287773,0.420321241912227,0.155242598661533,0.000630346840926905,0.147624330348209,0.0965000651465204,0.086473061367089,0.12790680404788,0.130341961103517,0.00196710152911757,0.0649545376936062,0.148340745378453,0.110368479541996,1.42753894759009,0.0157949646038045,1.83393094820765,0.413979749182254,0.0589486888360361,0.0678720674317296,0.0442108253457593,0.131121817278175,0.0365387457853382,0.0168751028322431,0.0442108253457593,0.366093420435433,0.00308884675769402,0.852266673566946,0.0714273236348366,0.132715346044724,0.00329111046875399,0.789461903580603,0.0312093817019672,0.681366448651368,0.0104526353142326,0.0164067793647608,0.0124299458379158,0.0166923597787139,0.670044754476321,0.0840103977456048,0.0762960487173458,0.325928942566203,0.0784818378065301,0.179829445327717,0.127492827213836,0.026346937631785,0.00947518536003681,0.677262995863995,0.114066567488366,1.85274720208705,0.127112452673322,0.0623256070064101,0.106120595258921,0.0449747937540372,0.301921895040769,0.0838811819084171,0.00315561947602539,0.0189773911010558,0.120165856383946,0.00972421846918379,0.24885947283598,0.0275149914177536,0.0166923597787139,0.4614888708907,0.222255573559327,0.4749052974546,0.0254058618940309,0.00606966185826972,0.827322983180659,1.5113569279239,0.759237674965495,1.12760799793578,0.0990147880697231,0.0672674435219996,0.596949718959995,0.115713176180213,0.00139918589257872,0.114066567488366,0.0104526353142326,0.160222861825975,0.116359392214721,0.0511957384793949,0.0197704517689435,0.534450682074606,0.0511205834228772,2.24669672978285,0.0509089323214273,0.0249964838592474,0.0160902603051206,0.0170812991270618,0.0211611751863212,0.126835008955256,1.81956858667671,0.0818233581757009,0.696458063029389,1.66883035840179,0.318279354387568,0.00606966185826972,1.16361236276525,0.123058776142951,0.242734811952473,1.82894429986613,0.0145213193944826,0.0446081133619268,0.00659116107410934,7.64673164169309,0.0614688361424385,0.200783076682715,0.127082372867785,0.0295083978353268,0.0451549731464553,0.062377621797737,0.510889114260664,0.0370373374431246,0.733220807624962,1.41454585401049,0.772466906198559,0.0161927835441409,0.0461348153468877,0.00121304115851171,0.123894859606519,0.0158634862926648,0.144969953327665,0.398201553352697,0.0106031621346388,0.838939681505618,0.0166923597787139,0.0563537107044377,0.242692670085453,0.0865962984195445,0.569228794705177,0.811119783314346,0.412590154340158,0.983306348200182,0.0450129006423602,0.0990147880697231,0.00883507889890769,0.71333933208293,3.8297034345307,0.0614688361424385,0.339639092055912,0.56316430549341,0.0237393202984525,0.06151247459663,0.00431164095251631,1.06843273894035,0.0104526353142326,0.00431164095251631,0.238855027868265,0.527932892062772,0.0104526353142326,0.0702572251429715,0.0211611751863212,0.00459848528213147,0.407118019355027,0.06811620739093,0.242692670085453,0.0536593860418449,0.0838811819084171,0.0293641058665195,2.35649627345399,0.0248045111492735,0.018213394299291,0.0549054579017661,1.07473807794826,0.00298562862325312,0.335263588908335,0.399640159717293,0.0056492655075572,0.983306348200182,0.47403262163327,0.307585845727944,0.0147363309021572,0.00117552329551341,0.0018835297482147,0.260577083666056,0.351365526453055,0.0865962984195445,0.0211611751863212,0.0118623681966753,0.34923111096301,0.0108307155591846,0.124560809529937,0.041642515680023,0.00920032085875758,0.13991393500292,0.0195272682659921,0.0545262690039174,0.139551597426331,0.0834034906019596,0.267268122237729,0.0767994132389366,0.103262806319046,0.0353007315049884,0.0853543878004371,0.00754325657883153,0.398264454570971,0.015338392640947,0.0207523199348193,0.114066567488366,0.0979520601416119,0.90621722768677,0.0672674435219996,0.0672674435219996,0.410765223052957,0.00593828021807046,0.0600873322836296,0.0744324950640675,0.00136212981132986,0.218273979582299,3.52730622459711,0.269749005971493,0.174822974370871,0.0122447960612203,0.167227212152352,0.252295270665418,0.0623256070064101,0.218315004292681,0.0104526353142326,0.00460997542369454,0.28179445315511,0.0248045111492735,4.28269097748201,0.15780710056914,0.0509113179148502,0.0671524936478646,0.0104526353142326,0.0689510588577876,0.318279354387568,2.80295393810061,0.915265249077549,0.952782358557534,0.0648209704704071,0.0516439205950703,0.132715346044724,0.0402056440408849,0.652080678478522,3.69092413270053,0.384451721329113,0.266628846630413,0.0197048238177622,0.0158634862926648,0.0833471713831507,0.106774017517753,0.296295551042119,0.0615350227233221,0.0672674435219996,4.88592007480629,0.127082372867785,0.0247487854387873,0.176647343233519,0.34255287393985,11.4116480165989,0.127741389958166,0.863957793479752,2.88348278215581,0.00031765305898337,0.0036929116085014,0.0706672089775578,0.398264454570971,0.050779317312943,1.16018308085989,0.0580026618595194,0.0234933792595118,0.0248072680145259,0.0118003362744535,0.239907307577072,3.91256171229082,0.0450129006423602,0.216704140792974,0.0880904578378599,0.0536593860418449,0.396645837929196,0.00454757381358763,0.103221773004438,0.999202836828873,0.313490101280747,1.70337394248222,0.0672674435219996,0.039951814594055,0.0298681146805114,0.0867046605295849,0.43344367334862,0.0168094838384131,1.6750653847722,0.425559465026273,0.0862675424748999,2.14262610757867,0.412590154340158,0.534986947383463,0.00906073420687269,0.304232836009593,0.157601579104025,1.46701900688956,0.599116152359974,0.239589405634418,0.0509113179148502,0.286932727875795,0.302759077691639,0.0118003362744535,0.139772387784669,0.176647343233519,0.203508139916298,0.184572302455561,0.106282034046892,0.0104526353142326,0.00883507889890769,0.0104526353142326,2.95201102151312,3.24807556221579,0.162279905666064,0.0166923597787139,0.0189773911010558,0.0865962984195445,0.1026859616258,0.160490047961719,0.00121304115851171,0.14688205927712,0.0672674435219996,0.318820057256344,0.117997882705024,0.205347012399778,0.00431164095251631,0.407662897684194,0.00407996707515448,1.8425675979387,3.05288422140308,0.0833813436043561,0.161882422645581,0.277823633722508,0.00593828021807046,0.0056492655075572,0.0838811819084171,0.00569817735165309,0.0353007315049884,2.10196275758761,5.93037984211076,0.0733973846470418,0.179708478603025,0.00315561947602539,0.0481498192638804,0.285741913890512,0.0861908744219117,0.619946507828552,0.363348817458538,0.0538566549074897,0.0252575824864749,0.0217902084804504,0.0873851019999457,0.332457453097218,0.0854771605495596,0.018213394299291,0.344992489960381,0.0954495555153234,0.0400753596802403,0.172391925833378,0.0652882672422695,0.0158634862926648,0.294462991629555,0.169369509795821,0.1637483621332,0.273785007619018,0.0164067793647608,14.12459755485,0.291924562374098,0.669228429146024,0.166955620277339,0.0509089323214273,0.00401548166356793,1.43993703494127,12.2682216630714,0.0293641058665195,0.0217902084804504,0.0135603957562288,0.00973201219475736,0.0722995605337732,0.0818233581757009,0.017899566106878,0.0955158988491402,0.028873429718069,0.0946349501008994,0.0695971501675616,0.0234933792595118,0.00298562862325312,0.161996617373057,0.0204109004922536,0.115597204501953,0.0248045111492735,0.00883507889890769,0.112506122298816,0.4749052974546,0.0118003362744535,0.0234933792595118,0.341889577893,0.0142690548867,0.0622997709104631,0.0614688361424385,0.0653641229937024,0.906464415110111,0.0164067793647608,0.0194044368633364,0.084929341663334,0.0711145100325839,0.0206588798271408,1.72954978567044,0.0672674435219996,0.0205929766871262,0.10827065906627,0.328432089316972,2.1174003114224,0.0859849785928735,0.0798668339486119,0.747061605620297,0.00031765305898337,0.199841863296602,0.0149236544925937,0.175789107303533,0.115713176180213,0.0135603957562288,1.00022216874956,1.84235365134298,0.159955258527847,0.0372751825812681,0.0804943338538985,0.0217902084804504,0.170720957578179,0.14872925706803,0.335263588908335,0.549036014309833,0.11257503223522,0.0315022628655003,0.723409143501444,0.0158634862926648,0.026346937631785,0.0684394952173052,0.165394713354411,0.026346937631785,0.534986947383463,1.43993703494127,0.120265187631754,0.0442675565992241,0.00606966185826972,0.0604456738943895,0.122217162201907,0.0281881093361584,0.118158133474985,0.0182955290652585,0.180958083293105,0.200444144814504,0.0481498192638804,0.031855754521557,0.0416258592695452,0.220582915864296,0.774329935269415,0.0234933792595118,0.254385584751704,15.1375867596922,0.0538825670683027,0.172718602450644,0.116546719657008,0.026346937631785,0.100951116740171,0.039774259322378,0.0195276971424756,0.230374177552424,0.0672674435219996,0.495797110231909,0.132252086222209,0.0984476320602317,0.15511231908647,0.440614822866909,0.0147363309021572,1.77685276051159,0.823738843653089,0.0658825337796571,0.0190172573069676,0.0838811819084171,2.79735631068676,6.41692831814497,0.0925248104359321,0.31696833193685,0.00407996707515448,0.028873429718069,0.104257587805067,0.176604495064169,0.0951798824116844,0.0182955290652585,0.0494464413743751,0.673883655461331,14.3152538927215,2.58752712613323,0.045031647498376,0.0702572251429715,0.119453263291254,0.132715346044724,0.491822904754046,0.0615350227233221,0.0168094838384131,0.388643386366342,0.00947518536003681,0.00298562862325312,0.0672674435219996,0.0134742627195686,0.146982674275998,0.00407996707515448,0.0804943338538985,0.0168094838384131,0.0375975226228227,0.142631983206131,0.0188941321505588,16.9791461024352,0.216889984338719,0.00385631494084208,0.320522853032132,0.034366002325725,0.287210597864711,0.00196710152911757,0.302759077691639,0.408404278262663,0.0614688361424385,0.186842172927274,0.00434189191494265,0.00659116107410934,0.296895070260511,0.0614688361424385,0.176647343233519,0.122456569618017,0.163821784062743,0.328432089316972,0.0104526353142326,0.203690243410467,0.00883507889890769,0.177830892049282,0.332011330904214,0.388643386366342,0.0369651588949079,0.0880904578378599,0.144969953327665,0.0200925978977868,0.504484930372734,0.0623256070064101,0.0158634862926648,0.070841533977155,0.753497149129608,0.00459848528213147,0.00196710152911757,0.180898268771905,0.00139918589257872,1.01132996467137,0.14107584819544,0.0248045111492735,1.48404736472065,0.435319409721699,0.0653269443253174,0.0104526353142326,0.0256352826026638,0.460357155308212,0.158897545279111,0.395759724424602,0.0561139370689358,0.132420405705277,0.127741389958166,0.0316361073828435,0.141966427290487,0.0732364782185092,0.187341164750132,0.0158634862926648,0.151186263156311,0.0205929766871262,0.097825723998313,0.00606966185826972,0.95491021292783,0.151236535247784,0.0973847797736891,0.0442675565992241,0.13393311960292,0.0356099939614014,1.86480236110726,0.102446216999783,0.0511957384793949,0.0137578450631309,0.164096866263574,0.235885177423294,0.0854771605495596,5.09247708559928,0.0855239831172985,0.606675266200296,0.0652882672422695,0.0385214867161939,0.0358044923638088,0.0023534462217772,0.109462044918209,0.0579637945817318,0.0363519060066446,0.603290031236948,0.154874628505491,0.11660273484972,0.00329111046875399,0.376152599299968,0.0627324484858831,0.593388325884775,0.16197689883158,0.0197048238177622,0.0589486888360361,0.556633631943226,0.338265663430834,0.0182955290652585,0.0819983600707709,0.00385631494084208,0.127112452673322,0.00935623776165214,1.66609894835959,0.0204109004922536,0.0604456738943895,0.242734811952473,0.147497384945501,0.660633482616011,0.0295083978353268,0.112375649207978,0.158872345450678,0.221383115827005,0.908072601399471,1.29531021700938,0.00394511854196675,0.0164067793647608,0.00329111046875399,0.165394713354411,0.159898986010081,0.159955258527847,0.114446778696734,0.0897302419562838,0.00593828021807046,0.377944688328723,0.0168094838384131,0.766067324558473,0.325928942566203,0.254547025768231,0.215895834297239,1.08262837411284,0.06151247459663,0.0339590335548997,0.861903347868282,2.60870450477857,0.00407996707515448,0.0114905144772177,0.0739806170536602,0.0106031621346388,0.267254374669945,0.055086724641985,0.0796105337802797,0.0368490065677169,0.00593828021807046,0.0485392254700822,0.0104526353142326,0.113938020039937,0.0693807584889081,0.171714971769673,0.831553019550386,0.313538031623998,0.697216633115848,0.519917706386989,0.0468791300281877,7.97701580393569,0.0307572237431331,0.114066567488366,0.296291478485074,0.114066567488366,0.0270303960634119,0.0854771605495596,10.2094842505378,0.224470313622975,0.0364774306440582,0.0204109004922536,0.123418114107032,0.0281881093361584,0.125051568964389,0.0880904578378599,0.0833775144840563,0.689959176761919,0.0281881093361584,0.00366698754491492,0.021169488761108,0.144969953327665,0.0780274377942296,0.00516427843486195,0.192660491908561,1.74877377617655,4.45048097418933,0.272741791895483,6.26586826834013,6.61507221371342,0.207262326188889,0.227820185459639,0.00973201219475736,0.0402056440408849,0.0672674435219996,0.0257029326308624,0.460025208666674,0.663425625872115,0.54317107327208,2.35526620489374,0.0234933792595118,0.0108307155591846,0.117679888038899,0.00431164095251631,0.220879106936892,0.0672674435219996,0.0373833566867464,0.506444801044179,0.0715473711912393,2.56903457301212,2.21269566567625,0.504484930372734,0.0166923597787139,0.0599202751946987,0.169691702769797,0.505136440277799,0.0189188080507413,0.529434064200115,0.0253805703473298,0.124084143327283,0.00561411833693983,5.60389231953702,0.0356391500831134,0.351365526453055,0.11257503223522,0.802428535184632,0.199841863296602,0.368015360218303,0.0370373374431246,0.0206588798271408,0.00407996707515448,0.203690243410467,0.0458301838355025,20.5392666893437,0.207214877406833,0.00880087425033379,1.18158195544378,0.179829445327717,0.534986947383463,1.72712346255123,0.181872908667897,0.03537453040137,0.0375060843610187,0.24353366187228,0.593115429137124,0.0500443931820051,0.0670227649342383,0.00407996707515448,0.0136027177129889,0.0544495745810078,0.00606966185826972,0.122944688288505,0.0400753596802403,0.0377855186980461,0.0166923597787139,0.335352361410472,0.0211611751863212,0.135838918788729,0.0104526353142326,1.10217269897057,0.364850270824192,0.00947518536003681,0.0281881093361584,0.7184932362531,0.00606966185826972,0.0168094838384131,0.460357155308212,0.0515699503778721,0.0614688361424385,0.173455098927895,0.0166923597787139,0.0158634862926648,4.44661700509788,0.104079114187546,0.896960793527102,1.23250300359772,4.37343671619527,1.90953796787754,0.269681893107705,0.0784701086209545,0.723409143501444,7.19349239771284,0.534223497007966,0.257974418335539,0.0777800747953343,0.302724821164582,0.117547265407849,0.00141021433832185,0.420321241912227,0.017899566106878,0.280578545139398,0.0978783847614536,1.55131518705359,0.267268122237729,0.010755650529379,0.0166923597787139,0.0762534009412713,0.0848488080712362,0.041642515680023,3.07343488232969,0.357458104081827,0.509287949512919,0.489166081321539,0.29721610124596,2.26990749389777,0.328171148286734,0.0889080927701457,0.0449183172606626,0.331913239441346,0.00358117086348743,0.0538566549074897,0.163875678553274,1.70290451370788,0.0056492655075572,0.0295998638073874,0.0350432059123405,0.250437607260711,0.167227212152352,0.184572302455561,0.222749765123598,0.0375060843610187,0.0615350227233221,0.0854980610680273,0.819579684037794,0.0901956666439285,0.0364774306440582,0.147555826816082,0.178816622135615,0.0416258592695452,0.409061425552064,0.00947518536003681,0.00419614325645256,0.363051347649499,0.0070660955044122,0.160490047961719,0.569228794705177,0.109910230318687,0.29721610124596,0.215813500466117,0.0221622331632297,0.277512516244089,0.0826797667647909,0.105208262580619,0.0690082726024044,0.480621462365362,0.357458104081827,0.1117341894595,0.0248045111492735,0.132559921801293,0.828048045597703,0.0166923597787139,0.0721113079456428,0.0166923597787139,0.0818863834804944,0.0771332687539687,0.175124664174564,1.70283436129619,0.759721440138285,0.028873429718069,1.92099498997252,0.00272520147235411,0.194214674956053,0.0166923597787139,0.0030054913774704,0.0697195790921209,0.359253881461244,0.0104526353142326,0.207462448041871,0.00462157892865075,0.203381535491855,2.41883446633665,0.310163489465506,0.160490047961719,0.123058776142951,0.114066567488366,0.270872524317462,0.0702572251429715,0.100942193736438,0.0130541836704991,1.81510269470582,0.00946692262568576,0.0106031621346388,0.102224479852055,0.0166923597787139,1.92099498997252,0.0166923597787139,0.312176782074709,1.33299773664646,0.838939681505618,1.25663616730034,0.646869662348049,13.9243718113797,0.100626175904096,0.0200925978977868,0.125376737316837,0.16197689883158,0.0941211497349001,0.0446081133619268,0.220245119617206,0.0563537107044377,0.0206588798271408,0.100626175904096,12.3954032822907,0.0428138877325034,0.0567407770865506,0.208322218241635,0.13276848348873,0.36627878345694,0.0450129006423602,0.0349217380812796,0.0275149914177536,4.97709383083028,0.0145213193944826,0.00659116107410934,0.0896599668422461,0.328432089316972,0.122944688288505,2.55752601514943,0.0865962984195445,0.0283021768645347,1.05158296189547,2.78452721892123,0.127112452673322,0.324044829076066,0.0444272643730453,0.0464017913773774,0.00459848528213147,0.314401139539687,0.0896599668422461,0.187017552926824,0.556860216755851,2.07877242517985,0.567107760103152,0.00136212981132986,6.26586826834013,0.0854771605495596,0.061449438940265,0.00845139272583984,0.0965000651465204,0.0979520601416119,0.0319768447348491,0.781211470313931,0.0816403774495707,0.872104041613047,0.00329111046875399,0.039951814594055,0.177183261384205,0.0168094838384131,0.0023534462217772,0.00407996707515448,0.120748776837183,0.0880904578378599,0.00947518536003681,0.639358191145854,0.0972301811406793,0.708207966970726,0.270492530181306,0.115713176180213,0.00659116107410934,0.00298562862325312,6.79417623332282,0.295361631588783,0.223111197998437,1.44363652139823,0.263555246954439,1.1155284869259,8.07231716765735,0.182638250549365,0.393295904189277,0.0131517532401528,0.59406639971811,0.59406639971811,0.0826797667647909,0.0757730526987372,0.0197704517689435,0.00358117086348743,0.00968881239431921,0.0221622331632297,6.67868950051557,0.00385631494084208,0.1359132967779,0.0197048238177622,0.0275149914177536,1.15237642212938,0.091311383826482,0.154590008104649,0.127112452673322,0.0323115001686479,0.0962088048460175,0.0614688361424385,0.0648209704704071,0.00947518536003681,0.102446216999783,0.0281881093361584,0.733799319238283,0.428736611015552,0.695905405252045,0.0780274377942296,0.0979520601416119,0.0672674435219996,0.00659116107410934,0.116359392214721,0.216904587190577,0.0436931412956842,0.0147363309021572,0.57738345911058,0.0479261967394173,0.0375060843610187,0.193794080389875,0.19407527342166,5.41413153633167,0.0189773911010558,0.124496247530311,0.00606966185826972,0.563044886499033,0.0428138877325034,0.856105571133433,0.107080952056579,0.419462067012117,0.0626706940572397,0.0467529612935936,1.66585006896796,0.00385631494084208,0.0978062116951104,0.0375060843610187,0.0573281145128735,0.0511957384793949,0.00593828021807046,0.0672674435219996,0.0614688361424385,0.0858586070512467,7.78726307360474,0.133743058944992,1.99123182285677,0.00593828021807046,0.0234933792595118,0.0382919160356111,0.234386139209632,0.00459848528213147,0.0941027396022316,0.772466906198559,0.0402122274036898,0.0589486888360361,0.0206588798271408,0.5355160017489,11.5881746504514,1.00330919146326,0.00817736899861541,0.275053816248871,0.0102545440838995,0.0463351197509933,0.135375822880032,2.59573294099667,0.146764443869745,0.00782002579161583,0.00329111046875399,0.0158634862926648,0.558025382523306,0.362177050086721,0.879885022353824,0.0221622331632297,0.0294599736810421,0.0960897372373451,0.138504957656793,0.37842496506115,0.0234933792595118,0.0375060843610187,0.200636011298592,0.00659116107410934,0.0316361073828435,0.181190396534799,0.0509089323214273,0.198397576263175,0.692312854722668,0.127112452673322,0.0536593860418449,0.0494943416052741,1.75801595243402,0.0364774306440582,0.0614688361424385,0.127492827213836,1.47131513677247,1.50763367048553,0.03537453040137,0.010565234498871,0.399071280990619,0.0645938043462783,0.13465706616156,0.619699920957624,0.0294108857145262,0.95184407633718,0.0209226495669109,0.0326795945322925,0.353596715042238,1.18158195544378,0.0195276971424756,0.0106031621346388,0.166611779311744,0.569300183142978,2.99273953794751,0.135161957826904,0.220582915864296,0.410171541217819,0.0135603957562288,0.0248072680145259,0.18272273423756,0.0652882672422695,0.14872925706803,0.194214674956053,0.0690070682116648,0.48664559628603,0.143543163200891,0.0564530643128249,0.010798078778355,0.114066567488366,0.0615350227233221,0.0890324487110214,0.191960923483952,0.00606966185826972,0.229636920602312,0.0372752573181502,4.41156490577509,0.631452838652344,0.0023534462217772,0.382518492292416,0.0371827179078236,0.00516427843486195,0.194214674956053,0.091311383826482,0.0259934121928891,0.0518396544368133,0.015338392640947,0.0580026618595194,0.710585557984506,2.21269566567625,0.323981847619161,0.050779317312943,0.193863255710462,0.0994520499286496,0.032758357455683,0.187133153861838,0.110608502004934,0.147849009907857,0.0799541881680441,0.0672674435219996,0.045553521417846,0.0702572251429715,0.662119227052322,0.0642829960411013,0.0234479477314513,0.250751986494635,0.302481776185465,3.14498196064316,9.20539383799202,0.0118003362744535,0.0485392254700822,0.17073973588619,0.0664206568967991,0.11257503223522,3.35291085457109,0.32470819810687,0.284570532230096,0.0655408566197993,0.0164067793647608,0.363348817458538,0.692997948692464,0.0195276971424756,32.7145038898649,0.018213394299291,0.562661050419632,0.00459848528213147,0.00401548166356793,0.0615350227233221,0.178816622135615,0.0247487854387873,0.0232712094423213,0.0369651588949079,0.0289181750853702,0.748507442622767,0.131001225702531,0.139461583975561,0.0799541881680441,8.78848315508989,0.0854771605495596,0.0379269966281466,0.176642004796691,0.739914160767582,0.0141000991332968,0.36735933625874,0.0147363309021572,1.01395594661858,0.267497458680833,5.25459442237312,0.193863255710462,0.0965000651465204,0.823848864570809,0.193334566978367,0.220879106936892,0.051071341334625,0.709636137017505,3.13287247198881,0.0197704517689435,0.362805752016718,0.0197704517689435,0.0155702375890131,0.0812983042046511,0.00793295388501706,0.160583477377983,0.425373720054506,0.0450129006423602,0.0158634862926648,0.11257503223522,0.0854771605495596,0.672241385914354,0.584580940020264,0.571041744945995,0.558025382523306,0.0536593860418449,0.0202307610424076,0.0167186168192371,1.04922910789765,0.177037993178653,1.02012226293847,0.0623256070064101,0.0615350227233221,0.676970125739374,0.00593828021807046,0.0451549731464553,0.028873429718069,0.0580026618595194,0.65770987889104,0.00593828021807046,0.0017348219626119,4.95818313727607,0.112633692284027,0.00481868046262364,0.310499378838883,1.28452153699458,0.0442108253457593,0.0804943338538985,0.00230106010368019,0.0166923597787139,0.692997948692464,0.0614688361424385,0.269043412475993,0.371069390674875,0.132715346044724,0.283607175800663,2.0343983655893,0.253295754378762,0.276518464457158,1.16553235552538,0.0440351249654628,0.0965000651465204,0.772031147554754,0.294462991629555,0.202441690777921,0.018213394299291,0.127082372867785,0.00454757381358763,3.17218998284575,1.63601070256898,0.290384458127712,0.0536593860418449,0.160297931024884,0.0623256070064101,0.0686147573237967,1.63085802398507,0.0450154327996536,1.16933326792904,2.04937063295103,6.87287284592103,0.0889080927701457,4.63114648154876,0.00718858733121906,1.26377650870486,19.7661579882844,0.140139117126687,0.0458301838355025,0.52661118184281,9.61230501226108,0.0200925978977868,0.0402056440408849,0.0890324487110214,0.15780710056914,0.00459848528213147,0.0218767093611828,0.994904779254316,0.198665166604565,0.00329111046875399,0.0189188080507413,0.54052729630661,0.00315561947602539,0.034366002325725,0.424661214887011,0.0166923597787139,0.266622874173771,0.922136212712862,0.213647439158468,0.127112452673322,0.0615350227233221,0.154699480913639,0.0653987493258338,0.0979520601416119,0.0293641058665195,0.652435135349233,0.358011388376295,0.0217902084804504,0.133367645199529,0.120891445029521,0.139580531528287,0.321416229334828,0.125051568964389,0.226551875534636,1.2186756352481,0.0450129006423602,0.276518464457158,0.0804943338538985,2.78173320384583,0.0369651588949079,0.0375060843610187,0.181872908667897,0.0215769217855509,0.209190645794384,0.114766438533409,0.0444282422240755,0.0106031621346388,0.00606966185826972,0.0762534009412713,0.131019485648435,0.080040595786052,0.0431138364279397,1.87434685734114,2.02600177165827,0.00459848528213147,0.362896457419757,0.0893353318370075,0.00298562862325312,0.0142690548867,0.044900200029293,1.22724834017576,0.119453263291254,0.114066567488366,0.0784557389321442,0.086473061367089,0.157120641777891,0.0972884686457311,0.141829045667774,0.0854771605495596,0.258856019391655,0.026346937631785,0.0922029316571254,0.0168094838384131,0.377445717304472,0.881562402130479,15.207019127477,0.224996968048284,0.0432898659173311,1.00022216874956,0.00593828021807046,0.0521577164702833,0.179903226159086,0.102719278506068,0.291924562374098,0.0135603957562288,0.516576311902631,0.208322218241635,0.752038294712209,0.0417681070583219,0.0589486888360361,0.213615280460692,0.901587534234326,0.0159185958779729,0.0834034906019596,0.00473174806234623,0.0394506369694468,0.0979520601416119,0.0898788814722309,0.64583867411378,0.793267084368073,1.1155284869259,0.710929212503727,0.104770984859427,0.15261163394772,0.0275149914177536,0.0701296865066609,0.00815315331791984,0.0649545376936062,0.0217902084804504,0.297828514755134,0.139151858384974,0.104257587805067,0.016936726755383,0.107354964735127,0.623393631273757,0.697827833071276,0.271794447053332,0.187133153861838,0.0987762894302276,0.294462991629555,0.134007410175259,0.110608502004934,0.0760852880259302,1.04031903540814,3.14376154936558,1.74607300865188,0.0921245689971997,0.0142339139889482,0.0402056440408849,0.015338392640947,0.0767741931631283,0.0379269966281466,0.0854771605495596,0.562661050419632,29.3209198855437,0.228683342823997,0.00385631494084208,0.139551597426331,0.159955258527847,0.225893350019311,0.00947518536003681,0.00891067980873313,0.0686831843255189,0.00569817735165309,0.00900911140759986,0.00161138485580163,0.354955723552566,8.01750396936567,0.552216831869672,0.351365526453055,2.22165518113966,0.127112452673322,12.1563352506112,4.95005566778172,0.125392135575007,0.0197704517689435,0.106178081739245,0.0589486888360361,0.0419258260310568,0.783385026727992,0.028091858780483,0.600453152665349,0.0784701086209545,0.0256352826026638,0.0614688361424385,0.677447449227901,0.154699480913639,0.0280354687584875,0.392164636690708,0.0461414541080635,1.79977855698426,0.72396183243304,0.254385584751704,0.0536593860418449,0.118040280298015,0.0898788814722309,0.140441827618829,0.0496720635814372,3.09600772170997,0.534986947383463,0.0653269443253174,1.07756792744276,0.00385631494084208,0.0204109004922536,0.0421820176991698,0.376503541901391,0.746390556891458,0.0080119599543022,0.0965000651465204,0.563037699333491,0.0131517532401528,0.254599496072089,0.230331082855626,0.00141021433832185,0.534986947383463,0.0370373374431246,0.0563537107044377,0.117064180344618,2.31206943509226,7.46270443382008,0.0152817399384943,0.522239114467398,0.108462620284522,0.108187611798949,0.0250824135508519,0.968409336058904,3.37926472722503,0.127112452673322,0.0234933792595118,0.207329829782868,0.316993299438617,0.0544495745810078,0.339830011781286,0.0509089323214273,0.00569817735165309,0.169244350078357,0.021169488761108,0.122456569618017,0.0672674435219996,0.0596106501921475,0.632221359277129,4.86448582190475,0.176822923993521,0.0104526353142326,9.2401461510752,0.105275190435028,0.220582915864296,2.14623847011808,0.381068134290834,0.248021236835145,3.623814095536,0.731830118142868,0.0645938043462783,0.0056492655075572,0.0536593860418449,0.230374177552424,0.0589486888360361,0.133101625794071,0.0451736261219523,0.0242652955432981,0.0141000991332968,0.592912995949684,0.0200925978977868,0.0990217824913059,0.0336641489431937,0.475607525941823,0.239589405634418,0.00298562862325312,0.573964973214923,0.0711581260335338,1.4159585467273,0.147497384945501,0.0197704517689435,0.0237393202984525,0.0464017913773774,0.0149236544925937,0.0940929920643764,0.215895834297239,0.00947518536003681,0.11056174765258,5.03083620167645,0.114066567488366,0.00385631494084208,0.00883507889890769,0.609295899690258,0.00407996707515448,0.133187348953427,1.46701900688956,0.0785688473429434,0.113612820733828,0.71445289000866,0.0130013793455995,0.716357492454833,0.0723862417869145,0.0688140767070753,0.0672674435219996,0.107509225564609,0.493261463737139,1.50922229818231,0.10965467486682,2.30161730519302,0.0838811819084171,0.100739627812261,0.119837468423119,0.0197704517689435,0.0168518802392531,0.0400753596802403,2.10605389990405,2.57482448741981,0.0375060843610187,0.125069310896634,1.64391579580838,0.00198975967419686,0.713558289750622,0.954396692555429,0.0256352826026638,1.68757584383539,1.31036804576223,7.02059507735066,67.7493980298725,0.357458104081827,0.325881386097665,0.0432898659173311,0.0245112648384883,0.00139918589257872,0.290384458127712,0.0221622331632297,0.619634504239108,0.62885710337542,0.305141178101242,0.0141000991332968,0.0464017913773774,0.028873429718069,0.124332655687698,0.37011790758766,0.0340984319390846,0.125148618462469,0.0402056440408849,0.527664667520168,0.0828678802633485,0.0435811396153694,0.0647065170226978,0.0744213950415091,1.20963538018143,0.564182570279465,1.86588482324686,0.079435255590737,2.20613549125272,0.00754063054493942,0.550341686100982,0.018166276459895,0.714261552513639,6.53148759356459,0.00454888400368057,0.0645938043462783,0.117547265407849,0.215813500466117,0.0526219675418347,0.0784818378065301,0.0563537107044377,0.105208262580619,0.08584195103585,0.0197704517689435,0.178816622135615,0.010755650529379,0.193794080389875,0.0179759732781862,0.0134742627195686,0.0217902084804504,0.021976713239791,0.0333026105921905,0.316993299438617,0.0614688361424385,0.154699480913639,0.277512516244089,0.0234933792595118,0.0402056440408849,0.0853543878004371,0.0339799027559752,0.405093305006978,0.00947518536003681,0.114066567488366,0.0622997709104631,0.0217902084804504,0.0604456738943895,0.000345389407912768,0.00947518536003681,0.0197048238177622,0.0080119599543022,0.0118003362744535,0.892849993410572,0.0436931412956842,0.00947518536003681,0.00141412862086026,0.0391938617044803,0.00136212981132986,0.193794080389875,0.0422529947164534,1.35995780342297,0.546112377572161,0.0521577164702833,0.139580531528287,0.358941946191684,0.588436608366706,0.0295727025259229,0.0479261967394173,34.8743793120609,17.8570087393054,0.079435255590737,0.294209144505384,0.0380208847945878,0.220582915864296,0.0168094838384131,0.232651006717618,0.167227212152352,0.0195276971424756,0.00385631494084208,0.0375060843610187,0.0818233581757009,0.272666347113849,0.193794080389875,0.0610643916579056,0.792710506616295,0.0440351249654628,0.0195276971424756,0.677262995863995,0.00106898696268826,0.0356391500831134,0.303235832750071,0.124496247530311,0.418672752987534,0.00329111046875399,0.233107723866566,14.5730710257323,0.00116380427287773,0.289120623600522,0.0614688361424385,0.356654192452932,0.304692206097218,0.0218015032544538,0.0589486888360361,5.00646154854412,0.476479463240617,0.512330110705102,0.0248072680145259,0.118230199542716,0.445635278711868,1.74730249053248,0.130073992263818,0.483680168167945,0.114152776790755,0.643829418560291,0.531958932652289,0.0681953556596605,0.0408362763240822,0.00447892063826242,0.0277073537847336,0.0880904578378599,0.406288495256855,0.00298562862325312,0.147588660948933,0.0739806170536602,0.110567818774538,0.010755650529379,0.00947518536003681,0.207329829782868,0.461922296554563,0.552216831869672,0.0158634862926648,0.15261163394772,0.159018166067558,1.56109605617001,0.0366642317113751,0.00298562862325312,0.116546719657008,0.493261463737139,0.00947518536003681,1.6500516131931,8.25013045006797,0.362814867265735,1.15237500841377,0.118158133474985,0.0653491919517554,0.0711581260335338,0.276449007674386,0.010755650529379,6.38719460971422,0.015338392640947,0.432405787609527,0.0530406981121325,0.129028584304363,1.72063278558047,0.140879977786088,0.512920144939503,0.491146081353113,0.496124363480676,0.0553250879628495,0.0614688361424385,0.366199086548791,0.140139117126687,0.0538566549074897,0.13004656096305,0.0400753596802403,0.114353957333331,0.0804943338538985,0.0145213193944826,0.0623843738112641,0.446382889181637,0.289840175023475,0.11147905894624,0.167908100667173,0.13276848348873,1.13003004351555,0.393928072034445,0.0309123152511477,0.0671524936478646,0.601703072225002,4.84165109865088,0.0372752573181502,0.0568055645398992,1.14494857581857,0.00946692262568576,0.663275368136992,0.0840103977456048,0.0618751050272665,0.0488140139804341,0.432978330630874,0.0874040765097886,0.194932071339973,3.09054653413688,26.0707791339303,0.0762960487173458,0.0231168783910513,0.205651842854809,0.0435566654250721,0.280323531233836,0.443072085072397,0.00329111046875399,0.0168094838384131,0.0125400876763405,0.00148341115078733,0.00121304115851171,0.141829045667774,0.00460997542369454,0.549163491021169,0.15261163394772,0.00606966185826972,0.0536593860418449,0.43933034515539,0.127112452673322,0.328171148286734,0.122378034288297,0.0234933792595118,0.151186263156311,0.443219048533731,0.118282737570121,0.114066567488366,0.00116380427287773,0.62885710337542,0.11257503223522,3.34036182990055,0.381727119703066,2.55787855534371,0.113612820733828,0.364850270824192,0.47419934597336,1.36795161441189,0.592912995949684,0.00460997542369454,0.159941977547238,0.512679211265931,0.00718858733121906,0.0293641058665195,0.00815315331791984,0.0395768064216012,2.44077516514408,0.114766438533409,0.0253285940718115,0.00459848528213147,1.63644302241281,2.78647044675219,1.33144247141756,2.61350933288483,0.0356391500831134,2.17709960955772,2.44343515670071,0.125051568964389,0.130751309049599,0.114066567488366,0.180958083293105,0.147555826816082,0.0890324487110214,0.0987422917741626,0.286999379376735,0.0671524936478646,0.224996968048284,0.215905176032228,0.112506122298816,3.51333542111365,1.41846318324499,0.558434412804878,1.2607153105325,0.266622874173771,0.0589486888360361,0.0865962984195445,0.0549054579017661,0.0357231972671091,0.0481498192638804,0.218315004292681,0.028873429718069,0.0293641058665195,0.277512516244089,0.0370373374431246,0.0850333259913629,0.186671303761123,0.00454888400368057,0.0946349501008994,0.287702887750983,0.0356844989582654,0.00782002579161583,0.0711581260335338,0.125051568964389,0.364269810653563,0.0275149914177536,0.018213394299291,0.218273979582299,0.0604456738943895,0.413046732256739,0.0916409699409624,0.0130013793455995,0.03537453040137,0.0197048238177622,0.600292755078881,1.57563671015629,5.65036006936757,0.342432287531116,0.239199267515393,0.0277840733794765,0.0147363309021572,0.333215359769113,0.0446081133619268,0.00606966185826972,0.0441861856363517,0.00411683845347271,0.689683378520733,5.12205037458532,0.158593500542026,0.20010080630113,0.103443184645079,0.114066567488366,0.409106491975956,0.375322060857722,1.07473807794826,0.0100659223145108,0.124496247530311,5.49084327430958,0.122944688288505,0.242734811952473,0.798645942842311,0.00329111046875399,0.151236535247784,0.138955065096102,0.0168094838384131,0.0234479477314513,0.0375060843610187,0.10232134426748,0.0365387457853382,0.228892054006225,0.15261163394772,0.00606966185826972,0.015338392640947,0.0971362738478571,0.11257503223522,0.0212220249015296,0.0256352826026638,0.102791677567528,0.134500410888056,0.0197704517689435,0.578459814274898,4.19389982507273,0.00759819506386321,0.793122984323734,0.13276848348873,0.01349860600608,0.626165782074011,0.0280354687584875,0.915265249077549,0.12409527185678,2.1181521132051,0.000281394652558477,0.284570532230096,0.0168094838384131,1.27531052337609,0.235680397959205,1.47832294829237,0.0028537161017384,0.0187370465834964,0.0979520601416119,1.47259077984334,0.0343640697906325,0.0308553045328201,0.230331082855626,0.0234933792595118,0.0941211497349001,0.0463351197509933,0.30637523218776,0.399071280990619,0.127776024196659,0.00459848528213147,0.113612820733828,0.10353914402543,0.0211611751863212,1.05147716944473,0.00329111046875399,0.0796105337802797,0.0278937932733824,0.258856019391655,3.33640983156088,0.191758364845341,0.655347208295686,1.31508566502527,0.0391938617044803,0.00298562862325312,0.00593828021807046,0.480621462365362,0.00411683845347271,7.23001704150166,0.0596106501921475,0.306740880560016,3.25736497607709,0.163821784062743,1.44363652139823,0.0197048238177622,0.158872345450678,0.129371360167829,0.0234933792595118,0.0257987873052996,0.574311767879177,0.044900200029293,0.0118003362744535,0.341149441010209,0.028495276092218,0.122456569618017,18.0683950698369,1.71014685308443,0.0380208847945878,0.00385631494084208,0.0206588798271408,0.131374407254324,0.0218767093611828,0.0370373374431246,0.141829045667774,2.75837998709723,0.0168094838384131,0.0933654710249695,0.0141000991332968,3.14032948089238,1.90968145752524,0.31696833193685,0.00407996707515448,0.187133153861838,0.0748577078109076,0.0645304689742235,0.390927568486095,0.0284753346801675,0.21804495800474,0.0563537107044377,0.000812948016292426,1.64659296072613,0.480621462365362,1.2406883724025,0.0197048238177622,0.00606966185826972,2.11992242161474,0.0760852880259302,0.0402056440408849,0.0750295576116174,0.0834034906019596,0.243590516752951,0.512213976065635,0.0628421673191005,0.00298562862325312,0.0702572251429715,0.0553263138474479,0.438774130635394,0.00718858733121906,0.389554862023999,0.33110634916855,0.225445845543885,0.0702572251429715,0.214293458369392,0.225799201816166,0.0649545376936062,0.0444282422240755,0.0509089323214273,0.0344255500471701,0.129028584304363,0.110693361465161,0.0653641229937024,11.3797880187201,0.00431164095251631,0.0812983042046511,0.0157531875731702,0.0570257778453027,0.124084143327283,0.156347279432297,0.0536593860418449,0.11056174765258,0.086992843136258,0.0965000651465204,0.1359132967779,0.346847478772388,0.0188542634863658,0.721734284275401,0.174422167810016,0.0248072680145259,2.88614865307913,0.36466617530845,0.0342001133295079,0.105208262580619,0.157854238902655,6.92923987674356,0.144969953327665,0.00593828021807046,0.0395768064216012,0.729798808830232,0.127112452673322,0.367380393955958,0.00459848528213147,0.0356103344810315,0.0652882672422695,0.0356103344810315,0.0954495555153234,1.71300938102735,0.0368401375339735,35.7613513479023,2.61628293141982,0.187133153861838,0.00577742582270165,0.00473174806234623,0.00298562862325312,3.60733657222729,0.267268122237729,0.327855572596143,0.0300422025754072,0.116546719657008,2.26727094406629,0.0652882672422695,0.260836285894463,0.047451881650676,0.00298562862325312,2.5072384406032,0.0990147880697231,0.548316455055722,0.0580026618595194,0.0695971501675616,0.00407996707515448,0.0796105337802797,1.88070524070508,0.0536593860418449,0.0197048238177622,0.631851823429748,13.3709840690697,0.0166923597787139,0.0248072680145259,1.50090365543082,0.311547848305952,0.0953686806252274,0.0467563303602578,0.0158634862926648,0.0614688361424385,0.0594047185404571,0.0702572251429715,0.10232134426748,0.270872524317462,0.0689510588577876,0.208322218241635,0.0212573040441202,0.116504743482068,0.0416070136836051,0.0614688361424385,0.606675266200296,0.03537453040137,2.64003365770061,0.0379033786577836,1.07388914041018,0.686872049649894,5.11403892472243,0.0408362763240822,0.081225155070545,0.026346937631785,0.57738345911058,0.000876057544547143,0.388918547630115,2.54741012130271,8.96169124608967,0.118040280298015,0.480621462365362,0.294209144505384,0.332905181108188,0.16197689883158,0.0275149914177536,0.0843911641814317,0.122944688288505,0.0444282422240755,0.335352361410472,10.0436246941716,0.549036014309833,0.127112452673322,0.0623616639636408,0.000988228391909021,0.0614688361424385,1.84235365134298,0.109462044918209,1.12586614249647,2.63647958858745,0.142513035515441,0.460357155308212,2.08656490842702,0.393226438443169,0.0652882672422695,0.0562382573975696,0.251049302928911,5.09014627366869,0.014661865093209,0.0104526353142326,0.00329111046875399,0.0536593860418449,0.0663963811889072,0.13276848348873,0.489866827637269,0.0444282422240755,0.391722540789193,0.628534319468648,0.0324334366906665,0.00031765305898337,1.34864408807354,0.0356103344810315,0.0460309008872369,0.493261463737139,0.0647295615861959,2.65967188753088,0.634920015229307,0.454719075434713,0.0159185958779729,1.06695125253939,0.0339799027559752,0.0036929116085014,0.0874710816595967,2.43186501213404,0.242449105026232,0.286351167534854,14.0690996901466,0.132715346044724,0.598378880693123,0.901587534234326,0.0344255500471701,0.225523022083862,0.163839447445812,0.689134005066903,0.0402056440408849,0.0293641058665195,0.375641850125064,0.0157949646038045,0.0136027177129889,0.0521577164702833,0.0468799402113693,0.0364519868070057,0.0368401375339735,0.0708418517605244,0.108314342098055,0.0168094838384131,0.00407996707515448,0.0563537107044377,0.127746232871427,0.0623843738112641,0.0278937932733824,0.0312093817019672,0.139580531528287,0.0896599668422461,0.126835008955256,0.418055814795551,0.0711581260335338,0.220083516426305,0.0221622331632297,0.0106031621346388,0.990489518150543,0.010798078778355,0.0536593860418449,0.00298562862325312,0.00815315331791984,0.0435566654250721,1.14622761985752,0.076383150017142,0.0880904578378599,4.33392914242096,0.0747212588098682,0.0896257407819922,0.0705746203268183,0.105208262580619,0.0716689639488025,0.10393050620414,0.0168094838384131,0.0164067793647608,0.00407996707515448,0.0796413838387279,0.362177050086721,0.248221427873007,0.124496247530311,0.231179028699573,0.0479314891784595,1.01515083339461,0.0702572251429715,0.0451736261219523,0.00659116107410934,0.146232686812539,0.766549632363785,0.0116453971485285,0.041642515680023,0.0293641058665195,0.152047798370628,0.0865962984195445,0.0166923597787139,0.118422861714971,0.00935623776165214,2.34166367387658,0.37011790758766,0.729798808830232,0.0413083051753416,0.104770984859427,0.00883507889890769,0.0402056440408849,0.0536593860418449,0.0293641058665195,0.11257503223522,0.14872925706803,1.17780531452575,0.373904691191459,0.057492475836355,0.0375060843610187,0.0166923597787139,0.17792457015105,1.10893383768568,0.48924763036347,0.017407444778782,0.00407996707515448,0.00315561947602539,0.0391298319760489,0.0149236544925937,0.0236237653649776,0.11147905894624,0.00606966185826972,0.0623256070064101,0.0147363309021572,0.0994520499286496,0.270065314754182,0.127112452673322,0.0449747937540372,0.0755580722852818,1.61333491089658,7.04035458479123,0.00385631494084208,0.0416258592695452,0.128770266148453,0.0195276971424756,0.00141021433832185,0.618041802098337,0.00419614325645256,0.251894667473625,0.270872524317462,0.0217902084804504,0.0195276971424756,2.00634714758166,0.00947518536003681,0.0518396544368133,0.0164067793647608,0.127112452673322,0.168620218911856,0.0104526353142326,0.119453263291254,0.220879106936892,0.0218767093611828,0.00329111046875399,0.0702572251429715,0.0248045111492735,0.0105157735116567,0.00459848528213147,0.819145477471472,0.0442108253457593,0.13373995139241,0.0375060843610187,0.0690082726024044,0.0843911641814317,0.159941977547238,1.5189137666983,0.0435811396153694,0.0854771605495596,0.106251583884537,2.38687628186109,1.30791419214591,0.00606966185826972,0.106774017517753,0.325117440250423,0.0107767401614758,0.0599202751946987,0.122944688288505,0.0135316751001583,0.134625400292063,0.0951798824116844,0.0375060843610187,0.0854771605495596,0.0166923597787139,0.118158133474985,0.10232134426748,0.0377855186980461,0.238855027868265,0.29721610124596,25.381019909786,0.127492827213836,8.33132352080373,0.287210597864711,0.25671482047013,0.0217902084804504,0.677262995863995,1.05496587971326,0.00883507889890769,0.168582470580305,0.0106031621346388,0.0104526353142326,0.487864267488757,0.124496247530311,0.146727455715937,0.00947518536003681,0.122944688288505,0.159898986010081,0.0104526353142326,0.220083516426305,0.046582877889561,0.0793391320060809,0.00659116107410934,0.0182955290652585,0.10312114630122,0.979969235588701,0.492174181697456,0.0159185958779729,0.0182955290652585,0.0672674435219996,5.59715132673827,0.243590516752951,1.73563291909374,0.0614688361424385,0.275547622392463,0.0965000651465204,0.0302847822582646,0.0796105337802797,0.100626175904096,0.207329829782868,0.00116380427287773,0.0511957384793949,0.0446081133619268,0.0197704517689435,0.0458301838355025,0.0168094838384131,0.0168094838384131,0.126144350068723,0.139551597426331,0.0275149914177536,0.271317295866564,0.0281881093361584,8.78153594192412,0.220083516426305,0.0221622331632297,0.0568806219311672,0.534986947383463,0.00121304115851171,0.0907443590065623,0.0965000651465204,0.0293641058665195,0.0242461588666874,0.180616849423213,0.0840969254626935,0.00454888400368057,0.104257587805067,0.0672674435219996,0.0922029316571254,0.0689510588577876,0.159898986010081,0.00407996707515448,0.290140878765139,3.75358344586581,6.11157968198377,0.173726946534251,0.131121045511493,3.53280442716001,0.144190642300856,0.208322218241635,0.16581544056764,0.0234933792595118,0.193082830101439,0.239589405634418,0.0504855775522382,0.550957089234625,0.0799541881680441,0.117547265407849,0.0343414645022923,0.011249800747522,0.0645938043462783,0.0776891094684477,0.0692840141297325,0.36466617530845,0.050779317312943,0.0166923597787139,0.171714971769673,1.78521779122648,0.00315561947602539,2.95387555831798,0.114066567488366,0.0353007315049884,0.0141000991332968,0.125051568964389,0.772971386813549,0.119637655192105,0.0741497824672452,0.0672674435219996,0.0614688361424385,0.00329111046875399,0.509287949512919,0.127112452673322,1.14533373239862,0.276536209097572,2.90440375381977,0.0197048238177622,0.0369607853007353,0.00883507889890769,1.05315992379101,0.0719642168944171,0.0903550141797481,0.0118732859916435,0.0144258920915731,0.76329275061048,0.0461414541080635,0.485818746342585,0.459189144717777,0.10520214126983,0.0281881093361584,0.0402056440408849,0.0124535686093941,0.968211406707937,0.449274994573416,0.144969953327665,0.0248072680145259,0.0179759732781862,0.139094474316629,0.00198975967419686,0.183750849493497,0.0294599736810421,0.497562729686413,1.66585006896796,0.0104526353142326,0.48664559628603,0.00659116107410934,0.240087043690842,0.111534732323082,0.0561749865281889,0.290140878765139,0.1117341894595,0.0893353318370075,0.14605728316933,0.0137578450631309,0.0380208847945878,0.0614688361424385,1.49783045679992,0.0107632805068111,2.07742810029321,0.179260381790001,0.229636920602312,0.028495276092218,0.0240426889317082,0.145505916471976,0.0281881093361584,0.0164067793647608,0.027281126693858,0.0451549731464553,0.269043412475993,0.0615350227233221,0.247380535212008,0.00298562862325312,0.00298562862325312,0.0295727025259229,0.139551597426331,0.00947518536003681,0.00905388074928039,0.017899566106878,0.114066567488366,9.70651552977048,0.0614688361424385,0.242692670085453,0.0343640697906325,1.01102372185409,0.725258242286904,1.32566649698957,1.3275176058935,0.115713176180213,0.0693807584889081,0.534986947383463,1.11144368164132,0.0377855186980461,0.0197048238177622,0.00459848528213147,0.377162630879585,0.160513639763373,0.162213499576052,0.107903145087254,1.72063278558047,0.44728811816046,0.00407996707515448,0.0674442387796625,0.00116380427287773,8.05983968004063,0.329025534407141,0.0277073537847336,0.182058971952635,0.0158634862926648,0.139551597426331,0.0141000991332968,0.0248072680145259,0.0855239831172985,0.00454888400368057,1.52412297030909,2.52038218430913,1.93662810126536,0.0166923597787139,0.211308039781596,0.676361667979867,0.615703275462965,0.941847002116691,0.120165856383946,0.13991393500292,0.045031647498376,0.0402056440408849,0.0281881093361584,0.00407996707515448,0.15225344922641,0.420179948907894,0.00659116107410934,0.00315561947602539,0.30464529903587,0.0695971501675616,0.0511957384793949,0.435338192435453,1.72063278558047,0.579439955128524,0.0672674435219996,0.829043109758129,0.0130013793455995,0.00190403025668718,0.262560904592156,0.112767511126604,0.0369651588949079,0.00272520147235411,0.0458301838355025,0.133101625794071,0.335263588908335,0.0739806170536602,0.0205929766871262,0.034661585609613,2.57009485908457,1.26326174883616,0.0736606829966013,0.147588660948933,0.0493749854546897,0.0889080927701457,0.0150014926423921,0.0411970508705327,0.00593828021807046,1.28543707714528,0.0195276971424756,0.132715346044724,0.619699920957624,0.266622874173771,0.210666145350451,0.252771231248908,0.125051568964389,0.0197704517689435,1.05496587971326,0.00407996707515448,0.133367645199529,0.00593828021807046,0.0702572251429715,0.472649120930349,0.314113536287398,2.1326473455997,0.0965000651465204,0.354955723552566,1.5609620976277,9.25157549294076,4.23368748584406,0.620995122081968,0.0536593860418449,0.0247487854387873,13.6017377017191,21.0775540943318,4.44789312162608,0.00883507889890769,0.0023534462217772,0.251049302928911,0.0368401375339735,0.0206385049319028,0.0289181750853702,0.0218536705005053,0.0990147880697231,0.289263993900311,0.00116380427287773,0.125051568964389,0.569300183142978,0.186509463249524,0.0650062642654958,0.357458104081827,0.218315004292681,0.0573281145128735,0.553746445241948,0.0818863834804944,0.0880904578378599,0.127112452673322,0.0212220249015296,0.116546719657008,0.0579469719086449,0.351365526453055,0.0880904578378599,0.154874628505491,0.016936726755383,0.0468973412283722,0.0780274377942296,0.0362734805403617,0.0159185958779729,0.0365387457853382,0.0843911641814317,0.0818233581757009,0.0965000651465204,0.0252325255677302,0.329025534407141,0.0375060843610187,0.0244685358898696,0.0162059917283721,0.0381459276249513,0.328047559396122,0.714487322930782,0.428515700451037,0.039774259322378,2.82295085960226,0.0589486888360361,0.177183261384205,0.0221622331632297,0.0118003362744535,0.667131023432279,0.21551608319756,0.041642515680023,0.413046732256739,0.00121304115851171,0.0672674435219996,0.811119783314346,0.492507260379483,0.116546719657008,0.00905388074928039,0.524956087889121,0.0130541836704991,0.0197048238177622,0.234815621699189,0.0481498192638804,0.0536593860418449,0.0217902084804504,0.0370373374431246,0.0653641229937024,0.332656622863167,0.220148126842577,0.179903226159086,0.20010080630113,0.0343414645022923,0.0614688361424385,0.276518464457158,0.0660586253442284,0.0245112648384883,0.534986947383463,0.0166923597787139,0.197279060457972,0.00593828021807046,0.449426240305483,0.00454888400368057,0.0451549731464553,0.472858395353483,0.144969953327665,0.0488140139804341,0.0195276971424756,1.68246267748174,0.44728811816046,0.0511957384793949,0.0221622331632297,0.258764129592,0.0788591939294461,0.0767994132389366,0.00121304115851171,2.82295085960226,0.0082119985779289,20.1039706559244,0.0804205837138711,0.000485568677475681,0.0762880291991772,0.217811910528249,0.764744878158135,0.135161957826904,0.00459848528213147,0.993418999893802,0.0195276971424756,0.276207910406349,1.41949612645821,0.483478322076573,0.329441263272636,0.0166923597787139,4.95818313727607,3.28424516693132,0.0275149914177536,1.00120204166983,0.0159185958779729,0.0500183361885817,0.0800417130893048,0.00901402907892245,0.0141000991332968,0.0672674435219996,0.126104296692911,0.399071280990619,0.132715346044724,0.239589405634418,0.00418774768751389,0.95184407633718,0.0521577164702833,0.0104526353142326,0.0139374711613066,1.92751424547221,0.06151247459663,0.338299794303057,0.0316361073828435,1.62945134548282,0.506586540081076,0.973583095517455,0.0204109004922536,0.0660054216157546,0.0545582764929851,3.43963295559426,1.01300126400627,0.0880904578378599,0.0479314891784595,0.641761703806222,1.23002970756969,0.263861312671935,0.00795127294477724,0.0118242839508101,0.159941977547238,0.0733973846470418,0.870348892879163,0.0023534462217772,0.139551597426331,0.240087043690842,0.0380208847945878,0.0529027543094091,0.00329111046875399,0.382930928186899,0.116546719657008,0.0660960450124907,0.0615350227233221,0.344312176574087,0.176647343233519,0.220879106936892,0.0196227163419745,1.8907020190211,0.0164067793647608,0.0890324487110214,0.774329935269415,0.00315561947602539,0.0994520499286496,3.58484246422342,0.0992192482586448,1.6307676966894,1.72713699121472,0.126519839533059,0.00431164095251631,0.162279905666064,0.295361631588783,4.40183707892892,0.692997948692464,0.628534319468648,0.00116380427287773,0.00705543581085461,0.655409432124841,0.0274985885485839,0.0872187672686948,0.00315561947602539,0.155394148119172,0.00454888400368057,0.235399829909775,0.246654502095659,0.0560823010583364,0.0840969254626935,1.85571823027536,0.312176782074709,0.016936726755383,0.00593828021807046,1.06059269484096,0.010565234498871,0.0293641058665195,0.0623843738112641,0.220083516426305,0.00116380427287773,0.287210597864711,0.0536593860418449,0.125376737316837,0.387670196806909,0.0593635254365486,1.99123182285677,0.127112452673322,0.25489053748679,0.204144785646028,0.0711581260335338,0.1359132967779,0.00315561947602539,2.70639498872232,0.677262995863995,0.0147363309021572,0.147624330348209,0.0479261967394173,0.00569817735165309,0.45715996835543,0.0551188580531276,0.031971786709729,0.220879106936892,0.220148126842577,0.526713195095954,0.0614688361424385,1.37571034541215,0.134113164488797,0.0218767093611828,0.0249891481114933,1.79134119106725,0.59406639971811,0.0365387457853382,0.13991393500292,0.0036929116085014,0.261133801354263,0.2558106850751,0.0903651086711145,0.0380208847945878,0.0070660955044122,0.00196710152911757,0.0672674435219996,0.381027913487168,0.0553263138474479,0.127112452673322,0.292738257657991,0.0579469719086449,0.0135316751001583,4.86876691490669,0.0234933792595118,0.0373833566867464,10.9654633482235,0.0435811396153694,0.239589405634418,0.0324598363689652,4.56580397095966,0.115597204501953,0.218714887124655,0.0352016040845777,0.154292790670578,0.0166923597787139,0.152890942598921,0.0450154327996536,1.46701900688956,0.0375060843610187,0.00606966185826972,0.1637483621332,0.201094287946858,1.27920780360741,0.485337682795176,1.26377650870486,0.018564102308703,0.0451549731464553,0.0221622331632297,2.43186501213404,1.83739078106241,0.148911154117599,0.822688502290722,0.31696833193685,0.0017348219626119,0.152047798370628,0.0293641058665195,0.681366448651368,0.144969953327665,0.00593828021807046,0.0236347992928947,0.0451549731464553,0.0563675539190095,2.82295085960226,0.132715346044724,1.91476480551791,0.00883507889890769,0.199725602217908,0.161501799635138,0.0865962984195445,0.497562729686413,4.64668137690425,0.00031765305898337,0.774329935269415,1.13359579347678,0.00569817735165309,0.0377062011016278,1.43704344592041,5.14089888595133,2.70639498872232,0.11257503223522,0.303083727796863,0.0189188080507413,0.294252249799152,1.08379621744191,2.67653903824566,2.77738477335047,0.0834034906019596,0.837851219781081,0.0270303960634119,0.845352377607299,0.103616792240542,0.619699920957624,0.824321111567357,0.379410084359023,0.811314589666861,0.00659116107410934,0.0278937932733824,0.099706439323769,0.709326288799336,0.924732063721248,0.0450129006423602,0.00117552329551341,0.349434387278569,0.534986947383463,0.477000980750578,0.0992192482586448,0.130751309049599,0.179903226159086,0.390467667477944,0.139551597426331,1.25554326206904,0.0511857792419379,0.0865962984195445,0.1359132967779,0.120748776837183,0.00473174806234623,0.026346937631785,2.04599603157991,0.0702572251429715,0.0234933792595118,5.833166645808,0.0234933792595118,0.696075744578418,0.0158634862926648,0.357458104081827,0.0470598702861263,0.10746395828625,1.51534304210223,0.0653364468487771,0.0589486888360361,0.177183261384205,0.309030472806492,2.25724812224896,0.0420257274415236,0.239589405634418,0.0589486888360361,0.131374407254324,0.492281931842033,0.028873429718069,0.0702572251429715,0.11147905894624,1.03662314903675,0.00947518536003681,0.0444282422240755,0.174422167810016,0.163839447445812,0.0206588798271408,0.0649391720650664,0.080040595786052,0.258253262867908,3.33996453269189,8.27345627401839,0.185418668023435,0.086473061367089,2.05261996110592,0.0702572251429715,0.520922167029769,0.133089224759337,0.0946349501008994,2.70639498872232,0.00407996707515448,0.0293641058665195,0.0166923597787139,0.00431164095251631,4.15356066647978,0.0104526353142326,0.182137587432888,0.0444282422240755,0.114766438533409,0.0730760340759955,0.0164067793647608,0.110572284773651,0.0812983042046511,0.620259236425479,0.00366698754491492,0.0767741931631283,0.0463351197509933,0.0356844989582654,0.0116773790911124,0.509391874698079,0.0645938043462783,3.03065085113373,0.0804943338538985,0.0247487854387873,0.628842586006387,0.217029851601869,0.0321153854128733,0.25569487652148,0.0444282422240755,0.640638791204819,0.0278937932733824,0.00116380427287773,0.132715346044724,0.0165094669358971,0.0281414757580683,0.0992192482586448,0.00606966185826972,0.466141521873863,0.180561623540819,0.114066567488366,15.8697878497523,0.175313891249716,0.0672674435219996,0.117148323093359,0.276207910406349,0.0464017913773774,0.0276319973730302,0.076890751303345,0.183685342799604,0.0672674435219996,0.670044754476321,0.0711581260335338,2.07868325425406,0.496124363480676,0.0200925978977868,0.139580531528287,0.226258928432434,0.0653269443253174,0.0277435643331768,1.2607153105325,0.246654502095659,0.0854771605495596,0.357458104081827,0.00272520147235411,0.0563537107044377,0.00883507889890769,0.141829045667774,0.0176786975345918,0.124496247530311,0.143753060707225,0.655409432124841,3.32583604430131,1.28192398142758,0.0159185958779729,0.0451549731464553,0.411301009545268,0.021976713239791,0.00839375177350195,0.00569817735165309,0.298647172606965,0.0252575824864749,0.125646978459109,0.0454863584200561,0.310482337696921,0.0197048238177622,0.00593828021807046,0.0136027177129889,0.185567600030243,0.143753060707225,0.00116380427287773,0.127112452673322,0.206621115785932,3.58484246422342,1.1249257922934,0.19407527342166,0.889929672691884,0.315882773888673,0.195473961730693,0.781086644742982,0.0538566549074897,0.198680521213169,0.130073992263818,0.00139918589257872,0.0528618138018548,0.267268122237729,1.79691671275973,0.0979520601416119,0.4749052974546,0.0604287866407293,0.225893350019311,0.0776999209165473,0.127112452673322,0.290384458127712,0.0865962984195445,0.0158634862926648,0.388643386366342,0.0589486888360361,0.0168094838384131,0.107869078610365,0.341889577893,0.0614688361424385,0.00407996707515448,0.0561139370689358,0.0946349501008994,0.506493728927312,0.320852092205766,0.0941211497349001,4.91092618232917,0.811007947826258,0.0672674435219996,1.75034207206869,0.0538566549074897,0.339072076955283,0.0804555364246732,0.222799243572054,0.432978330630874,0.127425489299559,0.171714971769673,0.425373720054506,0.999049795429896,0.410171541217819,1.91476480551791,0.169606571700878,0.762957391202675,0.4407270376245,4.14331845132528,0.00754325657883153,0.124579879373261,0.00385631494084208,0.00407996707515448,0.677262995863995,0.0212220249015296,1.99116706812396,0.507860687004746,0.261907276380634,0.268311092509908,5.78150311519852,0.0276319973730302,0.0375060843610187,0.420321241912227,0.0672674435219996,1.34804417990397,0.4749052974546,0.0413083051753416,0.677226289148063,0.0990147880697231,0.0159185958779729,0.0858246506532958,0.00176108913705591,0.0492310720780484,0.00883507889890769,2.46764344799579,0.00411683845347271,0.388643386366342,0.213342996857309,0.114066567488366,7.85836952988927,0.122944688288505,0.0979520601416119,0.0855239831172985,0.502773087378392,0.0568806219311672,2.52942317415856,0.357458104081827,0.488302390662843,1.26377650870486,0.716198104137063,0.0750295576116174,0.0898788814722309,0.282365305567232,0.24711189832656,0.322977377915035,0.28445023504853,0.337055644369106,4.7614867666801,0.0271936790271745,0.40942691722469,12.4889636183537,1.3482691773245,0.232651006717618,0.00946692262568576,0.0464017913773774,0.0323758120927884,14.1535080400722,0.0780274377942296,0.103262806319046,0.0444282422240755,0.489641467673164,0.0865962984195445,0.00121304115851171,0.650228047419585,0.276137607518688,0.549018115318202,0.0245237100835782,0.0104526353142326,0.118230199542716,0.524765189701827,0.0645938043462783,0.0171550175497202,0.0672674435219996,3.26200375102729,1.07473807794826,0.499260599776372,0.16197689883158,0.127112452673322,0.010755650529379,0.403847558861173,0.487864267488757,0.0579469719086449,0.0158634862926648,0.506444801044179,0.0168094838384131,0.0589486888360361,0.250751986494635,2.90440375381977,3.48298917192031,3.26116938735293,0.357458104081827,0.08264702096919,1.03747889450713,0.056436732778513,20.1066880699352,0.786351271158804,0.0865962984195445,1.18158195544378,0.390467667477944,11.33740992302,1.26874941034796,0.0941211497349001,3.02750359454795,0.0999374459332967,0.673153034015182,0.840914278716581,0.216205147099859,0.00759819506386321,0.00516427843486195,0.0141569450874245],"text":["log2FoldChange: 0.0022764412<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.9783827<br />gene: FBgn0000008","log2FoldChange: -0.2399189435<br />-log10(padj): 5.406049e-01<br />log10(baseMean): 3.6387441<br />gene: FBgn0000017","log2FoldChange: -0.1046739119<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 2.6218101<br />gene: FBgn0000018","log2FoldChange: 0.2108477918<br />-log10(padj): 2.525758e-02<br />log10(baseMean): 0.8066005<br />gene: FBgn0000024","log2FoldChange: -0.0917880714<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.9955124<br />gene: FBgn0000032","log2FoldChange: 0.4630237099<br />-log10(padj): 1.282792e-01<br />log10(baseMean): 1.1490539<br />gene: FBgn0000037","log2FoldChange: 0.3146273449<br />-log10(padj): 7.779423e-01<br />log10(baseMean): 4.9149118<br />gene: FBgn0000042","log2FoldChange: 0.6196801037<br />-log10(padj): 4.727725e+00<br />log10(baseMean): 4.4936276<br />gene: FBgn0000043","log2FoldChange: 0.4150249098<br />-log10(padj): 1.425243e-01<br />log10(baseMean): 1.4315848<br />gene: FBgn0000044","log2FoldChange: 0.1526724206<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 0.8127454<br />gene: FBgn0000045","log2FoldChange: 0.6042643296<br />-log10(padj): 2.437044e-01<br />log10(baseMean): 1.3745002<br />gene: FBgn0000046","log2FoldChange: 0.6822367168<br />-log10(padj): 1.524096e-01<br />log10(baseMean): 0.9672820<br />gene: FBgn0000047","log2FoldChange: -0.1354078502<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 3.3697501<br />gene: FBgn0000052","log2FoldChange: 0.2961466671<br />-log10(padj): 7.160896e-01<br />log10(baseMean): 3.3222792<br />gene: FBgn0000053","log2FoldChange: -0.1198062010<br />-log10(padj): 1.426320e-01<br />log10(baseMean): 2.8157666<br />gene: FBgn0000054","log2FoldChange: -0.0234143271<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.8341957<br />gene: FBgn0000057","log2FoldChange: 0.2057844963<br />-log10(padj): 3.608838e-01<br />log10(baseMean): 2.6072062<br />gene: FBgn0000063","log2FoldChange: 0.2996399969<br />-log10(padj): 1.380347e+00<br />log10(baseMean): 3.8985697<br />gene: FBgn0000064","log2FoldChange: 2.6797251700<br />-log10(padj): 4.515799e+01<br />log10(baseMean): 2.5343390<br />gene: FBgn0000071","log2FoldChange: -0.0558137619<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 2.3506309<br />gene: FBgn0000077","log2FoldChange: -1.0202991158<br />-log10(padj): 1.068433e+00<br />log10(baseMean): 1.4626368<br />gene: FBgn0000078","log2FoldChange: -1.0671340524<br />-log10(padj): 1.560562e+00<br />log10(baseMean): 2.2028092<br />gene: FBgn0000079","log2FoldChange: -0.0082016940<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 3.7358094<br />gene: FBgn0000083","log2FoldChange: -0.1485475371<br />-log10(padj): 1.143540e-01<br />log10(baseMean): 3.2294047<br />gene: FBgn0000084","log2FoldChange: 0.7537674156<br />-log10(padj): 3.320113e-01<br />log10(baseMean): 1.1008223<br />gene: FBgn0000092","log2FoldChange: 0.4530196399<br />-log10(padj): 1.425243e-01<br />log10(baseMean): 1.3240810<br />gene: FBgn0000094","log2FoldChange: 0.0071568333<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 3.4839862<br />gene: FBgn0000097","log2FoldChange: -0.1611240875<br />-log10(padj): 2.147990e-01<br />log10(baseMean): 4.4049500<br />gene: FBgn0000100","log2FoldChange: 0.2061149871<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.6996130<br />gene: FBgn0000108","log2FoldChange: -0.2397716491<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.1223052<br />gene: FBgn0000109","log2FoldChange: 0.0641408817<br />-log10(padj): 5.702578e-02<br />log10(baseMean): 2.7577583<br />gene: FBgn0000115","log2FoldChange: 1.2908845377<br />-log10(padj): 9.111233e+00<br />log10(baseMean): 2.8184906<br />gene: FBgn0000116","log2FoldChange: 0.1568306184<br />-log10(padj): 2.552433e-01<br />log10(baseMean): 4.0339332<br />gene: FBgn0000117","log2FoldChange: -0.1052624329<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 2.8085439<br />gene: FBgn0000119","log2FoldChange: 0.0820616573<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 0.9580928<br />gene: FBgn0000121","log2FoldChange: -0.4032167835<br />-log10(padj): 2.234896e+00<br />log10(baseMean): 2.8110289<br />gene: FBgn0000139","log2FoldChange: 0.0720127651<br />-log10(padj): 5.105255e-02<br />log10(baseMean): 2.9758871<br />gene: FBgn0000140","log2FoldChange: -0.0985671055<br />-log10(padj): 1.238255e-01<br />log10(baseMean): 3.2801011<br />gene: FBgn0000142","log2FoldChange: 0.4902494166<br />-log10(padj): 4.450481e+00<br />log10(baseMean): 3.1976411<br />gene: FBgn0000146","log2FoldChange: 0.4197778420<br />-log10(padj): 1.519504e+00<br />log10(baseMean): 2.6185134<br />gene: FBgn0000147","log2FoldChange: 0.2776169654<br />-log10(padj): 5.553243e-01<br />log10(baseMean): 3.9517242<br />gene: FBgn0000150","log2FoldChange: 0.1361608620<br />-log10(padj): 1.326801e-01<br />log10(baseMean): 2.7083709<br />gene: FBgn0000152","log2FoldChange: -0.1950188106<br />-log10(padj): 4.687913e-02<br />log10(baseMean): 1.3945099<br />gene: FBgn0000153","log2FoldChange: -0.1215316724<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 2.9523651<br />gene: FBgn0000163","log2FoldChange: -0.0159188027<br />-log10(padj): 2.152311e-03<br />log10(baseMean): 0.9055925<br />gene: FBgn0000166","log2FoldChange: -0.0293008601<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 3.8348623<br />gene: FBgn0000171","log2FoldChange: 0.1538117267<br />-log10(padj): 2.322870e-01<br />log10(baseMean): 3.1891419<br />gene: FBgn0000173","log2FoldChange: 0.3975751524<br />-log10(padj): 1.404418e-01<br />log10(baseMean): 1.4378684<br />gene: FBgn0000180","log2FoldChange: -0.1666503399<br />-log10(padj): 3.935021e-01<br />log10(baseMean): 4.1287777<br />gene: FBgn0000181","log2FoldChange: -0.0133228281<br />-log10(padj): 1.144841e-02<br />log10(baseMean): 3.4220363<br />gene: FBgn0000183","log2FoldChange: 0.0342052686<br />-log10(padj): 1.693673e-02<br />log10(baseMean): 3.1967832<br />gene: FBgn0000210","log2FoldChange: 0.0995483976<br />-log10(padj): 1.285425e-01<br />log10(baseMean): 3.5168585<br />gene: FBgn0000212","log2FoldChange: -0.2522723122<br />-log10(padj): 3.455020e-01<br />log10(baseMean): 2.3193550<br />gene: FBgn0000221","log2FoldChange: -0.1048446414<br />-log10(padj): 1.073550e-01<br />log10(baseMean): 2.8927421<br />gene: FBgn0000228","log2FoldChange: -0.1142404876<br />-log10(padj): 6.981422e-02<br />log10(baseMean): 2.9578337<br />gene: FBgn0000229","log2FoldChange: -0.0278354104<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.3947100<br />gene: FBgn0000239","log2FoldChange: 0.0515732599<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 0.9180261<br />gene: FBgn0000241","log2FoldChange: 0.1144497958<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 2.2059322<br />gene: FBgn0000242","log2FoldChange: -0.3976990903<br />-log10(padj): 1.478189e+00<br />log10(baseMean): 2.7077246<br />gene: FBgn0000244","log2FoldChange: 0.1683565666<br />-log10(padj): 6.037484e-02<br />log10(baseMean): 2.2465158<br />gene: FBgn0000246","log2FoldChange: -0.1627584556<br />-log10(padj): 2.535656e-01<br />log10(baseMean): 3.4253329<br />gene: FBgn0000247","log2FoldChange: -0.1014345268<br />-log10(padj): 1.307513e-01<br />log10(baseMean): 3.3524360<br />gene: FBgn0000250","log2FoldChange: 0.0798131712<br />-log10(padj): 5.490431e-02<br />log10(baseMean): 4.0125102<br />gene: FBgn0000253","log2FoldChange: -0.4507137775<br />-log10(padj): 2.099657e+00<br />log10(baseMean): 3.3393290<br />gene: FBgn0000256","log2FoldChange: 0.0573740618<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.8769572<br />gene: FBgn0000257","log2FoldChange: 0.0604293894<br />-log10(padj): 1.124980e-02<br />log10(baseMean): 3.0332426<br />gene: FBgn0000258","log2FoldChange: -0.0470845803<br />-log10(padj): 2.088100e-02<br />log10(baseMean): 3.3075404<br />gene: FBgn0000259","log2FoldChange: -0.1626711776<br />-log10(padj): 2.272806e-01<br />log10(baseMean): 3.2794777<br />gene: FBgn0000261","log2FoldChange: -0.1254405284<br />-log10(padj): 1.091934e-01<br />log10(baseMean): 3.3226760<br />gene: FBgn0000273","log2FoldChange: 0.1522382188<br />-log10(padj): 3.442555e-02<br />log10(baseMean): 1.8419563<br />gene: FBgn0000276","log2FoldChange: -0.4868834242<br />-log10(padj): 1.341132e-01<br />log10(baseMean): 1.5048844<br />gene: FBgn0000277","log2FoldChange: -0.6696273639<br />-log10(padj): 7.771917e-01<br />log10(baseMean): 1.6503281<br />gene: FBgn0000278","log2FoldChange: 0.4894408909<br />-log10(padj): 6.941868e-01<br />log10(baseMean): 1.9926101<br />gene: FBgn0000279","log2FoldChange: -0.1200691939<br />-log10(padj): 1.467644e-01<br />log10(baseMean): 3.5473132<br />gene: FBgn0000283","log2FoldChange: -0.5314148943<br />-log10(padj): 2.921475e+00<br />log10(baseMean): 2.6856219<br />gene: FBgn0000286","log2FoldChange: -0.1807471329<br />-log10(padj): 4.580490e-01<br />log10(baseMean): 3.5376605<br />gene: FBgn0000289","log2FoldChange: 0.6365628335<br />-log10(padj): 4.282691e+00<br />log10(baseMean): 4.7062732<br />gene: FBgn0000299","log2FoldChange: -0.0541038568<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 3.2155843<br />gene: FBgn0000307","log2FoldChange: 0.0379720572<br />-log10(padj): 2.023076e-02<br />log10(baseMean): 4.2242167<br />gene: FBgn0000308","log2FoldChange: 0.0178116750<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 1.7663560<br />gene: FBgn0000313","log2FoldChange: -0.0722090724<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.1573563<br />gene: FBgn0000317","log2FoldChange: 0.2543528287<br />-log10(padj): 4.904134e-01<br />log10(baseMean): 2.9574871<br />gene: FBgn0000318","log2FoldChange: 0.0247973032<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 4.1471477<br />gene: FBgn0000319","log2FoldChange: 0.1604157213<br />-log10(padj): 1.751247e-01<br />log10(baseMean): 2.4303435<br />gene: FBgn0000326","log2FoldChange: -0.1741356385<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 2.3564655<br />gene: FBgn0000330","log2FoldChange: 0.0069671832<br />-log10(padj): 1.707839e-03<br />log10(baseMean): 1.4036285<br />gene: FBgn0000337","log2FoldChange: 0.1921946711<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 3.2631389<br />gene: FBgn0000338","log2FoldChange: 0.0085667546<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.7527734<br />gene: FBgn0000339","log2FoldChange: 0.6415327638<br />-log10(padj): 1.458717e+00<br />log10(baseMean): 1.9731844<br />gene: FBgn0000346","log2FoldChange: 0.3845004223<br />-log10(padj): 1.970780e-01<br />log10(baseMean): 1.5355212<br />gene: FBgn0000351","log2FoldChange: 0.0416551846<br />-log10(padj): 3.580449e-02<br />log10(baseMean): 2.9229455<br />gene: FBgn0000352","log2FoldChange: -0.0940377337<br />-log10(padj): 1.084218e-01<br />log10(baseMean): 4.0485674<br />gene: FBgn0000370","log2FoldChange: 0.0894979702<br />-log10(padj): 7.260138e-02<br />log10(baseMean): 3.0969870<br />gene: FBgn0000376","log2FoldChange: 0.0887113089<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 2.9507453<br />gene: FBgn0000377","log2FoldChange: -0.1255576668<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 2.8652025<br />gene: FBgn0000382","log2FoldChange: 0.0213072126<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.7772598<br />gene: FBgn0000384","log2FoldChange: 0.4659709799<br />-log10(padj): 3.262004e+00<br />log10(baseMean): 3.0335142<br />gene: FBgn0000392","log2FoldChange: -0.1883427714<br />-log10(padj): 3.150560e-01<br />log10(baseMean): 2.7936343<br />gene: FBgn0000394","log2FoldChange: -0.3760813428<br />-log10(padj): 5.209222e-01<br />log10(baseMean): 2.6962417<br />gene: FBgn0000395","log2FoldChange: 0.0638145799<br />-log10(padj): 6.926412e-02<br />log10(baseMean): 3.3855056<br />gene: FBgn0000404","log2FoldChange: 0.5267527374<br />-log10(padj): 3.536977e+00<br />log10(baseMean): 3.3651436<br />gene: FBgn0000405","log2FoldChange: -2.0331198289<br />-log10(padj): 1.057315e+01<br />log10(baseMean): 2.4240719<br />gene: FBgn0000406","log2FoldChange: -0.1030249462<br />-log10(padj): 1.224566e-01<br />log10(baseMean): 3.6234121<br />gene: FBgn0000409","log2FoldChange: 0.2339693690<br />-log10(padj): 6.763617e-01<br />log10(baseMean): 3.4470235<br />gene: FBgn0000412","log2FoldChange: -0.0072723993<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 3.4328746<br />gene: FBgn0000413","log2FoldChange: -0.2418168740<br />-log10(padj): 8.619033e-01<br />log10(baseMean): 3.3693069<br />gene: FBgn0000414","log2FoldChange: -0.3704062413<br />-log10(padj): 4.300251e+00<br />log10(baseMean): 4.5865657<br />gene: FBgn0000416","log2FoldChange: 0.4404492162<br />-log10(padj): 7.259514e-02<br />log10(baseMean): 0.8288308<br />gene: FBgn0000422","log2FoldChange: 0.1955000808<br />-log10(padj): 2.400870e-01<br />log10(baseMean): 2.8439645<br />gene: FBgn0000426","log2FoldChange: 0.5272579179<br />-log10(padj): 3.583452e+00<br />log10(baseMean): 2.7922282<br />gene: FBgn0000447","log2FoldChange: 0.3668265497<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 0.8819376<br />gene: FBgn0000448","log2FoldChange: -0.0082550855<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 3.4312157<br />gene: FBgn0000454","log2FoldChange: -0.1628970097<br />-log10(padj): 1.356387e-01<br />log10(baseMean): 2.7288279<br />gene: FBgn0000455","log2FoldChange: -0.0762214261<br />-log10(padj): 7.382405e-02<br />log10(baseMean): 3.1890395<br />gene: FBgn0000464","log2FoldChange: -0.1368983854<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 3.4205247<br />gene: FBgn0000472","log2FoldChange: 0.0978319172<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 1.2084934<br />gene: FBgn0000473","log2FoldChange: -0.2239256203<br />-log10(padj): 5.139412e-01<br />log10(baseMean): 2.9453382<br />gene: FBgn0000479","log2FoldChange: -0.0448618448<br />-log10(padj): 2.937668e-02<br />log10(baseMean): 3.0072042<br />gene: FBgn0000482","log2FoldChange: 0.1263208635<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.0484462<br />gene: FBgn0000489","log2FoldChange: 0.8632144375<br />-log10(padj): 3.225730e+00<br />log10(baseMean): 2.3025819<br />gene: FBgn0000490","log2FoldChange: 0.0241025678<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 2.9846628<br />gene: FBgn0000499","log2FoldChange: 0.6882374602<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 0.8484053<br />gene: FBgn0000504","log2FoldChange: 0.1076992203<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.7782294<br />gene: FBgn0000520","log2FoldChange: 0.3313994622<br />-log10(padj): 1.284786e+00<br />log10(baseMean): 2.9500897<br />gene: FBgn0000524","log2FoldChange: -0.5788071296<br />-log10(padj): 1.269015e+00<br />log10(baseMean): 2.1361444<br />gene: FBgn0000527","log2FoldChange: 0.3396221118<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 0.8257409<br />gene: FBgn0000533","log2FoldChange: -0.7120071487<br />-log10(padj): 1.540445e+00<br />log10(baseMean): 2.2493706<br />gene: FBgn0000535","log2FoldChange: 0.0441241381<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.9402995<br />gene: FBgn0000536","log2FoldChange: -0.1219809397<br />-log10(padj): 6.881408e-02<br />log10(baseMean): 3.6908541<br />gene: FBgn0000541","log2FoldChange: 0.0412635857<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 2.2075044<br />gene: FBgn0000542","log2FoldChange: 0.1724117990<br />-log10(padj): 1.681616e-01<br />log10(baseMean): 2.8616846<br />gene: FBgn0000543","log2FoldChange: -0.1274289291<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 2.9719748<br />gene: FBgn0000546","log2FoldChange: 0.5315694508<br />-log10(padj): 1.144949e+00<br />log10(baseMean): 2.5337969<br />gene: FBgn0000547","log2FoldChange: -0.1920040465<br />-log10(padj): 4.194621e-01<br />log10(baseMean): 5.2927934<br />gene: FBgn0000556","log2FoldChange: 0.3084360723<br />-log10(padj): 1.657443e-01<br />log10(baseMean): 1.6408181<br />gene: FBgn0000557","log2FoldChange: 0.0170079831<br />-log10(padj): 1.077674e-02<br />log10(baseMean): 5.0312468<br />gene: FBgn0000559","log2FoldChange: -0.1331109506<br />-log10(padj): 2.016282e-01<br />log10(baseMean): 3.1641333<br />gene: FBgn0000562","log2FoldChange: 0.7437964245<br />-log10(padj): 3.052884e+00<br />log10(baseMean): 2.7796624<br />gene: FBgn0000565","log2FoldChange: 0.1441879040<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.9021767<br />gene: FBgn0000566","log2FoldChange: -1.2263293522<br />-log10(padj): 1.035312e+01<br />log10(baseMean): 2.4169308<br />gene: FBgn0000567","log2FoldChange: -0.2089768065<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 3.5598110<br />gene: FBgn0000568","log2FoldChange: 0.3120654585<br />-log10(padj): 4.830210e-01<br />log10(baseMean): 3.1529727<br />gene: FBgn0000575","log2FoldChange: -0.3255626519<br />-log10(padj): 1.384990e+00<br />log10(baseMean): 3.7363583<br />gene: FBgn0000578","log2FoldChange: 0.4689710035<br />-log10(padj): 2.147764e+00<br />log10(baseMean): 3.9418838<br />gene: FBgn0000579","log2FoldChange: -0.1560128130<br />-log10(padj): 2.395095e-01<br />log10(baseMean): 3.5375042<br />gene: FBgn0000581","log2FoldChange: -0.1719840155<br />-log10(padj): 1.658154e-01<br />log10(baseMean): 2.5313935<br />gene: FBgn0000588","log2FoldChange: -0.3956910622<br />-log10(padj): 1.115369e-01<br />log10(baseMean): 1.3325506<br />gene: FBgn0000594","log2FoldChange: -0.2110533280<br />-log10(padj): 5.121700e-01<br />log10(baseMean): 3.1402233<br />gene: FBgn0000611","log2FoldChange: -0.0487691613<br />-log10(padj): 1.975768e-02<br />log10(baseMean): 2.5825788<br />gene: FBgn0000615","log2FoldChange: 0.0646771946<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.9134933<br />gene: FBgn0000617","log2FoldChange: 0.3056558405<br />-log10(padj): 3.628965e-01<br />log10(baseMean): 2.1810403<br />gene: FBgn0000618","log2FoldChange: -0.1359237172<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 3.0481885<br />gene: FBgn0000629","log2FoldChange: -0.1224473479<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 1.3174257<br />gene: FBgn0000630","log2FoldChange: 1.0671425395<br />-log10(padj): 1.291920e+00<br />log10(baseMean): 1.3920851<br />gene: FBgn0000633","log2FoldChange: 0.3770972164<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 2.5469176<br />gene: FBgn0000634","log2FoldChange: 0.4625965771<br />-log10(padj): 2.763870e-01<br />log10(baseMean): 2.1863655<br />gene: FBgn0000635","log2FoldChange: -0.2838802456<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 1.6994489<br />gene: FBgn0000636","log2FoldChange: -0.1321239858<br />-log10(padj): 2.397553e-01<br />log10(baseMean): 3.0707810<br />gene: FBgn0000662","log2FoldChange: -0.1079404129<br />-log10(padj): 7.805635e-02<br />log10(baseMean): 3.1762319<br />gene: FBgn0000667","log2FoldChange: -0.1575954168<br />-log10(padj): 2.510493e-01<br />log10(baseMean): 3.3550285<br />gene: FBgn0000709","log2FoldChange: 0.0494658749<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 3.0229270<br />gene: FBgn0000711","log2FoldChange: -0.1182590853<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 3.3229365<br />gene: FBgn0000719","log2FoldChange: 0.0840230538<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.9686660<br />gene: FBgn0000721","log2FoldChange: -0.0737073545<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.3040411<br />gene: FBgn0000723","log2FoldChange: -0.0224874184<br />-log10(padj): 8.278629e-03<br />log10(baseMean): 2.1162962<br />gene: FBgn0000808","log2FoldChange: -0.0373281082<br />-log10(padj): 1.242995e-02<br />log10(baseMean): 2.4652776<br />gene: FBgn0000810","log2FoldChange: 0.5984927645<br />-log10(padj): 1.452021e+00<br />log10(baseMean): 2.0992425<br />gene: FBgn0000826","log2FoldChange: 0.2767584667<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 1.5310519<br />gene: FBgn0000927","log2FoldChange: 0.4701331763<br />-log10(padj): 2.180708e+00<br />log10(baseMean): 2.8309845<br />gene: FBgn0000928","log2FoldChange: 0.0468080402<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 3.3756190<br />gene: FBgn0000986","log2FoldChange: -0.2290921773<br />-log10(padj): 7.399142e-01<br />log10(baseMean): 3.0818410<br />gene: FBgn0000996","log2FoldChange: -0.0099259618<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.6472412<br />gene: FBgn0001075","log2FoldChange: -0.0061787043<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 3.0587941<br />gene: FBgn0001078","log2FoldChange: -0.0243260821<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 2.5760527<br />gene: FBgn0001079","log2FoldChange: 0.0915323550<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 2.2784891<br />gene: FBgn0001083","log2FoldChange: 0.3759237600<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 1.8365293<br />gene: FBgn0001084","log2FoldChange: 0.5797744121<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 0.9976110<br />gene: FBgn0001085","log2FoldChange: 0.0015834622<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 3.0559193<br />gene: FBgn0001086","log2FoldChange: 0.0855354617<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 3.2315861<br />gene: FBgn0001087","log2FoldChange: 0.1643070737<br />-log10(padj): 6.441954e-02<br />log10(baseMean): 1.7153972<br />gene: FBgn0001089","log2FoldChange: 0.6444577556<br />-log10(padj): 6.891340e-01<br />log10(baseMean): 1.6174388<br />gene: FBgn0001090","log2FoldChange: 0.7080132272<br />-log10(padj): 3.626949e+00<br />log10(baseMean): 3.6358383<br />gene: FBgn0001091","log2FoldChange: 0.3750702797<br />-log10(padj): 1.021690e+00<br />log10(baseMean): 3.9603945<br />gene: FBgn0001092","log2FoldChange: -0.2785332113<br />-log10(padj): 6.157033e-01<br />log10(baseMean): 2.6951627<br />gene: FBgn0001098","log2FoldChange: 0.0728440077<br />-log10(padj): 1.143540e-01<br />log10(baseMean): 3.8359952<br />gene: FBgn0001104","log2FoldChange: 0.0785344869<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.6188715<br />gene: FBgn0001105","log2FoldChange: -0.1420309146<br />-log10(padj): 2.374332e-01<br />log10(baseMean): 3.3537604<br />gene: FBgn0001108","log2FoldChange: -0.5226206771<br />-log10(padj): 3.248774e+00<br />log10(baseMean): 4.5914043<br />gene: FBgn0001114","log2FoldChange: 0.6720490025<br />-log10(padj): 2.453553e+00<br />log10(baseMean): 2.9984310<br />gene: FBgn0001122","log2FoldChange: -0.0277738709<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.9644250<br />gene: FBgn0001123","log2FoldChange: 0.6805438835<br />-log10(padj): 6.792821e+00<br />log10(baseMean): 3.1022875<br />gene: FBgn0001124","log2FoldChange: -0.1573748216<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 3.3826058<br />gene: FBgn0001125","log2FoldChange: -0.3496978831<br />-log10(padj): 9.508892e-01<br />log10(baseMean): 2.4300093<br />gene: FBgn0001128","log2FoldChange: -0.0604963396<br />-log10(padj): 3.690087e-02<br />log10(baseMean): 2.6858249<br />gene: FBgn0001133","log2FoldChange: -0.3370001761<br />-log10(padj): 4.548636e-02<br />log10(baseMean): 0.9258590<br />gene: FBgn0001134","log2FoldChange: -1.2543021397<br />-log10(padj): 1.198148e+01<br />log10(baseMean): 2.6937703<br />gene: FBgn0001137","log2FoldChange: -0.1275892533<br />-log10(padj): 1.208914e-01<br />log10(baseMean): 3.6016743<br />gene: FBgn0001139","log2FoldChange: 0.0628927716<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 2.5716941<br />gene: FBgn0001142","log2FoldChange: 0.1889840425<br />-log10(padj): 9.668193e-02<br />log10(baseMean): 2.6267605<br />gene: FBgn0001145","log2FoldChange: -0.0660552873<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.3079811<br />gene: FBgn0001149","log2FoldChange: -0.3115220356<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 1.6490748<br />gene: FBgn0001150","log2FoldChange: 0.0762504063<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 3.4525352<br />gene: FBgn0001168","log2FoldChange: -0.0373235728<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 3.1503837<br />gene: FBgn0001169","log2FoldChange: 0.1796177618<br />-log10(padj): 5.394101e-01<br />log10(baseMean): 2.9765401<br />gene: FBgn0001179","log2FoldChange: 0.0366824239<br />-log10(padj): 2.092265e-02<br />log10(baseMean): 2.6762831<br />gene: FBgn0001185","log2FoldChange: 0.0726149746<br />-log10(padj): 6.917231e-02<br />log10(baseMean): 3.4030530<br />gene: FBgn0001186","log2FoldChange: 0.2666094720<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 0.9707371<br />gene: FBgn0001187","log2FoldChange: 0.1937238806<br />-log10(padj): 5.208817e-01<br />log10(baseMean): 3.1298383<br />gene: FBgn0001189","log2FoldChange: 0.3465485945<br />-log10(padj): 2.874503e+00<br />log10(baseMean): 3.5165589<br />gene: FBgn0001197","log2FoldChange: -0.0696177619<br />-log10(padj): 6.371831e-02<br />log10(baseMean): 2.8561811<br />gene: FBgn0001202","log2FoldChange: 0.1366767412<br />-log10(padj): 5.657396e-02<br />log10(baseMean): 2.5906638<br />gene: FBgn0001203","log2FoldChange: 0.0073717981<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 2.6299251<br />gene: FBgn0001205","log2FoldChange: 0.4795783916<br />-log10(padj): 3.566237e+00<br />log10(baseMean): 2.7730631<br />gene: FBgn0001206","log2FoldChange: -0.1552628642<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 3.7932106<br />gene: FBgn0001215","log2FoldChange: -0.0070879595<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 4.4836637<br />gene: FBgn0001218","log2FoldChange: 0.1772467551<br />-log10(padj): 2.004441e-01<br />log10(baseMean): 4.9308293<br />gene: FBgn0001219","log2FoldChange: 0.0369689130<br />-log10(padj): 3.243344e-02<br />log10(baseMean): 3.7834063<br />gene: FBgn0001220","log2FoldChange: -0.0861042482<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 3.0205940<br />gene: FBgn0001222","log2FoldChange: 1.4858469129<br />-log10(padj): 2.177959e+01<br />log10(baseMean): 2.7773775<br />gene: FBgn0001224","log2FoldChange: 1.3474935958<br />-log10(padj): 1.598973e+01<br />log10(baseMean): 2.6235621<br />gene: FBgn0001225","log2FoldChange: 1.7187127632<br />-log10(padj): 1.730066e+01<br />log10(baseMean): 3.1218583<br />gene: FBgn0001226","log2FoldChange: 0.4944115894<br />-log10(padj): 2.903845e-01<br />log10(baseMean): 1.4940495<br />gene: FBgn0001227","log2FoldChange: 0.0967307185<br />-log10(padj): 5.636755e-02<br />log10(baseMean): 2.2740252<br />gene: FBgn0001230","log2FoldChange: 0.1721372492<br />-log10(padj): 3.632154e-01<br />log10(baseMean): 4.6542332<br />gene: FBgn0001233","log2FoldChange: -0.2388141563<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 3.0133267<br />gene: FBgn0001234","log2FoldChange: 0.1741026507<br />-log10(padj): 4.629834e-01<br />log10(baseMean): 3.1003296<br />gene: FBgn0001247","log2FoldChange: 0.2087320434<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 3.4641220<br />gene: FBgn0001248","log2FoldChange: -0.0333336195<br />-log10(padj): 7.820026e-03<br />log10(baseMean): 1.6101870<br />gene: FBgn0001250","log2FoldChange: -0.0962125191<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.2832493<br />gene: FBgn0001255","log2FoldChange: 0.6655690873<br />-log10(padj): 2.946300e+00<br />log10(baseMean): 3.9079370<br />gene: FBgn0001257","log2FoldChange: 0.9384553054<br />-log10(padj): 1.164001e+01<br />log10(baseMean): 3.1818968<br />gene: FBgn0001258","log2FoldChange: 0.2792165536<br />-log10(padj): 6.717160e-02<br />log10(baseMean): 1.2173072<br />gene: FBgn0001259","log2FoldChange: -0.2293286964<br />-log10(padj): 9.533248e-02<br />log10(baseMean): 1.6140318<br />gene: FBgn0001263","log2FoldChange: -0.0864807325<br />-log10(padj): 4.369314e-02<br />log10(baseMean): 2.1973913<br />gene: FBgn0001276","log2FoldChange: 0.2392818415<br />-log10(padj): 3.779447e-01<br />log10(baseMean): 2.7228641<br />gene: FBgn0001280","log2FoldChange: -0.1002042766<br />-log10(padj): 1.307513e-01<br />log10(baseMean): 3.0820645<br />gene: FBgn0001291","log2FoldChange: 0.0406680072<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.8428439<br />gene: FBgn0001296","log2FoldChange: -0.3914917358<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 4.0445941<br />gene: FBgn0001297","log2FoldChange: 0.1467785759<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 3.0539791<br />gene: FBgn0001301","log2FoldChange: -0.1391849346<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 3.6081382<br />gene: FBgn0001308","log2FoldChange: 0.7202791314<br />-log10(padj): 3.223997e-01<br />log10(baseMean): 1.1337901<br />gene: FBgn0001311","log2FoldChange: 0.1511410513<br />-log10(padj): 2.766857e-01<br />log10(baseMean): 4.0917080<br />gene: FBgn0001316","log2FoldChange: 0.5156129071<br />-log10(padj): 1.112677e-01<br />log10(baseMean): 1.0021344<br />gene: FBgn0001321","log2FoldChange: -0.0437326243<br />-log10(padj): 3.627348e-02<br />log10(baseMean): 3.3986583<br />gene: FBgn0001324","log2FoldChange: -0.1308224025<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.6438985<br />gene: FBgn0001330","log2FoldChange: -0.5129192907<br />-log10(padj): 4.882155e+00<br />log10(baseMean): 3.0624826<br />gene: FBgn0001332","log2FoldChange: 0.1250077042<br />-log10(padj): 1.809581e-01<br />log10(baseMean): 2.8292144<br />gene: FBgn0001337","log2FoldChange: -0.0439236537<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.5428575<br />gene: FBgn0001341","log2FoldChange: 0.1936237725<br />-log10(padj): 3.493608e-01<br />log10(baseMean): 3.1131983<br />gene: FBgn0001404","log2FoldChange: -0.1435587724<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 2.4299747<br />gene: FBgn0001491","log2FoldChange: -0.1279441216<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 3.3525251<br />gene: FBgn0001565","log2FoldChange: 0.1403704311<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 2.8978376<br />gene: FBgn0001612","log2FoldChange: -0.2049829221<br />-log10(padj): 6.417617e-01<br />log10(baseMean): 3.7742734<br />gene: FBgn0001624","log2FoldChange: -0.2440394516<br />-log10(padj): 5.274490e-01<br />log10(baseMean): 3.4593612<br />gene: FBgn0001941","log2FoldChange: -0.2196501220<br />-log10(padj): 3.762454e-01<br />log10(baseMean): 4.7526927<br />gene: FBgn0001942","log2FoldChange: -0.0380487199<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 3.8231951<br />gene: FBgn0001961","log2FoldChange: -0.0788190663<br />-log10(padj): 8.004060e-02<br />log10(baseMean): 3.2552134<br />gene: FBgn0001965","log2FoldChange: -1.8746962465<br />-log10(padj): 1.874347e+00<br />log10(baseMean): 1.1567382<br />gene: FBgn0001967","log2FoldChange: -0.1651100756<br />-log10(padj): 1.826135e-01<br />log10(baseMean): 2.7302634<br />gene: FBgn0001970","log2FoldChange: -0.0430071544<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.5328412<br />gene: FBgn0001974","log2FoldChange: 0.4792828154<br />-log10(padj): 3.143950e+00<br />log10(baseMean): 2.8597776<br />gene: FBgn0001977","log2FoldChange: 0.0174814905<br />-log10(padj): 1.032306e-02<br />log10(baseMean): 3.7657971<br />gene: FBgn0001978","log2FoldChange: -0.1996653335<br />-log10(padj): 3.982645e-01<br />log10(baseMean): 3.1523778<br />gene: FBgn0001986","log2FoldChange: 0.2878177870<br />-log10(padj): 6.081608e-01<br />log10(baseMean): 2.7670462<br />gene: FBgn0001987","log2FoldChange: -0.1848489400<br />-log10(padj): 3.285963e-01<br />log10(baseMean): 2.9168947<br />gene: FBgn0001989","log2FoldChange: -0.1182741447<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 2.6239503<br />gene: FBgn0001990","log2FoldChange: 0.2730756783<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 0.8208700<br />gene: FBgn0001992","log2FoldChange: 0.0761105871<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.4526466<br />gene: FBgn0001994","log2FoldChange: -0.0472725306<br />-log10(padj): 3.267959e-02<br />log10(baseMean): 2.7002351<br />gene: FBgn0001995","log2FoldChange: 0.1547940352<br />-log10(padj): 1.855676e-01<br />log10(baseMean): 2.4302860<br />gene: FBgn0002021","log2FoldChange: -0.0393665636<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 3.1215552<br />gene: FBgn0002022","log2FoldChange: 0.2228847777<br />-log10(padj): 3.424323e-01<br />log10(baseMean): 3.5229208<br />gene: FBgn0002031","log2FoldChange: -0.1496005598<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.8415140<br />gene: FBgn0002036","log2FoldChange: -0.1007983082<br />-log10(padj): 1.437531e-01<br />log10(baseMean): 3.4389354<br />gene: FBgn0002044","log2FoldChange: 0.1590972816<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 3.5962158<br />gene: FBgn0002069","log2FoldChange: -0.2549002991<br />-log10(padj): 2.396890e-01<br />log10(baseMean): 3.4954384<br />gene: FBgn0002121","log2FoldChange: -0.0132755048<br />-log10(padj): 4.818680e-03<br />log10(baseMean): 3.0421454<br />gene: FBgn0002174","log2FoldChange: -0.0327571453<br />-log10(padj): 2.803547e-02<br />log10(baseMean): 3.8646823<br />gene: FBgn0002183","log2FoldChange: -0.1036049487<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 2.5318587<br />gene: FBgn0002283","log2FoldChange: 0.2102528958<br />-log10(padj): 4.556229e-01<br />log10(baseMean): 3.2187837<br />gene: FBgn0002284","log2FoldChange: -0.1954878251<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.8879456<br />gene: FBgn0002306","log2FoldChange: 0.1369227780<br />-log10(padj): 7.211131e-02<br />log10(baseMean): 2.1877581<br />gene: FBgn0002354","log2FoldChange: -0.0823520323<br />-log10(padj): 1.182302e-01<br />log10(baseMean): 3.4756231<br />gene: FBgn0002413","log2FoldChange: -0.1086230496<br />-log10(padj): 9.728847e-02<br />log10(baseMean): 3.7692706<br />gene: FBgn0002431","log2FoldChange: 0.1448598032<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 1.5935815<br />gene: FBgn0002440","log2FoldChange: -0.0720747734<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.2366386<br />gene: FBgn0002441","log2FoldChange: 0.0625719822<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.3532783<br />gene: FBgn0002466","log2FoldChange: -0.1087189016<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.6178708<br />gene: FBgn0002521","log2FoldChange: -0.0438743629<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 3.7185835<br />gene: FBgn0002524","log2FoldChange: 0.0760703944<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 3.6845849<br />gene: FBgn0002525","log2FoldChange: 0.0043392864<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 4.9215749<br />gene: FBgn0002526","log2FoldChange: -0.7477496964<br />-log10(padj): 1.174257e+01<br />log10(baseMean): 4.5777235<br />gene: FBgn0002527","log2FoldChange: 0.0142504219<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 4.1422033<br />gene: FBgn0002528","log2FoldChange: -0.5347333663<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 0.8204710<br />gene: FBgn0002531","log2FoldChange: 0.0258933051<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.1029552<br />gene: FBgn0002542","log2FoldChange: 0.7713872923<br />-log10(padj): 1.534509e+00<br />log10(baseMean): 2.9585047<br />gene: FBgn0002543","log2FoldChange: -0.0401334720<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 3.0414514<br />gene: FBgn0002552","log2FoldChange: 0.4111078933<br />-log10(padj): 1.236393e-01<br />log10(baseMean): 1.2525260<br />gene: FBgn0002564","log2FoldChange: -0.0787446108<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 3.2316036<br />gene: FBgn0002566","log2FoldChange: 1.0105394698<br />-log10(padj): 2.501320e+00<br />log10(baseMean): 1.6881236<br />gene: FBgn0002567","log2FoldChange: 0.0312711238<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.4374569<br />gene: FBgn0002576","log2FoldChange: -0.0724050103<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 2.1809897<br />gene: FBgn0002577","log2FoldChange: -0.1422024052<br />-log10(padj): 2.167041e-01<br />log10(baseMean): 4.0403248<br />gene: FBgn0002579","log2FoldChange: -0.0981183280<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 4.5112893<br />gene: FBgn0002590","log2FoldChange: 0.1164828682<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 0.9768021<br />gene: FBgn0002592","log2FoldChange: -0.1151525303<br />-log10(padj): 1.401391e-01<br />log10(baseMean): 4.3727973<br />gene: FBgn0002593","log2FoldChange: -0.1091600153<br />-log10(padj): 8.492934e-02<br />log10(baseMean): 4.5238949<br />gene: FBgn0002607","log2FoldChange: -1.5131709678<br />-log10(padj): 3.596786e+00<br />log10(baseMean): 1.6360375<br />gene: FBgn0002609","log2FoldChange: -0.0433770801<br />-log10(padj): 2.091710e-02<br />log10(baseMean): 4.6474718<br />gene: FBgn0002622","log2FoldChange: -0.1569161142<br />-log10(padj): 1.101842e-01<br />log10(baseMean): 4.5624405<br />gene: FBgn0002626","log2FoldChange: -0.1766157295<br />-log10(padj): 3.663217e-01<br />log10(baseMean): 3.4395911<br />gene: FBgn0002638","log2FoldChange: 0.2327037793<br />-log10(padj): 4.865861e-01<br />log10(baseMean): 2.6139886<br />gene: FBgn0002641","log2FoldChange: -0.1966303427<br />-log10(padj): 3.054294e-01<br />log10(baseMean): 3.4890608<br />gene: FBgn0002643","log2FoldChange: 0.1005484917<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.8187505<br />gene: FBgn0002645","log2FoldChange: 0.1435526190<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.5320444<br />gene: FBgn0002652","log2FoldChange: -0.2024680604<br />-log10(padj): 1.365953e-01<br />log10(baseMean): 1.9489938<br />gene: FBgn0002673","log2FoldChange: 0.1081380270<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 2.4835195<br />gene: FBgn0002707","log2FoldChange: 0.3491418157<br />-log10(padj): 5.652886e-01<br />log10(baseMean): 2.2350096<br />gene: FBgn0002715","log2FoldChange: -0.1309077132<br />-log10(padj): 4.551061e-02<br />log10(baseMean): 1.6908533<br />gene: FBgn0002716","log2FoldChange: 0.0222113798<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.8963114<br />gene: FBgn0002719","log2FoldChange: -0.1955691281<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.7121699<br />gene: FBgn0002723","log2FoldChange: 0.1390109434<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 2.5024799<br />gene: FBgn0002733","log2FoldChange: -0.5306023314<br />-log10(padj): 1.123756e-01<br />log10(baseMean): 1.0455827<br />gene: FBgn0002734","log2FoldChange: -0.0900449960<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.7517299<br />gene: FBgn0002736","log2FoldChange: -1.1563222802<br />-log10(padj): 1.842354e+00<br />log10(baseMean): 1.6138922<br />gene: FBgn0002773","log2FoldChange: -0.0025901372<br />-log10(padj): 2.301060e-03<br />log10(baseMean): 2.9678802<br />gene: FBgn0002774","log2FoldChange: -0.2313349099<br />-log10(padj): 4.505691e-01<br />log10(baseMean): 2.6021328<br />gene: FBgn0002775","log2FoldChange: -0.6670419399<br />-log10(padj): 4.172380e+00<br />log10(baseMean): 3.0775416<br />gene: FBgn0002778","log2FoldChange: -0.3447818054<br />-log10(padj): 5.675943e-01<br />log10(baseMean): 3.5738844<br />gene: FBgn0002780","log2FoldChange: -0.0219634933<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 3.6414195<br />gene: FBgn0002781","log2FoldChange: -0.0456981574<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 3.5839885<br />gene: FBgn0002783","log2FoldChange: 0.1212221005<br />-log10(padj): 1.707397e-01<br />log10(baseMean): 3.2387351<br />gene: FBgn0002787","log2FoldChange: 0.2034919598<br />-log10(padj): 2.697490e-01<br />log10(baseMean): 2.6049126<br />gene: FBgn0002791","log2FoldChange: -2.1316844759<br />-log10(padj): 1.105260e+01<br />log10(baseMean): 2.0005638<br />gene: FBgn0002868","log2FoldChange: 0.2514885897<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 2.8244270<br />gene: FBgn0002872","log2FoldChange: 0.2190748110<br />-log10(padj): 3.960670e-01<br />log10(baseMean): 3.3093590<br />gene: FBgn0002873","log2FoldChange: -0.1749523558<br />-log10(padj): 2.509237e-01<br />log10(baseMean): 2.9225855<br />gene: FBgn0002878","log2FoldChange: 0.3496863721<br />-log10(padj): 1.969713e+00<br />log10(baseMean): 2.9943999<br />gene: FBgn0002891","log2FoldChange: -0.0062530456<br />-log10(padj): 3.495999e-03<br />log10(baseMean): 2.6002942<br />gene: FBgn0002899","log2FoldChange: 0.0552962763<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.2886165<br />gene: FBgn0002901","log2FoldChange: 0.0496953985<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.8382312<br />gene: FBgn0002905","log2FoldChange: -0.2725790786<br />-log10(padj): 5.626611e-01<br />log10(baseMean): 2.7241254<br />gene: FBgn0002906","log2FoldChange: -0.0923470496<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.3251564<br />gene: FBgn0002914","log2FoldChange: 0.1078254864<br />-log10(padj): 3.724778e-02<br />log10(baseMean): 1.8083861<br />gene: FBgn0002917","log2FoldChange: 0.1703155702<br />-log10(padj): 4.456353e-01<br />log10(baseMean): 4.1399727<br />gene: FBgn0002921","log2FoldChange: -0.8728755386<br />-log10(padj): 3.202619e-01<br />log10(baseMean): 1.2366311<br />gene: FBgn0002922","log2FoldChange: 0.1127771737<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.6842325<br />gene: FBgn0002924","log2FoldChange: 0.1160298347<br />-log10(padj): 5.010244e-02<br />log10(baseMean): 2.0080411<br />gene: FBgn0002926","log2FoldChange: -0.2554111823<br />-log10(padj): 1.719463e-01<br />log10(baseMean): 1.9251792<br />gene: FBgn0002931","log2FoldChange: -0.0269612836<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.8168423<br />gene: FBgn0002932","log2FoldChange: -1.1771093249<br />-log10(padj): 4.196183e-01<br />log10(baseMean): 0.8454646<br />gene: FBgn0002939","log2FoldChange: -1.0265673650<br />-log10(padj): 1.647640e+00<br />log10(baseMean): 1.7442023<br />gene: FBgn0002945","log2FoldChange: 0.1297766750<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.4593806<br />gene: FBgn0002948","log2FoldChange: 0.1253140548<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.0648455<br />gene: FBgn0002962","log2FoldChange: -0.1171945840<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 3.5686390<br />gene: FBgn0002968","log2FoldChange: -0.0897271885<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 2.2583117<br />gene: FBgn0002973","log2FoldChange: -0.0945143315<br />-log10(padj): 7.390460e-02<br />log10(baseMean): 2.5306786<br />gene: FBgn0002989","log2FoldChange: 0.0416630604<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.1581882<br />gene: FBgn0003008","log2FoldChange: 0.0732162427<br />-log10(padj): 7.909689e-03<br />log10(baseMean): 0.9439157<br />gene: FBgn0003011","log2FoldChange: -0.0419929428<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 4.0610948<br />gene: FBgn0003013","log2FoldChange: 0.1620215428<br />-log10(padj): 4.140014e-02<br />log10(baseMean): 1.4654194<br />gene: FBgn0003015","log2FoldChange: 0.1104815396<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.4715439<br />gene: FBgn0003016","log2FoldChange: 0.1444667714<br />-log10(padj): 2.255230e-01<br />log10(baseMean): 3.1260411<br />gene: FBgn0003022","log2FoldChange: -0.0492023723<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 1.2787206<br />gene: FBgn0003023","log2FoldChange: -0.1096088472<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 4.5810947<br />gene: FBgn0003031","log2FoldChange: 0.4119045681<br />-log10(padj): 2.557274e+00<br />log10(baseMean): 3.2092907<br />gene: FBgn0003041","log2FoldChange: -0.0951335541<br />-log10(padj): 5.794697e-02<br />log10(baseMean): 2.2943737<br />gene: FBgn0003042","log2FoldChange: -0.0124856735<br />-log10(padj): 6.861557e-03<br />log10(baseMean): 3.2534026<br />gene: FBgn0003044","log2FoldChange: 0.1499839299<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 3.6674743<br />gene: FBgn0003048","log2FoldChange: 0.9282034348<br />-log10(padj): 1.640585e+00<br />log10(baseMean): 2.0884450<br />gene: FBgn0003053","log2FoldChange: 0.0744872673<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 3.3793780<br />gene: FBgn0003062","log2FoldChange: -0.1390718490<br />-log10(padj): 5.902384e-02<br />log10(baseMean): 3.0819361<br />gene: FBgn0003067","log2FoldChange: -0.0602658749<br />-log10(padj): 2.327121e-02<br />log10(baseMean): 2.2469596<br />gene: FBgn0003068","log2FoldChange: -0.0253746499<br />-log10(padj): 1.109448e-02<br />log10(baseMean): 3.1489052<br />gene: FBgn0003071","log2FoldChange: 0.6416111404<br />-log10(padj): 6.030563e+00<br />log10(baseMean): 3.3351196<br />gene: FBgn0003074","log2FoldChange: 0.7125526112<br />-log10(padj): 9.474994e+00<br />log10(baseMean): 3.1756975<br />gene: FBgn0003076","log2FoldChange: 0.0947716105<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.8325969<br />gene: FBgn0003079","log2FoldChange: 0.0982567001<br />-log10(padj): 5.702578e-02<br />log10(baseMean): 2.8039468<br />gene: FBgn0003082","log2FoldChange: 0.3085920472<br />-log10(padj): 9.053519e-02<br />log10(baseMean): 1.9565690<br />gene: FBgn0003087","log2FoldChange: -0.4286904701<br />-log10(padj): 7.518625e-02<br />log10(baseMean): 0.9980538<br />gene: FBgn0003089","log2FoldChange: -0.2307816210<br />-log10(padj): 3.390534e-01<br />log10(baseMean): 3.1839098<br />gene: FBgn0003093","log2FoldChange: 0.0560458349<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.1622234<br />gene: FBgn0003116","log2FoldChange: -0.3619772370<br />-log10(padj): 1.239508e+00<br />log10(baseMean): 3.7556149<br />gene: FBgn0003117","log2FoldChange: 0.0386114277<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 3.8231751<br />gene: FBgn0003118","log2FoldChange: 0.2322327350<br />-log10(padj): 9.276150e-01<br />log10(baseMean): 3.3623487<br />gene: FBgn0003124","log2FoldChange: -0.0705335775<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.0921159<br />gene: FBgn0003134","log2FoldChange: 0.9184945044<br />-log10(padj): 1.707221e+01<br />log10(baseMean): 3.6550318<br />gene: FBgn0003137","log2FoldChange: -0.0111173106<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 3.3554806<br />gene: FBgn0003138","log2FoldChange: 0.1647066687<br />-log10(padj): 2.368948e-01<br />log10(baseMean): 2.8197353<br />gene: FBgn0003139","log2FoldChange: -0.1018491537<br />-log10(padj): 6.024283e-02<br />log10(baseMean): 2.5678196<br />gene: FBgn0003141","log2FoldChange: 0.7925587472<br />-log10(padj): 2.822587e-01<br />log10(baseMean): 1.0312760<br />gene: FBgn0003149","log2FoldChange: 0.1009568675<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 2.8922655<br />gene: FBgn0003159","log2FoldChange: 0.4102644736<br />-log10(padj): 4.199353e-01<br />log10(baseMean): 1.9260502<br />gene: FBgn0003162","log2FoldChange: 0.2040331095<br />-log10(padj): 1.618824e-01<br />log10(baseMean): 3.6954539<br />gene: FBgn0003165","log2FoldChange: -0.2107996419<br />-log10(padj): 8.006431e-01<br />log10(baseMean): 3.1647786<br />gene: FBgn0003169","log2FoldChange: -0.1561391895<br />-log10(padj): 1.705303e-01<br />log10(baseMean): 3.3088471<br />gene: FBgn0003175","log2FoldChange: 0.2498669887<br />-log10(padj): 4.852636e-01<br />log10(baseMean): 3.5772169<br />gene: FBgn0003177","log2FoldChange: 0.0914645248<br />-log10(padj): 6.826958e-02<br />log10(baseMean): 3.7886915<br />gene: FBgn0003178","log2FoldChange: 1.0157655002<br />-log10(padj): 5.470330e-01<br />log10(baseMean): 1.0541930<br />gene: FBgn0003187","log2FoldChange: -0.1470416674<br />-log10(padj): 1.397123e-01<br />log10(baseMean): 3.6194507<br />gene: FBgn0003189","log2FoldChange: 0.1913493040<br />-log10(padj): 3.137700e-01<br />log10(baseMean): 2.6810730<br />gene: FBgn0003200","log2FoldChange: -0.2066406913<br />-log10(padj): 2.623168e-01<br />log10(baseMean): 3.0782512<br />gene: FBgn0003204","log2FoldChange: -0.0664202197<br />-log10(padj): 7.954962e-02<br />log10(baseMean): 3.3703972<br />gene: FBgn0003205","log2FoldChange: 0.0235641256<br />-log10(padj): 1.376660e-02<br />log10(baseMean): 2.8870029<br />gene: FBgn0003206","log2FoldChange: 0.1378137983<br />-log10(padj): 2.857419e-01<br />log10(baseMean): 2.9979958<br />gene: FBgn0003209","log2FoldChange: 0.1274560631<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 3.1665457<br />gene: FBgn0003210","log2FoldChange: -0.0633471378<br />-log10(padj): 4.842354e-02<br />log10(baseMean): 2.8964464<br />gene: FBgn0003218","log2FoldChange: 0.2835113806<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 2.3661117<br />gene: FBgn0003227","log2FoldChange: -0.1734897232<br />-log10(padj): 3.657691e-01<br />log10(baseMean): 3.4754331<br />gene: FBgn0003231","log2FoldChange: 0.5572267004<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 1.2298725<br />gene: FBgn0003255","log2FoldChange: 0.2185400065<br />-log10(padj): 4.079775e-01<br />log10(baseMean): 2.8336607<br />gene: FBgn0003257","log2FoldChange: -0.1892298125<br />-log10(padj): 4.125902e-01<br />log10(baseMean): 4.1815215<br />gene: FBgn0003261","log2FoldChange: 0.0521424612<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 3.3716667<br />gene: FBgn0003268","log2FoldChange: 0.0666654543<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 4.4120824<br />gene: FBgn0003274","log2FoldChange: 0.0231340320<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 2.8099837<br />gene: FBgn0003275","log2FoldChange: 0.0516205200<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 3.3390251<br />gene: FBgn0003276","log2FoldChange: -0.0298786828<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 4.0286682<br />gene: FBgn0003277","log2FoldChange: -0.1003807256<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 2.8161678<br />gene: FBgn0003278","log2FoldChange: -0.2263783004<br />-log10(padj): 1.942969e-01<br />log10(baseMean): 4.8013498<br />gene: FBgn0003279","log2FoldChange: 0.5604861378<br />-log10(padj): 1.926605e-01<br />log10(baseMean): 1.2239375<br />gene: FBgn0003285","log2FoldChange: 0.0194500018<br />-log10(padj): 6.267132e-03<br />log10(baseMean): 3.0516043<br />gene: FBgn0003301","log2FoldChange: -0.3430263590<br />-log10(padj): 2.579139e-01<br />log10(baseMean): 1.9006092<br />gene: FBgn0003302","log2FoldChange: -0.4686671115<br />-log10(padj): 9.064644e-01<br />log10(baseMean): 2.3832132<br />gene: FBgn0003308","log2FoldChange: 0.0719216011<br />-log10(padj): 5.611394e-02<br />log10(baseMean): 3.0955110<br />gene: FBgn0003310","log2FoldChange: 0.5384988679<br />-log10(padj): 5.252143e+00<br />log10(baseMean): 3.1307068<br />gene: FBgn0003317","log2FoldChange: 0.2853209264<br />-log10(padj): 5.351595e-02<br />log10(baseMean): 1.0462766<br />gene: FBgn0003319","log2FoldChange: -0.1886120331<br />-log10(padj): 5.098456e-01<br />log10(baseMean): 3.1138044<br />gene: FBgn0003321","log2FoldChange: -0.3633539138<br />-log10(padj): 1.797972e+00<br />log10(baseMean): 2.9063778<br />gene: FBgn0003328","log2FoldChange: 0.0958975566<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.8024130<br />gene: FBgn0003330","log2FoldChange: -0.1608031502<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 3.1825446<br />gene: FBgn0003334","log2FoldChange: -0.0342977281<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 3.3875413<br />gene: FBgn0003345","log2FoldChange: 0.0444974340<br />-log10(padj): 3.829192e-02<br />log10(baseMean): 3.1236520<br />gene: FBgn0003346","log2FoldChange: 0.2669330174<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.0901699<br />gene: FBgn0003353","log2FoldChange: -1.7995270483<br />-log10(padj): 2.290530e+00<br />log10(baseMean): 1.2482694<br />gene: FBgn0003366","log2FoldChange: -0.1119078840<br />-log10(padj): 1.261444e-01<br />log10(baseMean): 3.4436792<br />gene: FBgn0003371","log2FoldChange: -0.0447591014<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.1180950<br />gene: FBgn0003386","log2FoldChange: 0.0676517796<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 0.8390864<br />gene: FBgn0003388","log2FoldChange: 0.7874622515<br />-log10(padj): 1.642705e+00<br />log10(baseMean): 1.7687176<br />gene: FBgn0003390","log2FoldChange: -0.1641007214<br />-log10(padj): 2.202475e-01<br />log10(baseMean): 2.9795552<br />gene: FBgn0003391","log2FoldChange: -0.1681309497<br />-log10(padj): 4.080994e-01<br />log10(baseMean): 3.4279844<br />gene: FBgn0003392","log2FoldChange: -0.3025552979<br />-log10(padj): 6.215127e-01<br />log10(baseMean): 3.5735458<br />gene: FBgn0003396","log2FoldChange: 0.3392853731<br />-log10(padj): 8.385468e-01<br />log10(baseMean): 2.3601856<br />gene: FBgn0003401","log2FoldChange: 0.0085868070<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.9309892<br />gene: FBgn0003410","log2FoldChange: -0.1108222229<br />-log10(padj): 1.037983e-01<br />log10(baseMean): 3.3512996<br />gene: FBgn0003415","log2FoldChange: 0.0396232504<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.0702323<br />gene: FBgn0003416","log2FoldChange: -0.0050331185<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.7445472<br />gene: FBgn0003423","log2FoldChange: -0.0502530167<br />-log10(padj): 3.383930e-02<br />log10(baseMean): 3.6043167<br />gene: FBgn0003425","log2FoldChange: 0.0031462433<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 2.1587567<br />gene: FBgn0003435","log2FoldChange: 0.1304154217<br />-log10(padj): 9.183439e-02<br />log10(baseMean): 2.7145086<br />gene: FBgn0003444","log2FoldChange: 1.9817749729<br />-log10(padj): 6.789698e+00<br />log10(baseMean): 1.5599686<br />gene: FBgn0003447","log2FoldChange: 0.0704148405<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 2.7127120<br />gene: FBgn0003449","log2FoldChange: -0.0734925463<br />-log10(padj): 5.916838e-02<br />log10(baseMean): 2.8261223<br />gene: FBgn0003459","log2FoldChange: -0.0141758595<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.2080502<br />gene: FBgn0003462","log2FoldChange: -0.0578362620<br />-log10(padj): 3.727518e-02<br />log10(baseMean): 3.1255100<br />gene: FBgn0003463","log2FoldChange: 0.0307001163<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.6127105<br />gene: FBgn0003464","log2FoldChange: 0.1421540084<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 3.4493360<br />gene: FBgn0003475","log2FoldChange: 0.1274669688<br />-log10(padj): 1.345339e-01<br />log10(baseMean): 2.6530543<br />gene: FBgn0003479","log2FoldChange: 0.0091797256<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.7538771<br />gene: FBgn0003480","log2FoldChange: 0.1924957662<br />-log10(padj): 6.472956e-02<br />log10(baseMean): 1.5478273<br />gene: FBgn0003482","log2FoldChange: 0.0647376504<br />-log10(padj): 5.286181e-02<br />log10(baseMean): 2.8347163<br />gene: FBgn0003483","log2FoldChange: 0.0869310313<br />-log10(padj): 5.975085e-02<br />log10(baseMean): 3.6709267<br />gene: FBgn0003495","log2FoldChange: 0.1318536500<br />-log10(padj): 3.520160e-02<br />log10(baseMean): 1.5346672<br />gene: FBgn0003499","log2FoldChange: 2.1536936356<br />-log10(padj): 2.938737e+01<br />log10(baseMean): 2.4432016<br />gene: FBgn0003501","log2FoldChange: 0.7295210235<br />-log10(padj): 9.596541e+00<br />log10(baseMean): 3.0802324<br />gene: FBgn0003502","log2FoldChange: -0.4076231198<br />-log10(padj): 2.199410e+00<br />log10(baseMean): 3.7473743<br />gene: FBgn0003507","log2FoldChange: 0.1473311555<br />-log10(padj): 7.472126e-02<br />log10(baseMean): 1.9549518<br />gene: FBgn0003510","log2FoldChange: -0.0342008367<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.5411833<br />gene: FBgn0003511","log2FoldChange: -0.0526868639<br />-log10(padj): 3.197684e-02<br />log10(baseMean): 2.9943803<br />gene: FBgn0003512","log2FoldChange: 0.1249071015<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 3.6350948<br />gene: FBgn0003514","log2FoldChange: 0.0027190838<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 4.9548833<br />gene: FBgn0003517","log2FoldChange: 0.0396936123<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.3237786<br />gene: FBgn0003520","log2FoldChange: -0.6889484632<br />-log10(padj): 4.862840e+00<br />log10(baseMean): 3.1370833<br />gene: FBgn0003525","log2FoldChange: -0.4598172847<br />-log10(padj): 2.685650e-01<br />log10(baseMean): 1.4808086<br />gene: FBgn0003527","log2FoldChange: -0.1020522212<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 2.7975272<br />gene: FBgn0003545","log2FoldChange: -0.3428291515<br />-log10(padj): 2.315644e+00<br />log10(baseMean): 3.4228397<br />gene: FBgn0003557","log2FoldChange: -0.1155917895<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.9590570<br />gene: FBgn0003567","log2FoldChange: -0.3205111126<br />-log10(padj): 1.036394e+00<br />log10(baseMean): 2.9656298<br />gene: FBgn0003575","log2FoldChange: 0.0058475936<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.9530294<br />gene: FBgn0003598","log2FoldChange: 0.0718073734<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 3.5187801<br />gene: FBgn0003600","log2FoldChange: -0.0404147338<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 3.3782946<br />gene: FBgn0003607","log2FoldChange: -0.0882775964<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 3.2484468<br />gene: FBgn0003612","log2FoldChange: -0.2299481807<br />-log10(padj): 4.421245e-01<br />log10(baseMean): 3.0355194<br />gene: FBgn0003638","log2FoldChange: -0.3060663325<br />-log10(padj): 3.258814e-01<br />log10(baseMean): 2.7352666<br />gene: FBgn0003654","log2FoldChange: -0.3839771220<br />-log10(padj): 6.972166e-01<br />log10(baseMean): 2.1926428<br />gene: FBgn0003655","log2FoldChange: 0.1248249023<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 3.1756637<br />gene: FBgn0003656","log2FoldChange: 0.3051257485<br />-log10(padj): 4.929856e-02<br />log10(baseMean): 2.3777228<br />gene: FBgn0003659","log2FoldChange: -0.1367932674<br />-log10(padj): 2.319413e-01<br />log10(baseMean): 3.4391895<br />gene: FBgn0003660","log2FoldChange: 0.2162818891<br />-log10(padj): 4.243745e-01<br />log10(baseMean): 3.6606355<br />gene: FBgn0003676","log2FoldChange: 0.0370165596<br />-log10(padj): 2.342017e-02<br />log10(baseMean): 2.5263479<br />gene: FBgn0003687","log2FoldChange: -0.1063109764<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.7949572<br />gene: FBgn0003701","log2FoldChange: -0.3141341542<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 0.8272682<br />gene: FBgn0003710","log2FoldChange: 0.2222914909<br />-log10(padj): 2.808695e-01<br />log10(baseMean): 2.4396431<br />gene: FBgn0003714","log2FoldChange: -0.0060232787<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.8026690<br />gene: FBgn0003716","log2FoldChange: -0.0308294350<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 3.6148768<br />gene: FBgn0003717","log2FoldChange: -0.1289888860<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 3.5826818<br />gene: FBgn0003721","log2FoldChange: -0.0647848091<br />-log10(padj): 2.133138e-02<br />log10(baseMean): 2.1385613<br />gene: FBgn0003731","log2FoldChange: 0.2169019074<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 3.7773579<br />gene: FBgn0003732","log2FoldChange: -0.5168410060<br />-log10(padj): 9.035501e-02<br />log10(baseMean): 0.8950325<br />gene: FBgn0003733","log2FoldChange: 0.1609044065<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.1385635<br />gene: FBgn0003741","log2FoldChange: -0.0311860200<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.5849950<br />gene: FBgn0003742","log2FoldChange: -0.2244043795<br />-log10(padj): 6.917562e-01<br />log10(baseMean): 2.9353362<br />gene: FBgn0003744","log2FoldChange: 0.1349208360<br />-log10(padj): 1.812788e-02<br />log10(baseMean): 1.0849485<br />gene: FBgn0003747","log2FoldChange: 1.0732666309<br />-log10(padj): 1.997573e+01<br />log10(baseMean): 3.0398003<br />gene: FBgn0003748","log2FoldChange: 0.2866501466<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 1.3515613<br />gene: FBgn0003751","log2FoldChange: -0.4623149083<br />-log10(padj): 1.356962e+00<br />log10(baseMean): 3.5784134<br />gene: FBgn0003862","log2FoldChange: -0.1640198094<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 3.3773015<br />gene: FBgn0003866","log2FoldChange: -0.2894498872<br />-log10(padj): 9.579658e-01<br />log10(baseMean): 3.1773472<br />gene: FBgn0003870","log2FoldChange: -0.0845800867<br />-log10(padj): 1.101748e-01<br />log10(baseMean): 2.9861976<br />gene: FBgn0003882","log2FoldChange: 0.5160916582<br />-log10(padj): 1.315522e+00<br />log10(baseMean): 4.6096053<br />gene: FBgn0003884","log2FoldChange: 0.2066789829<br />-log10(padj): 4.046314e-01<br />log10(baseMean): 3.3374971<br />gene: FBgn0003885","log2FoldChange: 0.7591276401<br />-log10(padj): 2.011131e+00<br />log10(baseMean): 2.6173512<br />gene: FBgn0003886","log2FoldChange: 0.4212357478<br />-log10(padj): 1.011330e+00<br />log10(baseMean): 4.3833950<br />gene: FBgn0003887","log2FoldChange: 0.4656652563<br />-log10(padj): 1.675911e+00<br />log10(baseMean): 3.8350862<br />gene: FBgn0003888","log2FoldChange: -0.4971603136<br />-log10(padj): 1.014141e+00<br />log10(baseMean): 3.2154818<br />gene: FBgn0003890","log2FoldChange: -0.4352820350<br />-log10(padj): 2.017381e+00<br />log10(baseMean): 4.1153000<br />gene: FBgn0003891","log2FoldChange: -0.2487994240<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 2.9180255<br />gene: FBgn0003892","log2FoldChange: 0.0009441700<br />-log10(padj): 3.176531e-04<br />log10(baseMean): 0.9556001<br />gene: FBgn0003926","log2FoldChange: 0.9207668457<br />-log10(padj): 5.148328e-02<br />log10(baseMean): 0.9184584<br />gene: FBgn0003930","log2FoldChange: -0.1024461076<br />-log10(padj): 1.041875e-01<br />log10(baseMean): 4.3550845<br />gene: FBgn0003941","log2FoldChange: -0.0902102470<br />-log10(padj): 7.363229e-02<br />log10(baseMean): 4.3306465<br />gene: FBgn0003942","log2FoldChange: 0.4554613767<br />-log10(padj): 4.687913e-02<br />log10(baseMean): 3.8198200<br />gene: FBgn0003943","log2FoldChange: -0.2834265988<br />-log10(padj): 5.431859e-01<br />log10(baseMean): 2.3706801<br />gene: FBgn0003950","log2FoldChange: 0.1589082798<br />-log10(padj): 2.136474e-01<br />log10(baseMean): 3.5182100<br />gene: FBgn0003963","log2FoldChange: -0.0972504952<br />-log10(padj): 9.993745e-02<br />log10(baseMean): 3.1354845<br />gene: FBgn0003964","log2FoldChange: 0.0295367998<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 3.3239980<br />gene: FBgn0003969","log2FoldChange: -0.0189199769<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.2674637<br />gene: FBgn0003977","log2FoldChange: -0.0897755528<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 2.7258281<br />gene: FBgn0003978","log2FoldChange: -0.3410048893<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 1.6720561<br />gene: FBgn0003984","log2FoldChange: 0.8340690361<br />-log10(padj): 1.510186e+00<br />log10(baseMean): 1.8957198<br />gene: FBgn0003996","log2FoldChange: -0.4359127575<br />-log10(padj): 1.642791e-01<br />log10(baseMean): 2.0825246<br />gene: FBgn0003997","log2FoldChange: 0.4395470905<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 0.8765720<br />gene: FBgn0004028","log2FoldChange: 0.5760689183<br />-log10(padj): 1.179878e+00<br />log10(baseMean): 2.4030595<br />gene: FBgn0004034","log2FoldChange: -0.0426890817<br />-log10(padj): 9.060734e-03<br />log10(baseMean): 1.6687863<br />gene: FBgn0004047","log2FoldChange: -0.1292897696<br />-log10(padj): 1.139380e-01<br />log10(baseMean): 2.9925815<br />gene: FBgn0004049","log2FoldChange: 0.1239618715<br />-log10(padj): 9.780621e-02<br />log10(baseMean): 2.8932874<br />gene: FBgn0004050","log2FoldChange: -0.0369493003<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 3.1255261<br />gene: FBgn0004055","log2FoldChange: -0.2159394850<br />-log10(padj): 1.711819e-01<br />log10(baseMean): 2.9749301<br />gene: FBgn0004057","log2FoldChange: 0.4065389342<br />-log10(padj): 8.926247e-01<br />log10(baseMean): 3.4486796<br />gene: FBgn0004066","log2FoldChange: 0.0353275362<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.3702329<br />gene: FBgn0004087","log2FoldChange: 0.0431753764<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.5762213<br />gene: FBgn0004101","log2FoldChange: -0.1420598421<br />-log10(padj): 2.224519e-01<br />log10(baseMean): 3.6940838<br />gene: FBgn0004103","log2FoldChange: 0.0257004497<br />-log10(padj): 1.025454e-02<br />log10(baseMean): 2.6880565<br />gene: FBgn0004106","log2FoldChange: 0.0839717666<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.3456976<br />gene: FBgn0004107","log2FoldChange: 0.8416966185<br />-log10(padj): 5.118181e+00<br />log10(baseMean): 2.8198106<br />gene: FBgn0004108","log2FoldChange: -0.0190775003<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.4026951<br />gene: FBgn0004118","log2FoldChange: 0.0435519710<br />-log10(padj): 2.887147e-02<br />log10(baseMean): 2.7937212<br />gene: FBgn0004132","log2FoldChange: -0.2880262270<br />-log10(padj): 1.130030e+00<br />log10(baseMean): 3.0764162<br />gene: FBgn0004133","log2FoldChange: 0.0212271504<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 4.2261904<br />gene: FBgn0004145","log2FoldChange: -0.1297948135<br />-log10(padj): 1.504910e-01<br />log10(baseMean): 4.6721161<br />gene: FBgn0004167","log2FoldChange: 0.5418731149<br />-log10(padj): 2.778203e-01<br />log10(baseMean): 1.4200796<br />gene: FBgn0004169","log2FoldChange: -0.0099440917<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 3.7085401<br />gene: FBgn0004177","log2FoldChange: -0.0432431021<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 3.0644363<br />gene: FBgn0004179","log2FoldChange: -0.1973704769<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.0810571<br />gene: FBgn0004188","log2FoldChange: -0.4458571881<br />-log10(padj): 2.147764e+00<br />log10(baseMean): 3.1056147<br />gene: FBgn0004197","log2FoldChange: -0.3201896402<br />-log10(padj): 4.231086e-01<br />log10(baseMean): 3.6142168<br />gene: FBgn0004198","log2FoldChange: -0.1533666603<br />-log10(padj): 3.667551e-01<br />log10(baseMean): 3.0403219<br />gene: FBgn0004227","log2FoldChange: -0.0352670297<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 3.6924496<br />gene: FBgn0004237","log2FoldChange: 0.6449772453<br />-log10(padj): 1.013921e+00<br />log10(baseMean): 1.9241030<br />gene: FBgn0004359","log2FoldChange: -0.2426227580<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 0.8446379<br />gene: FBgn0004360","log2FoldChange: 0.1081485677<br />-log10(padj): 8.941119e-02<br />log10(baseMean): 3.5285327<br />gene: FBgn0004362","log2FoldChange: 0.0937162229<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.7961696<br />gene: FBgn0004363","log2FoldChange: 0.5590436490<br />-log10(padj): 7.530797e-01<br />log10(baseMean): 1.9384421<br />gene: FBgn0004364","log2FoldChange: 0.2175969107<br />-log10(padj): 4.312377e-01<br />log10(baseMean): 2.8325628<br />gene: FBgn0004367","log2FoldChange: 0.2198338822<br />-log10(padj): 4.038476e-01<br />log10(baseMean): 3.2786683<br />gene: FBgn0004368","log2FoldChange: -0.7871643397<br />-log10(padj): 7.649747e+00<br />log10(baseMean): 2.9207465<br />gene: FBgn0004369","log2FoldChange: 0.0198068557<br />-log10(padj): 1.347426e-02<br />log10(baseMean): 3.3091264<br />gene: FBgn0004370","log2FoldChange: 0.0299018186<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.5463675<br />gene: FBgn0004373","log2FoldChange: -0.0485443990<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.9604978<br />gene: FBgn0004374","log2FoldChange: 0.0165147358<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.2756825<br />gene: FBgn0004378","log2FoldChange: 0.1353886915<br />-log10(padj): 2.540774e-01<br />log10(baseMean): 2.8477808<br />gene: FBgn0004379","log2FoldChange: -0.0636498792<br />-log10(padj): 6.119574e-02<br />log10(baseMean): 2.8568697<br />gene: FBgn0004380","log2FoldChange: 0.0677430145<br />-log10(padj): 6.633266e-02<br />log10(baseMean): 2.7018822<br />gene: FBgn0004381","log2FoldChange: -0.3340708907<br />-log10(padj): 1.858000e+00<br />log10(baseMean): 3.3387944<br />gene: FBgn0004387","log2FoldChange: 0.2775345187<br />-log10(padj): 1.082628e+00<br />log10(baseMean): 3.4258940<br />gene: FBgn0004390","log2FoldChange: 0.0829574483<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 3.1627969<br />gene: FBgn0004391","log2FoldChange: -0.1887010611<br />-log10(padj): 3.251341e-01<br />log10(baseMean): 2.8819783<br />gene: FBgn0004395","log2FoldChange: 1.0786533428<br />-log10(padj): 5.934850e+00<br />log10(baseMean): 3.3844283<br />gene: FBgn0004396","log2FoldChange: -0.2041764598<br />-log10(padj): 4.048843e-01<br />log10(baseMean): 3.5028078<br />gene: FBgn0004397","log2FoldChange: -0.0981500246<br />-log10(padj): 8.760098e-02<br />log10(baseMean): 3.5345243<br />gene: FBgn0004399","log2FoldChange: 0.4324090136<br />-log10(padj): 4.731339e-01<br />log10(baseMean): 2.4087374<br />gene: FBgn0004400","log2FoldChange: 0.1102453780<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 3.8519671<br />gene: FBgn0004401","log2FoldChange: -0.0460153529<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 4.1542829<br />gene: FBgn0004403","log2FoldChange: -0.1963874033<br />-log10(padj): 2.850408e-01<br />log10(baseMean): 3.6185640<br />gene: FBgn0004404","log2FoldChange: 0.1502102578<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 2.8494300<br />gene: FBgn0004406","log2FoldChange: -0.1590246963<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 4.6008338<br />gene: FBgn0004413","log2FoldChange: 0.0480995481<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 3.1534489<br />gene: FBgn0004419","log2FoldChange: 0.5587043636<br />-log10(padj): 1.024462e-01<br />log10(baseMean): 0.8344450<br />gene: FBgn0004431","log2FoldChange: 0.2139521211<br />-log10(padj): 4.740326e-01<br />log10(baseMean): 3.8905885<br />gene: FBgn0004432","log2FoldChange: 0.2445811202<br />-log10(padj): 1.672272e-01<br />log10(baseMean): 2.9720008<br />gene: FBgn0004435","log2FoldChange: -0.2824449231<br />-log10(padj): 2.091891e-01<br />log10(baseMean): 3.2075035<br />gene: FBgn0004436","log2FoldChange: -0.1702459283<br />-log10(padj): 2.762079e-01<br />log10(baseMean): 3.3766721<br />gene: FBgn0004449","log2FoldChange: -0.3250322504<br />-log10(padj): 1.284522e+00<br />log10(baseMean): 3.5727298<br />gene: FBgn0004456","log2FoldChange: -0.1346400604<br />-log10(padj): 1.657443e-01<br />log10(baseMean): 2.6478046<br />gene: FBgn0004462","log2FoldChange: 0.0358798204<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 2.8246829<br />gene: FBgn0004463","log2FoldChange: 0.0671969905<br />-log10(padj): 3.627348e-02<br />log10(baseMean): 2.8351150<br />gene: FBgn0004465","log2FoldChange: 0.9137950764<br />-log10(padj): 6.019373e+00<br />log10(baseMean): 3.2624182<br />gene: FBgn0004507","log2FoldChange: -0.0271872081<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.1704424<br />gene: FBgn0004509","log2FoldChange: -0.0252254411<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.7725056<br />gene: FBgn0004510","log2FoldChange: 0.3240299980<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 1.1496611<br />gene: FBgn0004511","log2FoldChange: 0.4050046052<br />-log10(padj): 1.103206e+00<br />log10(baseMean): 2.7006467<br />gene: FBgn0004512","log2FoldChange: -0.2123493466<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 3.9061116<br />gene: FBgn0004551","log2FoldChange: -0.2099598835<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.5546184<br />gene: FBgn0004556","log2FoldChange: -0.1527564854<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.4524519<br />gene: FBgn0004569","log2FoldChange: -0.1536435927<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 3.5180523<br />gene: FBgn0004574","log2FoldChange: 0.0264255082<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.1729523<br />gene: FBgn0004581","log2FoldChange: -0.0042359281<br />-log10(padj): 3.005491e-03<br />log10(baseMean): 3.4867324<br />gene: FBgn0004583","log2FoldChange: 0.4030055125<br />-log10(padj): 1.802137e+00<br />log10(baseMean): 3.2637258<br />gene: FBgn0004584","log2FoldChange: 0.0401653942<br />-log10(padj): 2.227551e-02<br />log10(baseMean): 3.6209133<br />gene: FBgn0004587","log2FoldChange: 0.7696349545<br />-log10(padj): 8.014000e-01<br />log10(baseMean): 1.6830748<br />gene: FBgn0004595","log2FoldChange: 0.5120713383<br />-log10(padj): 1.733231e+00<br />log10(baseMean): 2.4025754<br />gene: FBgn0004597","log2FoldChange: -0.1169510212<br />-log10(padj): 1.433527e-01<br />log10(baseMean): 3.5644807<br />gene: FBgn0004598","log2FoldChange: 0.0578558148<br />-log10(padj): 4.675296e-02<br />log10(baseMean): 3.2477228<br />gene: FBgn0004603","log2FoldChange: -0.3347276178<br />-log10(padj): 5.901324e-01<br />log10(baseMean): 4.1291940<br />gene: FBgn0004606","log2FoldChange: -0.4737346372<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 1.0439646<br />gene: FBgn0004607","log2FoldChange: 0.0115489638<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 2.6149889<br />gene: FBgn0004611","log2FoldChange: -0.0247158631<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.9029433<br />gene: FBgn0004624","log2FoldChange: 0.0353053831<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.1094553<br />gene: FBgn0004625","log2FoldChange: 0.1434065709<br />-log10(padj): 1.038634e-01<br />log10(baseMean): 3.1026179<br />gene: FBgn0004629","log2FoldChange: -0.3300289719<br />-log10(padj): 8.014000e-01<br />log10(baseMean): 2.8370388<br />gene: FBgn0004635","log2FoldChange: 0.1186994977<br />-log10(padj): 1.332728e-01<br />log10(baseMean): 3.7440110<br />gene: FBgn0004636","log2FoldChange: 0.0070787569<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.1501431<br />gene: FBgn0004638","log2FoldChange: 0.2496489447<br />-log10(padj): 4.504400e-01<br />log10(baseMean): 3.1067472<br />gene: FBgn0004643","log2FoldChange: 0.7042184104<br />-log10(padj): 7.918610e-01<br />log10(baseMean): 1.6968238<br />gene: FBgn0004646","log2FoldChange: -0.1284634234<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 3.6747926<br />gene: FBgn0004647","log2FoldChange: 0.0229362096<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.1086384<br />gene: FBgn0004648","log2FoldChange: -0.7381456228<br />-log10(padj): 2.754742e+00<br />log10(baseMean): 2.1327419<br />gene: FBgn0004650","log2FoldChange: -0.6745731428<br />-log10(padj): 2.766012e+00<br />log10(baseMean): 3.2917428<br />gene: FBgn0004652","log2FoldChange: -0.0094628975<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.3182245<br />gene: FBgn0004654","log2FoldChange: -0.1494401782<br />-log10(padj): 1.817064e-01<br />log10(baseMean): 3.3050242<br />gene: FBgn0004655","log2FoldChange: -0.1259054134<br />-log10(padj): 7.986683e-02<br />log10(baseMean): 3.9136612<br />gene: FBgn0004656","log2FoldChange: -0.1773638455<br />-log10(padj): 4.955374e-01<br />log10(baseMean): 3.8313767<br />gene: FBgn0004657","log2FoldChange: -0.2761945737<br />-log10(padj): 9.889301e-01<br />log10(baseMean): 3.3879748<br />gene: FBgn0004687","log2FoldChange: 0.0884138019<br />-log10(padj): 9.789393e-02<br />log10(baseMean): 3.4257886<br />gene: FBgn0004698","log2FoldChange: -0.5796130306<br />-log10(padj): 1.049229e+00<br />log10(baseMean): 2.2640679<br />gene: FBgn0004797","log2FoldChange: -0.0281803734<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.8662251<br />gene: FBgn0004811","log2FoldChange: 0.1002961331<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 3.8449903<br />gene: FBgn0004828","log2FoldChange: 0.0490359045<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.4213821<br />gene: FBgn0004832","log2FoldChange: 0.1351639772<br />-log10(padj): 1.223780e-01<br />log10(baseMean): 3.1827459<br />gene: FBgn0004834","log2FoldChange: -0.2092438698<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 2.5496240<br />gene: FBgn0004837","log2FoldChange: -0.1040671158<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 3.9961206<br />gene: FBgn0004838","log2FoldChange: -1.4213425584<br />-log10(padj): 5.214040e-01<br />log10(baseMean): 0.9456155<br />gene: FBgn0004839","log2FoldChange: -0.2116228317<br />-log10(padj): 1.624874e-01<br />log10(baseMean): 2.1121175<br />gene: FBgn0004852","log2FoldChange: 0.0422616058<br />-log10(padj): 2.157692e-02<br />log10(baseMean): 2.5826923<br />gene: FBgn0004855","log2FoldChange: 0.0769039466<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 3.1119100<br />gene: FBgn0004856","log2FoldChange: -0.6216809717<br />-log10(padj): 6.373511e-01<br />log10(baseMean): 1.5918843<br />gene: FBgn0004858","log2FoldChange: -0.3650544369<br />-log10(padj): 1.624874e-01<br />log10(baseMean): 2.3307647<br />gene: FBgn0004859","log2FoldChange: 0.1185860631<br />-log10(padj): 6.568839e-02<br />log10(baseMean): 2.6601714<br />gene: FBgn0004860","log2FoldChange: -0.2975725249<br />-log10(padj): 4.168122e-01<br />log10(baseMean): 2.6918843<br />gene: FBgn0004861","log2FoldChange: -0.0924528396<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.0774706<br />gene: FBgn0004864","log2FoldChange: -0.8965595257<br />-log10(padj): 1.608910e+00<br />log10(baseMean): 1.7216061<br />gene: FBgn0004865","log2FoldChange: 0.0090612618<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 4.6008683<br />gene: FBgn0004867","log2FoldChange: -0.1877425907<br />-log10(padj): 4.287800e-01<br />log10(baseMean): 3.5194918<br />gene: FBgn0004868","log2FoldChange: 0.7349160919<br />-log10(padj): 1.956721e+00<br />log10(baseMean): 1.9696500<br />gene: FBgn0004870","log2FoldChange: 0.4096166792<br />-log10(padj): 1.139055e+00<br />log10(baseMean): 2.9937653<br />gene: FBgn0004872","log2FoldChange: -0.0834813669<br />-log10(padj): 1.084915e-01<br />log10(baseMean): 3.5889243<br />gene: FBgn0004873","log2FoldChange: -0.0666466890<br />-log10(padj): 4.897514e-02<br />log10(baseMean): 3.5493740<br />gene: FBgn0004875","log2FoldChange: 0.1453747455<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 3.0626043<br />gene: FBgn0004876","log2FoldChange: -0.4599034919<br />-log10(padj): 3.882313e-01<br />log10(baseMean): 1.6918807<br />gene: FBgn0004882","log2FoldChange: -0.1841746012<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 0.8506759<br />gene: FBgn0004885","log2FoldChange: 0.0796145306<br />-log10(padj): 6.765428e-02<br />log10(baseMean): 3.3863989<br />gene: FBgn0004888","log2FoldChange: 0.0887235933<br />-log10(padj): 7.965921e-02<br />log10(baseMean): 3.2437951<br />gene: FBgn0004889","log2FoldChange: -0.8186737414<br />-log10(padj): 8.155252e+00<br />log10(baseMean): 2.6806243<br />gene: FBgn0004893","log2FoldChange: -0.6775882064<br />-log10(padj): 1.315560e-01<br />log10(baseMean): 0.8425482<br />gene: FBgn0004896","log2FoldChange: -0.0713370301<br />-log10(padj): 6.861476e-02<br />log10(baseMean): 2.9024121<br />gene: FBgn0004901","log2FoldChange: 0.0091732657<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.8838112<br />gene: FBgn0004903","log2FoldChange: -0.0695004217<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 3.8646828<br />gene: FBgn0004907","log2FoldChange: 0.0953828828<br />-log10(padj): 3.802180e-02<br />log10(baseMean): 2.1591734<br />gene: FBgn0004908","log2FoldChange: -0.1589771499<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 3.2197694<br />gene: FBgn0004913","log2FoldChange: -0.1505128521<br />-log10(padj): 2.057948e-01<br />log10(baseMean): 3.0554388<br />gene: FBgn0004914","log2FoldChange: 0.1252601014<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.7426629<br />gene: FBgn0004915","log2FoldChange: 0.4380905769<br />-log10(padj): 4.172975e-01<br />log10(baseMean): 1.9884522<br />gene: FBgn0004919","log2FoldChange: -0.1894561111<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 3.3064844<br />gene: FBgn0004921","log2FoldChange: -0.1974571275<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 4.6871299<br />gene: FBgn0004922","log2FoldChange: -0.0426100656<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 3.5354435<br />gene: FBgn0004924","log2FoldChange: 0.0259097095<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.5909910<br />gene: FBgn0004925","log2FoldChange: 0.0864261957<br />-log10(padj): 1.062543e-01<br />log10(baseMean): 3.5872670<br />gene: FBgn0004926","log2FoldChange: 0.1373312393<br />-log10(padj): 1.078691e-01<br />log10(baseMean): 2.4824572<br />gene: FBgn0004957","log2FoldChange: 0.0176912689<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.9584482<br />gene: FBgn0005198","log2FoldChange: 0.0323537904<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.3839974<br />gene: FBgn0005278","log2FoldChange: -0.1497732520<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 2.9773209<br />gene: FBgn0005322","log2FoldChange: 0.2232337359<br />-log10(padj): 3.826523e-01<br />log10(baseMean): 2.8643703<br />gene: FBgn0005355","log2FoldChange: -0.2631350373<br />-log10(padj): 5.064937e-01<br />log10(baseMean): 2.9048496<br />gene: FBgn0005386","log2FoldChange: -0.1096728977<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 3.2660682<br />gene: FBgn0005410","log2FoldChange: 0.0528867744<br />-log10(padj): 4.675296e-02<br />log10(baseMean): 3.1089637<br />gene: FBgn0005411","log2FoldChange: 0.3075268700<br />-log10(padj): 6.583371e-01<br />log10(baseMean): 2.6126858<br />gene: FBgn0005427","log2FoldChange: -0.1296384198<br />-log10(padj): 8.502386e-02<br />log10(baseMean): 4.1975506<br />gene: FBgn0005533","log2FoldChange: -0.0298537732<br />-log10(padj): 2.332938e-02<br />log10(baseMean): 3.5957314<br />gene: FBgn0005536","log2FoldChange: 0.3582365702<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 1.2375965<br />gene: FBgn0005558","log2FoldChange: -0.3471188342<br />-log10(padj): 5.272292e-02<br />log10(baseMean): 0.8173753<br />gene: FBgn0005561","log2FoldChange: 0.0873835694<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.6882130<br />gene: FBgn0005563","log2FoldChange: 0.2747334777<br />-log10(padj): 9.264361e-01<br />log10(baseMean): 4.3254224<br />gene: FBgn0005585","log2FoldChange: 0.0809172792<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 1.6617833<br />gene: FBgn0005592","log2FoldChange: -0.6109591194<br />-log10(padj): 2.355266e+00<br />log10(baseMean): 4.5245258<br />gene: FBgn0005593","log2FoldChange: -0.0428952950<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 3.0158872<br />gene: FBgn0005596","log2FoldChange: 0.0955168133<br />-log10(padj): 8.881446e-02<br />log10(baseMean): 2.8758906<br />gene: FBgn0005612","log2FoldChange: -0.2127702954<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 0.8724382<br />gene: FBgn0005613","log2FoldChange: -0.1624722170<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 1.8061157<br />gene: FBgn0005614","log2FoldChange: -0.0193957433<br />-log10(padj): 1.548214e-02<br />log10(baseMean): 2.9631265<br />gene: FBgn0005616","log2FoldChange: -0.2595877294<br />-log10(padj): 5.465525e-01<br />log10(baseMean): 3.1282746<br />gene: FBgn0005617","log2FoldChange: -0.3305481269<br />-log10(padj): 8.477958e-02<br />log10(baseMean): 1.2303954<br />gene: FBgn0005619","log2FoldChange: -0.1662164884<br />-log10(padj): 1.736242e-01<br />log10(baseMean): 3.0100804<br />gene: FBgn0005624","log2FoldChange: -0.1246997474<br />-log10(padj): 4.732637e-02<br />log10(baseMean): 2.0348180<br />gene: FBgn0005629","log2FoldChange: 0.0612607580<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 4.0613533<br />gene: FBgn0005630","log2FoldChange: 0.1090337438<br />-log10(padj): 1.238647e-01<br />log10(baseMean): 3.2597016<br />gene: FBgn0005631","log2FoldChange: -0.0783576284<br />-log10(padj): 9.053519e-02<br />log10(baseMean): 3.9217463<br />gene: FBgn0005632","log2FoldChange: 0.1164376134<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 4.3075067<br />gene: FBgn0005634","log2FoldChange: -0.6321104834<br />-log10(padj): 1.110429e+00<br />log10(baseMean): 2.6047953<br />gene: FBgn0005636","log2FoldChange: 0.6674247035<br />-log10(padj): 6.380171e+00<br />log10(baseMean): 3.1980472<br />gene: FBgn0005640","log2FoldChange: -0.0729517278<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.9911585<br />gene: FBgn0005642","log2FoldChange: 0.0043754435<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.1104809<br />gene: FBgn0005648","log2FoldChange: 0.1421175068<br />-log10(padj): 3.019219e-01<br />log10(baseMean): 3.3982833<br />gene: FBgn0005649","log2FoldChange: 0.2003078141<br />-log10(padj): 3.154508e-01<br />log10(baseMean): 3.3910740<br />gene: FBgn0005655","log2FoldChange: 1.0494607162<br />-log10(padj): 4.916846e+00<br />log10(baseMean): 2.1642362<br />gene: FBgn0005660","log2FoldChange: 0.2123593186<br />-log10(padj): 8.535439e-02<br />log10(baseMean): 2.5134542<br />gene: FBgn0005666","log2FoldChange: -0.1736077421<br />-log10(padj): 9.336547e-02<br />log10(baseMean): 2.0354657<br />gene: FBgn0005670","log2FoldChange: -0.0091616881<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.6521392<br />gene: FBgn0005671","log2FoldChange: -0.0668517286<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.6573876<br />gene: FBgn0005672","log2FoldChange: -0.0733064970<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.6706131<br />gene: FBgn0005674","log2FoldChange: 0.0177017580<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.4902125<br />gene: FBgn0005683","log2FoldChange: -0.3336968849<br />-log10(padj): 1.564542e+00<br />log10(baseMean): 2.9379546<br />gene: FBgn0005694","log2FoldChange: 0.0788639551<br />-log10(padj): 6.470652e-02<br />log10(baseMean): 2.4042170<br />gene: FBgn0005695","log2FoldChange: -0.1937497526<br />-log10(padj): 3.056808e-01<br />log10(baseMean): 2.8662789<br />gene: FBgn0005696","log2FoldChange: -0.6729197640<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 1.2367558<br />gene: FBgn0005771","log2FoldChange: -0.0742453609<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.1452582<br />gene: FBgn0005777","log2FoldChange: 0.0137794474<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.7855860<br />gene: FBgn0008635","log2FoldChange: 0.5069821709<br />-log10(padj): 2.479816e+00<br />log10(baseMean): 2.4711967<br />gene: FBgn0008649","log2FoldChange: -0.2381890468<br />-log10(padj): 3.792354e-01<br />log10(baseMean): 3.0800974<br />gene: FBgn0008654","log2FoldChange: -0.1846575688<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 1.5701096<br />gene: FBgn0010015","log2FoldChange: 0.0693210541<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 1.7557119<br />gene: FBgn0010038","log2FoldChange: 0.1764862248<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 2.8781435<br />gene: FBgn0010039","log2FoldChange: -0.7101890644<br />-log10(padj): 1.868422e-01<br />log10(baseMean): 0.9356111<br />gene: FBgn0010040","log2FoldChange: 0.4252907295<br />-log10(padj): 5.913022e-01<br />log10(baseMean): 1.9590711<br />gene: FBgn0010041","log2FoldChange: -0.3002085420<br />-log10(padj): 2.254458e-01<br />log10(baseMean): 2.0174752<br />gene: FBgn0010043","log2FoldChange: -0.1037658697<br />-log10(padj): 1.261058e-01<br />log10(baseMean): 3.4092301<br />gene: FBgn0010051","log2FoldChange: -0.0320195950<br />-log10(padj): 5.431473e-03<br />log10(baseMean): 1.3122809<br />gene: FBgn0010052","log2FoldChange: -0.5190257810<br />-log10(padj): 1.020120e+00<br />log10(baseMean): 2.2506186<br />gene: FBgn0010053","log2FoldChange: -0.0496542843<br />-log10(padj): 2.579879e-02<br />log10(baseMean): 4.4246067<br />gene: FBgn0010078","log2FoldChange: 0.1069176782<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.9178693<br />gene: FBgn0010083","log2FoldChange: 1.0477236071<br />-log10(padj): 4.154926e+00<br />log10(baseMean): 2.0356516<br />gene: FBgn0010097","log2FoldChange: -0.1904962931<br />-log10(padj): 3.321188e-01<br />log10(baseMean): 3.7413057<br />gene: FBgn0010100","log2FoldChange: -0.0481854548<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.5912533<br />gene: FBgn0010110","log2FoldChange: 0.0207255877<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 3.6454117<br />gene: FBgn0010113","log2FoldChange: 0.0468012970<br />-log10(padj): 5.956492e-02<br />log10(baseMean): 3.4898796<br />gene: FBgn0010173","log2FoldChange: 0.2970865183<br />-log10(padj): 1.448946e+00<br />log10(baseMean): 3.0663141<br />gene: FBgn0010194","log2FoldChange: 0.0043347727<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 4.6097403<br />gene: FBgn0010198","log2FoldChange: 0.1211174753<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 3.0420495<br />gene: FBgn0010213","log2FoldChange: 0.0075830820<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.2214005<br />gene: FBgn0010215","log2FoldChange: -0.0386052892<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.8428096<br />gene: FBgn0010217","log2FoldChange: -0.1196091595<br />-log10(padj): 1.376726e-01<br />log10(baseMean): 2.7895385<br />gene: FBgn0010220","log2FoldChange: -0.5406584060<br />-log10(padj): 1.792584e+00<br />log10(baseMean): 2.7339013<br />gene: FBgn0010222","log2FoldChange: -1.4379677613<br />-log10(padj): 6.213690e+00<br />log10(baseMean): 1.8156027<br />gene: FBgn0010223","log2FoldChange: -0.4894078672<br />-log10(padj): 5.091373e+00<br />log10(baseMean): 4.1079157<br />gene: FBgn0010225","log2FoldChange: 0.2271851870<br />-log10(padj): 2.397917e-01<br />log10(baseMean): 2.2137909<br />gene: FBgn0010226","log2FoldChange: 0.4515868591<br />-log10(padj): 1.611258e+00<br />log10(baseMean): 3.1232682<br />gene: FBgn0010228","log2FoldChange: -0.1335728178<br />-log10(padj): 3.121768e-01<br />log10(baseMean): 3.4079932<br />gene: FBgn0010235","log2FoldChange: 0.2887065608<br />-log10(padj): 1.108588e+00<br />log10(baseMean): 3.3053760<br />gene: FBgn0010238","log2FoldChange: 0.0789125299<br />-log10(padj): 2.803547e-02<br />log10(baseMean): 2.0152535<br />gene: FBgn0010240","log2FoldChange: -0.0542387165<br />-log10(padj): 4.762189e-03<br />log10(baseMean): 0.8546143<br />gene: FBgn0010241","log2FoldChange: -0.3605455447<br />-log10(padj): 1.203287e+00<br />log10(baseMean): 3.8451823<br />gene: FBgn0010246","log2FoldChange: -0.1536427937<br />-log10(padj): 2.113080e-01<br />log10(baseMean): 3.5342445<br />gene: FBgn0010256","log2FoldChange: 0.5600838169<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 0.9115863<br />gene: FBgn0010258","log2FoldChange: -0.1385329828<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 1.1243696<br />gene: FBgn0010263","log2FoldChange: -0.0959756191<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 4.4476040<br />gene: FBgn0010265","log2FoldChange: 0.1304533682<br />-log10(padj): 9.954149e-02<br />log10(baseMean): 2.5329751<br />gene: FBgn0010269","log2FoldChange: 0.1298462229<br />-log10(padj): 1.986805e-01<br />log10(baseMean): 3.6026989<br />gene: FBgn0010278","log2FoldChange: -0.1756509253<br />-log10(padj): 2.748603e-01<br />log10(baseMean): 3.5775542<br />gene: FBgn0010280","log2FoldChange: 0.2414120847<br />-log10(padj): 7.482848e-01<br />log10(baseMean): 3.1267281<br />gene: FBgn0010282","log2FoldChange: -0.0719597584<br />-log10(padj): 3.627348e-02<br />log10(baseMean): 2.2585532<br />gene: FBgn0010287","log2FoldChange: 0.0737909682<br />-log10(padj): 7.104959e-02<br />log10(baseMean): 3.1288300<br />gene: FBgn0010288","log2FoldChange: -0.2259355394<br />-log10(padj): 4.749414e-01<br />log10(baseMean): 2.7549011<br />gene: FBgn0010292","log2FoldChange: -0.3826765020<br />-log10(padj): 1.993047e+00<br />log10(baseMean): 3.6908078<br />gene: FBgn0010300","log2FoldChange: -0.0082074907<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.7094204<br />gene: FBgn0010303","log2FoldChange: 0.1953638753<br />-log10(padj): 2.113080e-01<br />log10(baseMean): 2.5739697<br />gene: FBgn0010309","log2FoldChange: 0.2270745041<br />-log10(padj): 8.837435e-02<br />log10(baseMean): 1.6943402<br />gene: FBgn0010313","log2FoldChange: 0.5833144743<br />-log10(padj): 1.711317e+00<br />log10(baseMean): 2.5418596<br />gene: FBgn0010314","log2FoldChange: 0.0248994445<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.8095842<br />gene: FBgn0010315","log2FoldChange: -0.7596444366<br />-log10(padj): 2.898914e+00<br />log10(baseMean): 2.6687345<br />gene: FBgn0010316","log2FoldChange: -0.1619150916<br />-log10(padj): 3.124717e-01<br />log10(baseMean): 3.8323249<br />gene: FBgn0010328","log2FoldChange: 0.3870139663<br />-log10(padj): 2.548905e-01<br />log10(baseMean): 1.6599295<br />gene: FBgn0010329","log2FoldChange: -0.0379839694<br />-log10(padj): 3.085530e-02<br />log10(baseMean): 3.4859707<br />gene: FBgn0010333","log2FoldChange: 0.1398937149<br />-log10(padj): 1.811904e-01<br />log10(baseMean): 2.8734241<br />gene: FBgn0010339","log2FoldChange: -0.2852988858<br />-log10(padj): 1.809581e-01<br />log10(baseMean): 2.0639662<br />gene: FBgn0010340","log2FoldChange: -0.0633455149<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 3.7489446<br />gene: FBgn0010341","log2FoldChange: 0.0398360876<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 2.9587056<br />gene: FBgn0010342","log2FoldChange: 0.0446868574<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 3.5822239<br />gene: FBgn0010348","log2FoldChange: 0.0598628625<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.9263776<br />gene: FBgn0010349","log2FoldChange: -0.1575509000<br />-log10(padj): 1.332800e-01<br />log10(baseMean): 2.8212612<br />gene: FBgn0010350","log2FoldChange: -0.0083129576<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.5473256<br />gene: FBgn0010352","log2FoldChange: -0.0420190873<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.3088736<br />gene: FBgn0010355","log2FoldChange: -0.0473758580<br />-log10(padj): 2.903413e-02<br />log10(baseMean): 2.6701174<br />gene: FBgn0010356","log2FoldChange: -0.2547282797<br />-log10(padj): 7.143887e-01<br />log10(baseMean): 3.3157823<br />gene: FBgn0010379","log2FoldChange: -0.0546742291<br />-log10(padj): 3.269632e-02<br />log10(baseMean): 3.5535945<br />gene: FBgn0010380","log2FoldChange: 0.1705855274<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.0135229<br />gene: FBgn0010381","log2FoldChange: -0.5652307555<br />-log10(padj): 3.720895e+00<br />log10(baseMean): 3.0720515<br />gene: FBgn0010382","log2FoldChange: -0.6357710507<br />-log10(padj): 4.084043e-01<br />log10(baseMean): 1.4378002<br />gene: FBgn0010383","log2FoldChange: 0.0161265150<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.7546322<br />gene: FBgn0010388","log2FoldChange: -0.4959405798<br />-log10(padj): 8.973024e-02<br />log10(baseMean): 0.9478021<br />gene: FBgn0010389","log2FoldChange: 0.1506217639<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 3.3875399<br />gene: FBgn0010391","log2FoldChange: 0.8928770462<br />-log10(padj): 2.418314e+00<br />log10(baseMean): 3.0117158<br />gene: FBgn0010395","log2FoldChange: 0.1098046035<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.7839260<br />gene: FBgn0010397","log2FoldChange: 0.0219008372<br />-log10(padj): 7.383364e-03<br />log10(baseMean): 2.0984962<br />gene: FBgn0010398","log2FoldChange: 0.1340346118<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 2.9967032<br />gene: FBgn0010406","log2FoldChange: 0.4102354069<br />-log10(padj): 2.588560e-01<br />log10(baseMean): 1.6509766<br />gene: FBgn0010407","log2FoldChange: -0.1192585444<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 4.3247543<br />gene: FBgn0010408","log2FoldChange: -0.0928837399<br />-log10(padj): 6.554086e-02<br />log10(baseMean): 4.4166367<br />gene: FBgn0010409","log2FoldChange: 0.0941490565<br />-log10(padj): 4.490020e-02<br />log10(baseMean): 4.4881631<br />gene: FBgn0010410","log2FoldChange: -0.0825845282<br />-log10(padj): 4.612193e-02<br />log10(baseMean): 4.4038494<br />gene: FBgn0010411","log2FoldChange: -0.1238167395<br />-log10(padj): 6.990564e-02<br />log10(baseMean): 4.6509149<br />gene: FBgn0010412","log2FoldChange: 0.0838010108<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 3.2446513<br />gene: FBgn0010415","log2FoldChange: 0.0762685171<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.1249307<br />gene: FBgn0010416","log2FoldChange: 0.0527725683<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 2.6839718<br />gene: FBgn0010417","log2FoldChange: -0.0052919716<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.4236308<br />gene: FBgn0010421","log2FoldChange: 0.0867836467<br />-log10(padj): 8.054418e-02<br />log10(baseMean): 2.9680125<br />gene: FBgn0010422","log2FoldChange: 0.0625100273<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.6834029<br />gene: FBgn0010426","log2FoldChange: 0.1013703060<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 4.2436093<br />gene: FBgn0010434","log2FoldChange: -0.2272414784<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 1.7887195<br />gene: FBgn0010435","log2FoldChange: -0.0406249103<br />-log10(padj): 2.063850e-02<br />log10(baseMean): 2.8701124<br />gene: FBgn0010438","log2FoldChange: -0.2209486103<br />-log10(padj): 5.380087e-01<br />log10(baseMean): 2.9038388<br />gene: FBgn0010441","log2FoldChange: 0.5026567917<br />-log10(padj): 5.217135e+00<br />log10(baseMean): 3.6792728<br />gene: FBgn0010470","log2FoldChange: -0.7434340637<br />-log10(padj): 6.349200e-01<br />log10(baseMean): 1.4486443<br />gene: FBgn0010473","log2FoldChange: 1.6583340876<br />-log10(padj): 1.052941e+00<br />log10(baseMean): 0.9703208<br />gene: FBgn0010482","log2FoldChange: -0.1586177820<br />-log10(padj): 2.368080e-01<br />log10(baseMean): 4.1057922<br />gene: FBgn0010488","log2FoldChange: 0.2900993665<br />-log10(padj): 3.476674e-01<br />log10(baseMean): 2.1644149<br />gene: FBgn0010497","log2FoldChange: 0.3129005726<br />-log10(padj): 6.632754e-01<br />log10(baseMean): 2.3320504<br />gene: FBgn0010501","log2FoldChange: 0.0450691666<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 2.8034379<br />gene: FBgn0010504","log2FoldChange: -0.2274292873<br />-log10(padj): 5.445113e-01<br />log10(baseMean): 3.2823739<br />gene: FBgn0010516","log2FoldChange: -0.4518543497<br />-log10(padj): 7.199215e-01<br />log10(baseMean): 2.1283395<br />gene: FBgn0010520","log2FoldChange: 0.2360510193<br />-log10(padj): 2.466545e-01<br />log10(baseMean): 2.2525261<br />gene: FBgn0010531","log2FoldChange: -0.1958341574<br />-log10(padj): 3.319802e-01<br />log10(baseMean): 3.3119460<br />gene: FBgn0010548","log2FoldChange: -0.5157175012<br />-log10(padj): 2.870223e+00<br />log10(baseMean): 2.6182750<br />gene: FBgn0010549","log2FoldChange: 0.3004559933<br />-log10(padj): 1.277343e+00<br />log10(baseMean): 3.5050115<br />gene: FBgn0010551","log2FoldChange: -0.0915954508<br />-log10(padj): 5.112058e-02<br />log10(baseMean): 3.5643482<br />gene: FBgn0010575","log2FoldChange: -0.2339259494<br />-log10(padj): 7.479593e-01<br />log10(baseMean): 3.1936029<br />gene: FBgn0010583","log2FoldChange: 0.0714065506<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 3.0609494<br />gene: FBgn0010590","log2FoldChange: -0.6591484369<br />-log10(padj): 7.578823e+00<br />log10(baseMean): 3.2137421<br />gene: FBgn0010591","log2FoldChange: -0.0671080210<br />-log10(padj): 7.472126e-02<br />log10(baseMean): 3.2471481<br />gene: FBgn0010602","log2FoldChange: -0.1786385362<br />-log10(padj): 1.354695e-01<br />log10(baseMean): 2.2924269<br />gene: FBgn0010607","log2FoldChange: -0.0871430055<br />-log10(padj): 8.334717e-02<br />log10(baseMean): 3.6332855<br />gene: FBgn0010609","log2FoldChange: 0.0789623187<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 3.3529467<br />gene: FBgn0010611","log2FoldChange: 0.0966395860<br />-log10(padj): 9.219738e-02<br />log10(baseMean): 3.1237813<br />gene: FBgn0010612","log2FoldChange: 0.6367987163<br />-log10(padj): 3.571870e-01<br />log10(baseMean): 1.3520374<br />gene: FBgn0010620","log2FoldChange: 0.1497262010<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 3.9842154<br />gene: FBgn0010621","log2FoldChange: 0.2697439652<br />-log10(padj): 3.630513e-01<br />log10(baseMean): 2.3511729<br />gene: FBgn0010622","log2FoldChange: 0.1964908772<br />-log10(padj): 3.524375e-01<br />log10(baseMean): 3.1318281<br />gene: FBgn0010638","log2FoldChange: 0.0214777014<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.8541636<br />gene: FBgn0010651","log2FoldChange: 0.0051587272<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 3.3535152<br />gene: FBgn0010660","log2FoldChange: 0.0423710440<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.2846548<br />gene: FBgn0010704","log2FoldChange: -0.1735311100<br />-log10(padj): 1.907013e-01<br />log10(baseMean): 2.3248764<br />gene: FBgn0010709","log2FoldChange: 0.2403617223<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 3.0304166<br />gene: FBgn0010741","log2FoldChange: -0.0969154885<br />-log10(padj): 1.275663e-01<br />log10(baseMean): 3.2561010<br />gene: FBgn0010747","log2FoldChange: -0.0285116374<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.9608069<br />gene: FBgn0010750","log2FoldChange: -0.0198726939<br />-log10(padj): 7.504414e-03<br />log10(baseMean): 3.4184338<br />gene: FBgn0010762","log2FoldChange: -0.4724276481<br />-log10(padj): 2.174740e+00<br />log10(baseMean): 2.5312859<br />gene: FBgn0010768","log2FoldChange: -0.2134753336<br />-log10(padj): 4.606839e-01<br />log10(baseMean): 2.7060653<br />gene: FBgn0010770","log2FoldChange: -0.2020375198<br />-log10(padj): 5.165763e-01<br />log10(baseMean): 3.0214214<br />gene: FBgn0010772","log2FoldChange: -0.0971283303<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 3.2066961<br />gene: FBgn0010774","log2FoldChange: 0.4456964821<br />-log10(padj): 2.259148e+00<br />log10(baseMean): 3.5557749<br />gene: FBgn0010786","log2FoldChange: 0.0571539309<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 3.1815173<br />gene: FBgn0010803","log2FoldChange: -0.1038981647<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.9800721<br />gene: FBgn0010808","log2FoldChange: 0.2212496139<br />-log10(padj): 6.935278e-01<br />log10(baseMean): 2.9624650<br />gene: FBgn0010812","log2FoldChange: 0.2073118986<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 3.3042149<br />gene: FBgn0010825","log2FoldChange: -0.0669215443<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.6917309<br />gene: FBgn0010830","log2FoldChange: -0.2134691222<br />-log10(padj): 5.232524e-01<br />log10(baseMean): 3.4562491<br />gene: FBgn0010877","log2FoldChange: 0.8639529298<br />-log10(padj): 2.587527e+00<br />log10(baseMean): 2.2593260<br />gene: FBgn0010894","log2FoldChange: 0.2252482942<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 3.3411621<br />gene: FBgn0010905","log2FoldChange: -0.2719016896<br />-log10(padj): 7.459263e-01<br />log10(baseMean): 4.1265705<br />gene: FBgn0010909","log2FoldChange: -0.1576060484<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.7986403<br />gene: FBgn0010926","log2FoldChange: 0.4625886571<br />-log10(padj): 2.754742e+00<br />log10(baseMean): 3.2405952<br />gene: FBgn0011016","log2FoldChange: 0.1185710395<br />-log10(padj): 1.084915e-01<br />log10(baseMean): 2.5651982<br />gene: FBgn0011020","log2FoldChange: 0.0441108932<br />-log10(padj): 3.699260e-02<br />log10(baseMean): 3.1988631<br />gene: FBgn0011202","log2FoldChange: -0.1849012255<br />-log10(padj): 3.878179e-01<br />log10(baseMean): 3.1692986<br />gene: FBgn0011204","log2FoldChange: -0.2609428381<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.4723126<br />gene: FBgn0011205","log2FoldChange: -0.6903208261<br />-log10(padj): 2.377762e+00<br />log10(baseMean): 2.4379921<br />gene: FBgn0011206","log2FoldChange: 0.0605626452<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.8933717<br />gene: FBgn0011207","log2FoldChange: -0.0633745575<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 4.0787262<br />gene: FBgn0011211","log2FoldChange: -0.0336850136<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 3.7195354<br />gene: FBgn0011217","log2FoldChange: -0.1568237936<br />-log10(padj): 1.165721e-01<br />log10(baseMean): 3.8801188<br />gene: FBgn0011219","log2FoldChange: -0.1678284550<br />-log10(padj): 2.968951e-01<br />log10(baseMean): 3.0566922<br />gene: FBgn0011224","log2FoldChange: 0.1096043849<br />-log10(padj): 1.612492e-01<br />log10(baseMean): 2.9650279<br />gene: FBgn0011225","log2FoldChange: 0.0573129814<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.6700978<br />gene: FBgn0011227","log2FoldChange: -0.1693283428<br />-log10(padj): 3.042599e-01<br />log10(baseMean): 4.2362765<br />gene: FBgn0011230","log2FoldChange: 0.1773481298<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 2.7847952<br />gene: FBgn0011232","log2FoldChange: -0.2674840065<br />-log10(padj): 5.675943e-01<br />log10(baseMean): 3.3136622<br />gene: FBgn0011236","log2FoldChange: 0.3617649566<br />-log10(padj): 3.430572e-01<br />log10(baseMean): 2.0923219<br />gene: FBgn0011241","log2FoldChange: 0.7581918703<br />-log10(padj): 3.453840e+00<br />log10(baseMean): 2.4231171<br />gene: FBgn0011259","log2FoldChange: 2.0487403306<br />-log10(padj): 1.078272e+01<br />log10(baseMean): 2.3245493<br />gene: FBgn0011260","log2FoldChange: -0.1917635279<br />-log10(padj): 2.348156e-01<br />log10(baseMean): 4.5076152<br />gene: FBgn0011272","log2FoldChange: -0.7566044012<br />-log10(padj): 5.508715e+00<br />log10(baseMean): 2.8904502<br />gene: FBgn0011274","log2FoldChange: 0.0326931881<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 1.0537635<br />gene: FBgn0011276","log2FoldChange: 0.5659658788<br />-log10(padj): 1.217820e-01<br />log10(baseMean): 0.9440483<br />gene: FBgn0011280","log2FoldChange: -0.0883083488<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 4.6329959<br />gene: FBgn0011284","log2FoldChange: -0.1347633656<br />-log10(padj): 1.416564e-01<br />log10(baseMean): 2.7762844<br />gene: FBgn0011285","log2FoldChange: 0.2661357233<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.2728109<br />gene: FBgn0011286","log2FoldChange: 0.6741332915<br />-log10(padj): 3.550523e-01<br />log10(baseMean): 1.3956664<br />gene: FBgn0011288","log2FoldChange: -0.1862017945<br />-log10(padj): 5.591693e-01<br />log10(baseMean): 3.2887860<br />gene: FBgn0011289","log2FoldChange: 0.0933892582<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.7730360<br />gene: FBgn0011290","log2FoldChange: -0.0698774808<br />-log10(padj): 3.861693e-02<br />log10(baseMean): 2.3155996<br />gene: FBgn0011291","log2FoldChange: 0.0132458063<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.7994694<br />gene: FBgn0011297","log2FoldChange: -0.2413367182<br />-log10(padj): 4.456353e-01<br />log10(baseMean): 3.0791348<br />gene: FBgn0011300","log2FoldChange: 0.3565849633<br />-log10(padj): 1.331442e+00<br />log10(baseMean): 2.8061880<br />gene: FBgn0011305","log2FoldChange: 0.0012017107<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 2.6954194<br />gene: FBgn0011327","log2FoldChange: -0.2192193251<br />-log10(padj): 3.294413e-01<br />log10(baseMean): 2.4872782<br />gene: FBgn0011335","log2FoldChange: 0.0552562089<br />-log10(padj): 3.732572e-02<br />log10(baseMean): 3.5754379<br />gene: FBgn0011336","log2FoldChange: 0.0088276971<br />-log10(padj): 4.546311e-03<br />log10(baseMean): 3.2211542<br />gene: FBgn0011361","log2FoldChange: -0.0604741370<br />-log10(padj): 4.881401e-02<br />log10(baseMean): 2.9991103<br />gene: FBgn0011455","log2FoldChange: -0.0768147646<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.0217360<br />gene: FBgn0011474","log2FoldChange: 0.0214056425<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.6990494<br />gene: FBgn0011476","log2FoldChange: -0.1821279822<br />-log10(padj): 1.437531e-01<br />log10(baseMean): 3.0380941<br />gene: FBgn0011481","log2FoldChange: -0.0264510616<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 2.8532115<br />gene: FBgn0011509","log2FoldChange: 0.2293636311<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.5559329<br />gene: FBgn0011566","log2FoldChange: -0.1364758185<br />-log10(padj): 3.155402e-01<br />log10(baseMean): 3.3639121<br />gene: FBgn0011570","log2FoldChange: 0.2141335699<br />-log10(padj): 1.524096e-01<br />log10(baseMean): 3.5378386<br />gene: FBgn0011571","log2FoldChange: 0.1424544900<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 3.3012356<br />gene: FBgn0011573","log2FoldChange: -0.1821519499<br />-log10(padj): 8.906014e-02<br />log10(baseMean): 1.9788342<br />gene: FBgn0011576","log2FoldChange: 0.1295497923<br />-log10(padj): 1.666483e-01<br />log10(baseMean): 3.4370525<br />gene: FBgn0011577","log2FoldChange: -0.1652415183<br />-log10(padj): 2.400870e-01<br />log10(baseMean): 3.2699309<br />gene: FBgn0011584","log2FoldChange: -0.0748448554<br />-log10(padj): 6.485116e-02<br />log10(baseMean): 3.4297044<br />gene: FBgn0011586","log2FoldChange: 1.2602674434<br />-log10(padj): 7.898739e-01<br />log10(baseMean): 1.0994780<br />gene: FBgn0011589","log2FoldChange: 0.6026834230<br />-log10(padj): 9.228154e-01<br />log10(baseMean): 1.8424447<br />gene: FBgn0011591","log2FoldChange: 0.4915506977<br />-log10(padj): 1.934513e+00<br />log10(baseMean): 2.7034223<br />gene: FBgn0011592","log2FoldChange: 0.3111901426<br />-log10(padj): 1.670980e-01<br />log10(baseMean): 1.8230082<br />gene: FBgn0011596","log2FoldChange: 0.0591031173<br />-log10(padj): 5.508672e-02<br />log10(baseMean): 3.5070909<br />gene: FBgn0011604","log2FoldChange: 0.4499269119<br />-log10(padj): 3.605062e+00<br />log10(baseMean): 3.2483623<br />gene: FBgn0011606","log2FoldChange: -0.1084240056<br />-log10(padj): 1.852008e-01<br />log10(baseMean): 3.5646575<br />gene: FBgn0011638","log2FoldChange: 0.0077855096<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 3.2653827<br />gene: FBgn0011640","log2FoldChange: -0.0449143179<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.6339823<br />gene: FBgn0011642","log2FoldChange: -0.0633913130<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 3.0432310<br />gene: FBgn0011648","log2FoldChange: -0.7380151904<br />-log10(padj): 1.223780e-01<br />log10(baseMean): 0.8625322<br />gene: FBgn0011653","log2FoldChange: -0.0339138251<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.1692068<br />gene: FBgn0011655","log2FoldChange: -0.1458131809<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 2.9042929<br />gene: FBgn0011656","log2FoldChange: -0.3139673573<br />-log10(padj): 1.272113e+00<br />log10(baseMean): 2.7641810<br />gene: FBgn0011659","log2FoldChange: -0.0899231771<br />-log10(padj): 6.482097e-02<br />log10(baseMean): 2.5999685<br />gene: FBgn0011660","log2FoldChange: 0.1232685480<br />-log10(padj): 1.238949e-01<br />log10(baseMean): 4.3999611<br />gene: FBgn0011661","log2FoldChange: 0.0548351492<br />-log10(padj): 2.746491e-02<br />log10(baseMean): 3.1400958<br />gene: FBgn0011666","log2FoldChange: -0.1994330099<br />-log10(padj): 3.600233e-01<br />log10(baseMean): 3.0070505<br />gene: FBgn0011672","log2FoldChange: 0.9213025841<br />-log10(padj): 4.400458e+00<br />log10(baseMean): 2.0473545<br />gene: FBgn0011674","log2FoldChange: 0.6196361215<br />-log10(padj): 1.232709e+00<br />log10(baseMean): 2.2136223<br />gene: FBgn0011676","log2FoldChange: 0.2104382181<br />-log10(padj): 6.386177e-01<br />log10(baseMean): 3.2901312<br />gene: FBgn0011692","log2FoldChange: -0.3735385610<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 0.9850494<br />gene: FBgn0011695","log2FoldChange: -0.1696717932<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.4229437<br />gene: FBgn0011703","log2FoldChange: 0.1005887885<br />-log10(padj): 1.129479e-01<br />log10(baseMean): 3.4136625<br />gene: FBgn0011704","log2FoldChange: 0.0729537283<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 3.2124760<br />gene: FBgn0011705","log2FoldChange: 0.6998429024<br />-log10(padj): 4.767426e-01<br />log10(baseMean): 1.5003928<br />gene: FBgn0011706","log2FoldChange: -0.1461982740<br />-log10(padj): 1.691341e-01<br />log10(baseMean): 3.1721781<br />gene: FBgn0011708","log2FoldChange: 0.4002054367<br />-log10(padj): 4.333929e+00<br />log10(baseMean): 3.4103100<br />gene: FBgn0011710","log2FoldChange: 0.0609871135<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.0287369<br />gene: FBgn0011715","log2FoldChange: -0.5796983945<br />-log10(padj): 3.950488e+00<br />log10(baseMean): 4.1208609<br />gene: FBgn0011722","log2FoldChange: -0.0638558383<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 2.8418478<br />gene: FBgn0011725","log2FoldChange: 0.1177834246<br />-log10(padj): 7.240700e-02<br />log10(baseMean): 3.9221198<br />gene: FBgn0011726","log2FoldChange: 0.1264598161<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 2.8189075<br />gene: FBgn0011737","log2FoldChange: -0.2138206052<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 3.3514580<br />gene: FBgn0011739","log2FoldChange: 0.0571188205<br />-log10(padj): 5.091132e-02<br />log10(baseMean): 2.8528364<br />gene: FBgn0011740","log2FoldChange: 0.1354677747<br />-log10(padj): 1.798294e-01<br />log10(baseMean): 2.9314453<br />gene: FBgn0011741","log2FoldChange: 0.1658644505<br />-log10(padj): 3.585475e-01<br />log10(baseMean): 3.3363239<br />gene: FBgn0011742","log2FoldChange: 0.2317665693<br />-log10(padj): 8.399860e-01<br />log10(baseMean): 3.4954419<br />gene: FBgn0011744","log2FoldChange: 0.0803979570<br />-log10(padj): 9.085768e-02<br />log10(baseMean): 3.2093676<br />gene: FBgn0011745","log2FoldChange: -0.3364227956<br />-log10(padj): 4.868103e-02<br />log10(baseMean): 0.9300311<br />gene: FBgn0011746","log2FoldChange: -0.0766195887<br />-log10(padj): 2.623567e-02<br />log10(baseMean): 3.6549943<br />gene: FBgn0011747","log2FoldChange: 0.4357472542<br />-log10(padj): 1.860431e+00<br />log10(baseMean): 2.8273514<br />gene: FBgn0011754","log2FoldChange: -0.0825229503<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.7326431<br />gene: FBgn0011760","log2FoldChange: -0.2226966611<br />-log10(padj): 2.066211e-01<br />log10(baseMean): 2.3030664<br />gene: FBgn0011762","log2FoldChange: -0.0635229273<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 3.3005046<br />gene: FBgn0011763","log2FoldChange: 0.0163674637<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.2026777<br />gene: FBgn0011764","log2FoldChange: -0.2429771893<br />-log10(padj): 8.836604e-01<br />log10(baseMean): 3.7446840<br />gene: FBgn0011766","log2FoldChange: 0.1601529025<br />-log10(padj): 1.478490e-01<br />log10(baseMean): 3.2866481<br />gene: FBgn0011768","log2FoldChange: 0.4142426816<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 2.1458375<br />gene: FBgn0011769","log2FoldChange: 0.3790043544<br />-log10(padj): 9.569426e-01<br />log10(baseMean): 2.4400995<br />gene: FBgn0011770","log2FoldChange: 0.0005970575<br />-log10(padj): 8.760575e-04<br />log10(baseMean): 3.2151454<br />gene: FBgn0011771","log2FoldChange: 0.1414177976<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.7803307<br />gene: FBgn0011774","log2FoldChange: 0.0582516644<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.3950700<br />gene: FBgn0011785","log2FoldChange: 0.0228702202<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.8842014<br />gene: FBgn0011787","log2FoldChange: 0.1294528371<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.4664677<br />gene: FBgn0011802","log2FoldChange: 0.4376031550<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 2.6854040<br />gene: FBgn0011817","log2FoldChange: -0.0381836973<br />-log10(padj): 3.131544e-02<br />log10(baseMean): 2.7924618<br />gene: FBgn0011818","log2FoldChange: 0.4336069562<br />-log10(padj): 3.674904e+00<br />log10(baseMean): 3.7567033<br />gene: FBgn0011823","log2FoldChange: 0.1123171753<br />-log10(padj): 7.847011e-02<br />log10(baseMean): 3.0197685<br />gene: FBgn0011824","log2FoldChange: -0.1257922134<br />-log10(padj): 1.125061e-01<br />log10(baseMean): 3.0803265<br />gene: FBgn0011826","log2FoldChange: -0.0179426375<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 4.6740475<br />gene: FBgn0011828","log2FoldChange: 0.4437215031<br />-log10(padj): 5.593255e-01<br />log10(baseMean): 2.0166587<br />gene: FBgn0011829","log2FoldChange: -0.0486785658<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 2.7917035<br />gene: FBgn0011836","log2FoldChange: -0.0785889861<br />-log10(padj): 4.944644e-02<br />log10(baseMean): 3.4284284<br />gene: FBgn0011837","log2FoldChange: -0.3482688677<br />-log10(padj): 1.363202e+00<br />log10(baseMean): 3.2499830<br />gene: FBgn0012034","log2FoldChange: -0.1411362159<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 3.7315547<br />gene: FBgn0012036","log2FoldChange: 0.2456134910<br />-log10(padj): 2.722683e-01<br />log10(baseMean): 2.9205369<br />gene: FBgn0012037","log2FoldChange: 0.3839383938<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 1.2285903<br />gene: FBgn0012042","log2FoldChange: 0.0235471389<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 3.4739033<br />gene: FBgn0012049","log2FoldChange: 0.0567100943<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.2054515<br />gene: FBgn0012051","log2FoldChange: -0.0807328170<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 3.0645061<br />gene: FBgn0012058","log2FoldChange: -0.2297546530<br />-log10(padj): 3.244687e-01<br />log10(baseMean): 2.7904375<br />gene: FBgn0012066","log2FoldChange: -0.0064605265<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.9527698<br />gene: FBgn0013263","log2FoldChange: 0.0251056479<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 3.4268028<br />gene: FBgn0013269","log2FoldChange: -0.4907923995<br />-log10(padj): 4.725314e+00<br />log10(baseMean): 4.1469395<br />gene: FBgn0013272","log2FoldChange: 0.1121644542<br />-log10(padj): 2.891818e-02<br />log10(baseMean): 2.0784418<br />gene: FBgn0013303","log2FoldChange: -0.2441378101<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 3.5080072<br />gene: FBgn0013305","log2FoldChange: -0.0079746952<br />-log10(padj): 4.040816e-03<br />log10(baseMean): 4.3864009<br />gene: FBgn0013325","log2FoldChange: -0.2194917174<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 3.0617312<br />gene: FBgn0013334","log2FoldChange: 0.1994185420<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 3.4115226<br />gene: FBgn0013343","log2FoldChange: 0.4026071542<br />-log10(padj): 1.017956e+00<br />log10(baseMean): 2.7820616<br />gene: FBgn0013347","log2FoldChange: 0.2341783246<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 2.6000988<br />gene: FBgn0013432","log2FoldChange: 0.1888978556<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 2.0421232<br />gene: FBgn0013435","log2FoldChange: -0.1359923820<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.0269362<br />gene: FBgn0013442","log2FoldChange: 0.3955002754<br />-log10(padj): 6.937036e-02<br />log10(baseMean): 0.9905438<br />gene: FBgn0013469","log2FoldChange: 0.0075881986<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 2.4430622<br />gene: FBgn0013531","log2FoldChange: -0.4528872967<br />-log10(padj): 1.296073e+00<br />log10(baseMean): 2.9068295<br />gene: FBgn0013548","log2FoldChange: -0.0751709082<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.3924370<br />gene: FBgn0013563","log2FoldChange: -0.4848943666<br />-log10(padj): 5.415772e+00<br />log10(baseMean): 3.4037534<br />gene: FBgn0013576","log2FoldChange: 0.0874182490<br />-log10(padj): 9.723018e-02<br />log10(baseMean): 3.7376266<br />gene: FBgn0013591","log2FoldChange: -0.0350097371<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.5639281<br />gene: FBgn0013717","log2FoldChange: -0.2415073965<br />-log10(padj): 3.595662e-01<br />log10(baseMean): 2.4702314<br />gene: FBgn0013718","log2FoldChange: 0.4446745753<br />-log10(padj): 1.711693e+00<br />log10(baseMean): 2.5678886<br />gene: FBgn0013720","log2FoldChange: 0.0401382635<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 3.3734726<br />gene: FBgn0013726","log2FoldChange: 0.3824982931<br />-log10(padj): 4.896415e-01<br />log10(baseMean): 2.1324543<br />gene: FBgn0013732","log2FoldChange: -0.2663178184<br />-log10(padj): 7.343700e-01<br />log10(baseMean): 4.4648279<br />gene: FBgn0013733","log2FoldChange: -0.0668009170<br />-log10(padj): 5.091132e-02<br />log10(baseMean): 3.1234755<br />gene: FBgn0013746","log2FoldChange: -0.1757395501<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.5980970<br />gene: FBgn0013749","log2FoldChange: -0.1386292357<br />-log10(padj): 1.281670e-01<br />log10(baseMean): 3.2974215<br />gene: FBgn0013750","log2FoldChange: 0.1143167624<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.7583029<br />gene: FBgn0013753","log2FoldChange: 0.0079645723<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.7176339<br />gene: FBgn0013756","log2FoldChange: -0.0225403578<br />-log10(padj): 1.375887e-02<br />log10(baseMean): 3.4868758<br />gene: FBgn0013759","log2FoldChange: 0.1218000741<br />-log10(padj): 1.227327e-01<br />log10(baseMean): 2.5246646<br />gene: FBgn0013762","log2FoldChange: -0.6322573330<br />-log10(padj): 3.096008e+00<br />log10(baseMean): 4.4472024<br />gene: FBgn0013763","log2FoldChange: -0.0705501271<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 3.0133002<br />gene: FBgn0013764","log2FoldChange: 0.4963445061<br />-log10(padj): 4.300251e+00<br />log10(baseMean): 3.4335351<br />gene: FBgn0013765","log2FoldChange: -0.2923325225<br />-log10(padj): 6.462291e-01<br />log10(baseMean): 4.4853901<br />gene: FBgn0013770","log2FoldChange: 0.6369398476<br />-log10(padj): 1.223850e+00<br />log10(baseMean): 1.8618817<br />gene: FBgn0013771","log2FoldChange: 0.6307942110<br />-log10(padj): 1.387857e-01<br />log10(baseMean): 0.8649175<br />gene: FBgn0013772","log2FoldChange: 0.4013801396<br />-log10(padj): 7.499770e-01<br />log10(baseMean): 2.1754402<br />gene: FBgn0013773","log2FoldChange: -0.0881549900<br />-log10(padj): 8.129830e-02<br />log10(baseMean): 2.9125259<br />gene: FBgn0013799","log2FoldChange: -0.4394266285<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 1.1970108<br />gene: FBgn0013809","log2FoldChange: 0.3758351958<br />-log10(padj): 1.024186e+00<br />log10(baseMean): 2.6782922<br />gene: FBgn0013810","log2FoldChange: -0.8701172765<br />-log10(padj): 4.916846e+00<br />log10(baseMean): 2.5468058<br />gene: FBgn0013811","log2FoldChange: -0.2629789849<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 0.8265382<br />gene: FBgn0013812","log2FoldChange: 0.5631875600<br />-log10(padj): 2.613819e-01<br />log10(baseMean): 1.3199407<br />gene: FBgn0013813","log2FoldChange: 0.4159977967<br />-log10(padj): 4.208552e-01<br />log10(baseMean): 2.6233915<br />gene: FBgn0013948","log2FoldChange: -0.4352826351<br />-log10(padj): 2.836660e-01<br />log10(baseMean): 1.6064288<br />gene: FBgn0013953","log2FoldChange: 0.1853231159<br />-log10(padj): 3.818032e-01<br />log10(baseMean): 3.6732066<br />gene: FBgn0013954","log2FoldChange: -0.0318401709<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 3.3021012<br />gene: FBgn0013955","log2FoldChange: -0.8306620837<br />-log10(padj): 2.640659e-01<br />log10(baseMean): 1.0003968<br />gene: FBgn0013972","log2FoldChange: -0.1315235402<br />-log10(padj): 1.658154e-01<br />log10(baseMean): 3.5870583<br />gene: FBgn0013981","log2FoldChange: 0.1332624159<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 2.7592728<br />gene: FBgn0013983","log2FoldChange: -0.0734632795<br />-log10(padj): 3.580449e-02<br />log10(baseMean): 3.6697294<br />gene: FBgn0013984","log2FoldChange: -0.0478131617<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 3.4179823<br />gene: FBgn0013987","log2FoldChange: -0.3118218663<br />-log10(padj): 5.293197e-01<br />log10(baseMean): 3.4183138<br />gene: FBgn0013988","log2FoldChange: 0.3737374775<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 0.8740786<br />gene: FBgn0013995","log2FoldChange: 0.3840043701<br />-log10(padj): 1.590228e+00<br />log10(baseMean): 3.2870094<br />gene: FBgn0013997","log2FoldChange: -0.0037002371<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 3.0886137<br />gene: FBgn0013998","log2FoldChange: -0.0685013506<br />-log10(padj): 7.890842e-02<br />log10(baseMean): 3.3276112<br />gene: FBgn0014001","log2FoldChange: 0.1553273029<br />-log10(padj): 4.117913e-01<br />log10(baseMean): 4.2294073<br />gene: FBgn0014002","log2FoldChange: 0.0589291607<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 3.1740201<br />gene: FBgn0014006","log2FoldChange: -0.0942627067<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 3.1734462<br />gene: FBgn0014007","log2FoldChange: -0.2626842406<br />-log10(padj): 1.140944e+00<br />log10(baseMean): 3.2692816<br />gene: FBgn0014009","log2FoldChange: -0.1982815247<br />-log10(padj): 3.396391e-01<br />log10(baseMean): 3.4326310<br />gene: FBgn0014010","log2FoldChange: 0.1638892342<br />-log10(padj): 2.687088e-01<br />log10(baseMean): 3.0330137<br />gene: FBgn0014011","log2FoldChange: -0.1209801216<br />-log10(padj): 2.742267e-01<br />log10(baseMean): 3.4282051<br />gene: FBgn0014018","log2FoldChange: -0.1117989198<br />-log10(padj): 7.916925e-02<br />log10(baseMean): 4.0286303<br />gene: FBgn0014020","log2FoldChange: -0.0118237947<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 2.4182846<br />gene: FBgn0014022","log2FoldChange: 0.1057038482<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.7388930<br />gene: FBgn0014023","log2FoldChange: -0.0818722478<br />-log10(padj): 6.771908e-02<br />log10(baseMean): 3.1390914<br />gene: FBgn0014024","log2FoldChange: -0.0938824734<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 4.6170051<br />gene: FBgn0014026","log2FoldChange: -0.0956847685<br />-log10(padj): 6.301346e-02<br />log10(baseMean): 3.3597831<br />gene: FBgn0014028","log2FoldChange: 0.4867267563<br />-log10(padj): 1.394100e+00<br />log10(baseMean): 3.5336701<br />gene: FBgn0014029","log2FoldChange: 0.7220256065<br />-log10(padj): 3.684044e-01<br />log10(baseMean): 1.3941499<br />gene: FBgn0014031","log2FoldChange: 0.2101689331<br />-log10(padj): 2.273301e-01<br />log10(baseMean): 2.2853276<br />gene: FBgn0014032","log2FoldChange: -0.5173715145<br />-log10(padj): 1.572332e+00<br />log10(baseMean): 3.8125622<br />gene: FBgn0014033","log2FoldChange: 0.0768472289<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 3.7022460<br />gene: FBgn0014037","log2FoldChange: -0.0568960588<br />-log10(padj): 4.675633e-02<br />log10(baseMean): 3.4047090<br />gene: FBgn0014075","log2FoldChange: 0.1847625256<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 2.4151958<br />gene: FBgn0014092","log2FoldChange: 0.2705928061<br />-log10(padj): 9.588953e-01<br />log10(baseMean): 3.0107930<br />gene: FBgn0014127","log2FoldChange: -0.1461501499<br />-log10(padj): 2.140120e-01<br />log10(baseMean): 3.5280692<br />gene: FBgn0014133","log2FoldChange: 0.2129391861<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.1000213<br />gene: FBgn0014135","log2FoldChange: -0.3535205422<br />-log10(padj): 6.909588e-01<br />log10(baseMean): 4.3669087<br />gene: FBgn0014141","log2FoldChange: 0.5987404370<br />-log10(padj): 1.543149e+00<br />log10(baseMean): 3.5781366<br />gene: FBgn0014163","log2FoldChange: -0.1372476393<br />-log10(padj): 1.389968e-01<br />log10(baseMean): 3.7899317<br />gene: FBgn0014184","log2FoldChange: -0.1286698080<br />-log10(padj): 1.615927e-01<br />log10(baseMean): 3.6019076<br />gene: FBgn0014189","log2FoldChange: -0.1197550980<br />-log10(padj): 1.026860e-01<br />log10(baseMean): 2.8580962<br />gene: FBgn0014269","log2FoldChange: -0.0039001715<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 2.8032938<br />gene: FBgn0014340","log2FoldChange: 0.2504609130<br />-log10(padj): 3.978146e-01<br />log10(baseMean): 2.2965050<br />gene: FBgn0014342","log2FoldChange: -1.1873154135<br />-log10(padj): 7.476828e-01<br />log10(baseMean): 1.1525188<br />gene: FBgn0014343","log2FoldChange: -0.0023887419<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 2.9874226<br />gene: FBgn0014362","log2FoldChange: 0.0437367567<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 2.7648566<br />gene: FBgn0014366","log2FoldChange: -0.0520588203<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.0791907<br />gene: FBgn0014380","log2FoldChange: -0.1862069957<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 4.3819662<br />gene: FBgn0014388","log2FoldChange: 0.1822470269<br />-log10(padj): 3.196062e-01<br />log10(baseMean): 2.8843976<br />gene: FBgn0014391","log2FoldChange: -0.2051117181<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 1.4998320<br />gene: FBgn0014395","log2FoldChange: -0.0258838287<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.3039588<br />gene: FBgn0014411","log2FoldChange: -0.3587590800<br />-log10(padj): 1.729550e+00<br />log10(baseMean): 3.1320061<br />gene: FBgn0014417","log2FoldChange: -0.0942087623<br />-log10(padj): 5.254594e-02<br />log10(baseMean): 2.1179352<br />gene: FBgn0014427","log2FoldChange: -0.0053818742<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 3.5391357<br />gene: FBgn0014455","log2FoldChange: -0.0140962797<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 2.8563472<br />gene: FBgn0014467","log2FoldChange: -0.3205235455<br />-log10(padj): 1.484215e+00<br />log10(baseMean): 2.9244632<br />gene: FBgn0014469","log2FoldChange: -0.1884241171<br />-log10(padj): 3.179612e-01<br />log10(baseMean): 3.2101659<br />gene: FBgn0014857","log2FoldChange: 0.3316044521<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.0812469<br />gene: FBgn0014859","log2FoldChange: 0.0678020865<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.2230601<br />gene: FBgn0014861","log2FoldChange: 0.4836462661<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 0.9457154<br />gene: FBgn0014863","log2FoldChange: 0.0376624173<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 3.5851502<br />gene: FBgn0014868","log2FoldChange: 0.5609052245<br />-log10(padj): 3.740917e+00<br />log10(baseMean): 3.1962088<br />gene: FBgn0014869","log2FoldChange: -0.0363726220<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.9787104<br />gene: FBgn0014870","log2FoldChange: 0.0516459301<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.5934341<br />gene: FBgn0014877","log2FoldChange: 0.1853011459<br />-log10(padj): 3.455020e-01<br />log10(baseMean): 3.0379271<br />gene: FBgn0014879","log2FoldChange: -0.4341056380<br />-log10(padj): 3.417691e+00<br />log10(baseMean): 3.5635165<br />gene: FBgn0014906","log2FoldChange: 0.1134779963<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.4587309<br />gene: FBgn0014930","log2FoldChange: -0.3506289801<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 2.4286634<br />gene: FBgn0014931","log2FoldChange: -0.1411368785<br />-log10(padj): 1.786471e-01<br />log10(baseMean): 3.5085153<br />gene: FBgn0014963","log2FoldChange: -0.0742336578<br />-log10(padj): 7.947646e-02<br />log10(baseMean): 2.9632018<br />gene: FBgn0015000","log2FoldChange: 0.2238879314<br />-log10(padj): 5.725357e-01<br />log10(baseMean): 2.9817931<br />gene: FBgn0015014","log2FoldChange: 0.0063073483<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 3.8708888<br />gene: FBgn0015019","log2FoldChange: -0.0636546029<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 3.3957333<br />gene: FBgn0015024","log2FoldChange: -0.0119780492<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 1.9091426<br />gene: FBgn0015025","log2FoldChange: 0.2335338545<br />-log10(padj): 2.065045e-01<br />log10(baseMean): 3.0663242<br />gene: FBgn0015031","log2FoldChange: 0.2101928641<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 1.1031274<br />gene: FBgn0015034","log2FoldChange: 0.1188758940<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.8865007<br />gene: FBgn0015036","log2FoldChange: 0.0101153854<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.1711937<br />gene: FBgn0015037","log2FoldChange: 0.6607138404<br />-log10(padj): 6.654564e-01<br />log10(baseMean): 2.3198341<br />gene: FBgn0015040","log2FoldChange: 0.1317695823<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 2.7367740<br />gene: FBgn0015075","log2FoldChange: -0.1486423439<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 3.4095504<br />gene: FBgn0015218","log2FoldChange: 0.2121801606<br />-log10(padj): 7.716672e-02<br />log10(baseMean): 1.6933352<br />gene: FBgn0015221","log2FoldChange: 0.0326651241<br />-log10(padj): 2.121493e-02<br />log10(baseMean): 3.9936522<br />gene: FBgn0015222","log2FoldChange: 0.1520824767<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.7388123<br />gene: FBgn0015229","log2FoldChange: -0.0377751113<br />-log10(padj): 2.209430e-02<br />log10(baseMean): 2.7201050<br />gene: FBgn0015239","log2FoldChange: -0.2972383691<br />-log10(padj): 1.037479e+00<br />log10(baseMean): 2.7335006<br />gene: FBgn0015240","log2FoldChange: 0.2517136919<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 3.9879974<br />gene: FBgn0015245","log2FoldChange: -0.0395176306<br />-log10(padj): 2.945997e-02<br />log10(baseMean): 3.2588039<br />gene: FBgn0015247","log2FoldChange: -0.2269286195<br />-log10(padj): 4.511030e-01<br />log10(baseMean): 3.8503388<br />gene: FBgn0015268","log2FoldChange: 0.0124180191<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 3.0869071<br />gene: FBgn0015269","log2FoldChange: -0.2199356156<br />-log10(padj): 4.075456e-01<br />log10(baseMean): 2.5770842<br />gene: FBgn0015270","log2FoldChange: -0.1541322714<br />-log10(padj): 1.321434e-01<br />log10(baseMean): 2.3100384<br />gene: FBgn0015271","log2FoldChange: -0.1488820342<br />-log10(padj): 1.080771e-01<br />log10(baseMean): 2.8754537<br />gene: FBgn0015277","log2FoldChange: -0.0150820261<br />-log10(padj): 1.167738e-02<br />log10(baseMean): 3.2546570<br />gene: FBgn0015278","log2FoldChange: 0.0045766876<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.4026782<br />gene: FBgn0015279","log2FoldChange: 0.0735843063<br />-log10(padj): 7.084185e-02<br />log10(baseMean): 3.3028662<br />gene: FBgn0015282","log2FoldChange: 0.0677757974<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 3.0329277<br />gene: FBgn0015283","log2FoldChange: -0.2610498733<br />-log10(padj): 7.169114e-01<br />log10(baseMean): 2.7709715<br />gene: FBgn0015286","log2FoldChange: -0.3900044401<br />-log10(padj): 8.703489e-01<br />log10(baseMean): 3.8503939<br />gene: FBgn0015288","log2FoldChange: 0.1903687006<br />-log10(padj): 4.486032e-01<br />log10(baseMean): 2.9673237<br />gene: FBgn0015295","log2FoldChange: 0.1411255478<br />-log10(padj): 2.175378e-01<br />log10(baseMean): 3.1330170<br />gene: FBgn0015296","log2FoldChange: 0.0608432320<br />-log10(padj): 4.150994e-02<br />log10(baseMean): 2.6404178<br />gene: FBgn0015298","log2FoldChange: -0.0503661980<br />-log10(padj): 1.889413e-02<br />log10(baseMean): 2.8931060<br />gene: FBgn0015299","log2FoldChange: 0.1572147616<br />-log10(padj): 3.810849e-02<br />log10(baseMean): 1.3302349<br />gene: FBgn0015316","log2FoldChange: -0.0111231039<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 3.0553300<br />gene: FBgn0015320","log2FoldChange: 0.1086418826<br />-log10(padj): 1.182827e-01<br />log10(baseMean): 2.7926909<br />gene: FBgn0015321","log2FoldChange: -0.1192924203<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 3.6099297<br />gene: FBgn0015324","log2FoldChange: -0.0010128274<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 2.9345861<br />gene: FBgn0015331","log2FoldChange: 0.4195116370<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 0.9245782<br />gene: FBgn0015336","log2FoldChange: -0.0509289791<br />-log10(padj): 3.647743e-02<br />log10(baseMean): 2.8669381<br />gene: FBgn0015338","log2FoldChange: -0.0815940010<br />-log10(padj): 7.502956e-02<br />log10(baseMean): 2.8239710<br />gene: FBgn0015359","log2FoldChange: -0.2884443171<br />-log10(padj): 1.230030e+00<br />log10(baseMean): 2.9442399<br />gene: FBgn0015360","log2FoldChange: -0.9942605403<br />-log10(padj): 8.394678e+00<br />log10(baseMean): 2.5758676<br />gene: FBgn0015371","log2FoldChange: 0.0311929801<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.5850166<br />gene: FBgn0015372","log2FoldChange: 0.0639650487<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 2.8394998<br />gene: FBgn0015374","log2FoldChange: -0.4106680939<br />-log10(padj): 1.238000e+00<br />log10(baseMean): 2.5277205<br />gene: FBgn0015376","log2FoldChange: 0.2279539246<br />-log10(padj): 3.540780e-01<br />log10(baseMean): 2.8447879<br />gene: FBgn0015379","log2FoldChange: 0.0355678843<br />-log10(padj): 3.158478e-02<br />log10(baseMean): 3.4566619<br />gene: FBgn0015391","log2FoldChange: -0.1593156658<br />-log10(padj): 1.414067e-01<br />log10(baseMean): 3.0240393<br />gene: FBgn0015393","log2FoldChange: 0.2129860703<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 3.1154566<br />gene: FBgn0015396","log2FoldChange: 0.2765942690<br />-log10(padj): 2.704925e-01<br />log10(baseMean): 3.7067249<br />gene: FBgn0015399","log2FoldChange: 0.0222767673<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 2.8400906<br />gene: FBgn0015402","log2FoldChange: -0.2457562804<br />-log10(padj): 8.453524e-01<br />log10(baseMean): 3.5618071<br />gene: FBgn0015477","log2FoldChange: 0.2084478482<br />-log10(padj): 5.471766e-01<br />log10(baseMean): 3.4589884<br />gene: FBgn0015509","log2FoldChange: 0.0106588602<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.5591628<br />gene: FBgn0015513","log2FoldChange: 0.3809322026<br />-log10(padj): 8.577200e-01<br />log10(baseMean): 2.3450696<br />gene: FBgn0015520","log2FoldChange: -0.1746298941<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 4.1882929<br />gene: FBgn0015521","log2FoldChange: -0.8896156795<br />-log10(padj): 3.262004e+00<br />log10(baseMean): 2.1340395<br />gene: FBgn0015522","log2FoldChange: -0.1106722859<br />-log10(padj): 1.129479e-01<br />log10(baseMean): 3.3096636<br />gene: FBgn0015527","log2FoldChange: 0.5313422806<br />-log10(padj): 6.651122e+00<br />log10(baseMean): 3.2471645<br />gene: FBgn0015541","log2FoldChange: 0.2000300080<br />-log10(padj): 2.978285e-01<br />log10(baseMean): 3.4091099<br />gene: FBgn0015542","log2FoldChange: 0.0476443552<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 1.0551036<br />gene: FBgn0015543","log2FoldChange: 0.0836193423<br />-log10(padj): 6.881408e-02<br />log10(baseMean): 2.5695607<br />gene: FBgn0015544","log2FoldChange: 0.0372018767<br />-log10(padj): 2.972559e-02<br />log10(baseMean): 2.8133308<br />gene: FBgn0015546","log2FoldChange: -0.1684912571<br />-log10(padj): 1.105723e-01<br />log10(baseMean): 2.3094553<br />gene: FBgn0015553","log2FoldChange: -0.4401965402<br />-log10(padj): 2.424491e-01<br />log10(baseMean): 1.4824149<br />gene: FBgn0015558","log2FoldChange: 0.3038959017<br />-log10(padj): 1.020122e+00<br />log10(baseMean): 2.7638963<br />gene: FBgn0015562","log2FoldChange: 0.0683864764<br />-log10(padj): 2.538057e-02<br />log10(baseMean): 2.5848074<br />gene: FBgn0015565","log2FoldChange: 0.0570868205<br />-log10(padj): 4.032775e-02<br />log10(baseMean): 3.2881370<br />gene: FBgn0015567","log2FoldChange: 1.0667326408<br />-log10(padj): 8.422748e+00<br />log10(baseMean): 2.3229778<br />gene: FBgn0015568","log2FoldChange: 0.2938078573<br />-log10(padj): 7.412531e-01<br />log10(baseMean): 2.6155422<br />gene: FBgn0015569","log2FoldChange: -0.1029622785<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 2.1292755<br />gene: FBgn0015571","log2FoldChange: 0.5511863128<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 1.0190804<br />gene: FBgn0015572","log2FoldChange: 0.3917152084<br />-log10(padj): 1.086566e+00<br />log10(baseMean): 2.8541533<br />gene: FBgn0015575","log2FoldChange: -0.9240818698<br />-log10(padj): 4.129762e+00<br />log10(baseMean): 2.2483932<br />gene: FBgn0015576","log2FoldChange: -0.2977883284<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 2.5945437<br />gene: FBgn0015577","log2FoldChange: 0.2085389482<br />-log10(padj): 3.329052e-01<br />log10(baseMean): 2.3972393<br />gene: FBgn0015582","log2FoldChange: -0.0815674252<br />-log10(padj): 9.158368e-02<br />log10(baseMean): 3.7570098<br />gene: FBgn0015589","log2FoldChange: -0.3759325617<br />-log10(padj): 7.485074e-01<br />log10(baseMean): 3.0414279<br />gene: FBgn0015600","log2FoldChange: -0.1329163095<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 2.6844648<br />gene: FBgn0015602","log2FoldChange: 0.6237485979<br />-log10(padj): 2.238635e-01<br />log10(baseMean): 1.0926966<br />gene: FBgn0015609","log2FoldChange: -0.1520574312<br />-log10(padj): 3.150560e-01<br />log10(baseMean): 3.3698206<br />gene: FBgn0015610","log2FoldChange: -0.0704403605<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.9802249<br />gene: FBgn0015614","log2FoldChange: -0.1437282188<br />-log10(padj): 3.076228e-01<br />log10(baseMean): 3.2143912<br />gene: FBgn0015615","log2FoldChange: 0.3484223677<br />-log10(padj): 1.153113e+00<br />log10(baseMean): 2.6057318<br />gene: FBgn0015617","log2FoldChange: 0.0038501620<br />-log10(padj): 1.852736e-03<br />log10(baseMean): 2.0600457<br />gene: FBgn0015618","log2FoldChange: 0.1070369093<br />-log10(padj): 8.264702e-02<br />log10(baseMean): 2.7525942<br />gene: FBgn0015621","log2FoldChange: -0.1510762882<br />-log10(padj): 4.194621e-01<br />log10(baseMean): 3.8743106<br />gene: FBgn0015622","log2FoldChange: 0.0080260005<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.6802425<br />gene: FBgn0015623","log2FoldChange: -0.2602422310<br />-log10(padj): 3.279484e-01<br />log10(baseMean): 3.8365901<br />gene: FBgn0015624","log2FoldChange: 0.2211415710<br />-log10(padj): 4.027670e-01<br />log10(baseMean): 2.4476776<br />gene: FBgn0015625","log2FoldChange: -0.2737451106<br />-log10(padj): 7.680011e-01<br />log10(baseMean): 3.3303093<br />gene: FBgn0015657","log2FoldChange: -0.2667608589<br />-log10(padj): 3.258814e-01<br />log10(baseMean): 2.2236106<br />gene: FBgn0015663","log2FoldChange: -0.1721067962<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 3.1664469<br />gene: FBgn0015664","log2FoldChange: -0.2879883158<br />-log10(padj): 5.078607e-01<br />log10(baseMean): 3.0474228<br />gene: FBgn0015714","log2FoldChange: 0.2170819572<br />-log10(padj): 4.397975e-01<br />log10(baseMean): 3.6849955<br />gene: FBgn0015737","log2FoldChange: 0.0434771903<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.2214464<br />gene: FBgn0015754","log2FoldChange: -0.0636753316<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 4.4393882<br />gene: FBgn0015756","log2FoldChange: 0.0401252411<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.4320980<br />gene: FBgn0015765","log2FoldChange: -0.8669448828<br />-log10(padj): 3.473641e-01<br />log10(baseMean): 1.1143539<br />gene: FBgn0015766","log2FoldChange: -0.1392217453<br />-log10(padj): 2.400870e-01<br />log10(baseMean): 2.8708050<br />gene: FBgn0015772","log2FoldChange: -0.2939791106<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 1.4749916<br />gene: FBgn0015773","log2FoldChange: 0.3850550007<br />-log10(padj): 6.774474e-01<br />log10(baseMean): 2.1295756<br />gene: FBgn0015774","log2FoldChange: -0.1417625127<br />-log10(padj): 2.007831e-01<br />log10(baseMean): 3.6026399<br />gene: FBgn0015776","log2FoldChange: 0.5251995375<br />-log10(padj): 3.953634e+00<br />log10(baseMean): 3.0195331<br />gene: FBgn0015777","log2FoldChange: 0.2320354374<br />-log10(padj): 7.234091e-01<br />log10(baseMean): 3.9663626<br />gene: FBgn0015778","log2FoldChange: 0.0419413124<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.3994013<br />gene: FBgn0015781","log2FoldChange: 0.0115395963<br />-log10(padj): 6.267132e-03<br />log10(baseMean): 3.2378104<br />gene: FBgn0015789","log2FoldChange: -0.2397166729<br />-log10(padj): 5.970889e-01<br />log10(baseMean): 3.7274427<br />gene: FBgn0015790","log2FoldChange: -0.0990066151<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 3.1188973<br />gene: FBgn0015791","log2FoldChange: -0.3224983748<br />-log10(padj): 4.076629e-01<br />log10(baseMean): 2.2533437<br />gene: FBgn0015793","log2FoldChange: -0.0715292025<br />-log10(padj): 4.612193e-02<br />log10(baseMean): 2.7968238<br />gene: FBgn0015794","log2FoldChange: -0.4869849053<br />-log10(padj): 2.428087e+00<br />log10(baseMean): 3.3172857<br />gene: FBgn0015795","log2FoldChange: -0.1073384335<br />-log10(padj): 6.223514e-02<br />log10(baseMean): 2.9681508<br />gene: FBgn0015796","log2FoldChange: -0.1530781989<br />-log10(padj): 4.117913e-01<br />log10(baseMean): 3.2607030<br />gene: FBgn0015797","log2FoldChange: -0.0262259459<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.9273973<br />gene: FBgn0015799","log2FoldChange: -0.4247778309<br />-log10(padj): 2.822951e+00<br />log10(baseMean): 2.9137016<br />gene: FBgn0015801","log2FoldChange: -0.1292185720<br />-log10(padj): 2.737850e-01<br />log10(baseMean): 3.2704968<br />gene: FBgn0015803","log2FoldChange: 0.1638271804<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 3.2652443<br />gene: FBgn0015805","log2FoldChange: -0.2342812600<br />-log10(padj): 6.017031e-01<br />log10(baseMean): 3.7017005<br />gene: FBgn0015806","log2FoldChange: 0.1340781792<br />-log10(padj): 9.443517e-02<br />log10(baseMean): 2.8308851<br />gene: FBgn0015808","log2FoldChange: 0.0931093958<br />-log10(padj): 1.009422e-01<br />log10(baseMean): 2.9616700<br />gene: FBgn0015816","log2FoldChange: 0.1062201821<br />-log10(padj): 1.201659e-01<br />log10(baseMean): 3.0154127<br />gene: FBgn0015818","log2FoldChange: -0.1034203394<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.0817719<br />gene: FBgn0015828","log2FoldChange: 0.1006592004<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.2913480<br />gene: FBgn0015829","log2FoldChange: -0.1500688348<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 3.7071589<br />gene: FBgn0015834","log2FoldChange: -0.0521288566<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.2038916<br />gene: FBgn0015838","log2FoldChange: 0.1624822633<br />-log10(padj): 1.287703e-01<br />log10(baseMean): 2.7975969<br />gene: FBgn0015844","log2FoldChange: -0.6673442394<br />-log10(padj): 2.424491e-01<br />log10(baseMean): 1.1735395<br />gene: FBgn0015872","log2FoldChange: -0.4784167561<br />-log10(padj): 1.890835e+00<br />log10(baseMean): 2.8083794<br />gene: FBgn0015903","log2FoldChange: -0.0308099741<br />-log10(padj): 2.221399e-02<br />log10(baseMean): 2.8360801<br />gene: FBgn0015905","log2FoldChange: -0.1113857479<br />-log10(padj): 1.376726e-01<br />log10(baseMean): 3.3345744<br />gene: FBgn0015907","log2FoldChange: -0.1537714353<br />-log10(padj): 2.007831e-01<br />log10(baseMean): 3.2266420<br />gene: FBgn0015924","log2FoldChange: 0.1218396091<br />-log10(padj): 1.669714e-01<br />log10(baseMean): 2.8170125<br />gene: FBgn0015925","log2FoldChange: 0.3732771874<br />-log10(padj): 7.932671e-01<br />log10(baseMean): 2.3819382<br />gene: FBgn0015926","log2FoldChange: 0.1183850495<br />-log10(padj): 1.619829e-01<br />log10(baseMean): 3.2517020<br />gene: FBgn0015929","log2FoldChange: -0.2068741959<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 3.5030985<br />gene: FBgn0015949","log2FoldChange: 0.0142431011<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.0745409<br />gene: FBgn0016031","log2FoldChange: 0.5267732221<br />-log10(padj): 1.269015e+00<br />log10(baseMean): 2.6319115<br />gene: FBgn0016032","log2FoldChange: -0.0748834538<br />-log10(padj): 5.842375e-02<br />log10(baseMean): 2.6421707<br />gene: FBgn0016034","log2FoldChange: -0.0778073533<br />-log10(padj): 4.913937e-02<br />log10(baseMean): 2.3934742<br />gene: FBgn0016038","log2FoldChange: 0.2389302800<br />-log10(padj): 9.812372e-01<br />log10(baseMean): 3.0522872<br />gene: FBgn0016041","log2FoldChange: -0.1098641429<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.4999350<br />gene: FBgn0016054","log2FoldChange: 0.0081572666<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.5262165<br />gene: FBgn0016059","log2FoldChange: -0.2061354510<br />-log10(padj): 4.994296e-01<br />log10(baseMean): 2.8565593<br />gene: FBgn0016070","log2FoldChange: 0.5296388398<br />-log10(padj): 2.241911e+00<br />log10(baseMean): 4.5537611<br />gene: FBgn0016075","log2FoldChange: 0.1142893483<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 2.7178723<br />gene: FBgn0016076","log2FoldChange: -0.0207658396<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.5942576<br />gene: FBgn0016078","log2FoldChange: 0.6733157863<br />-log10(padj): 1.182512e+00<br />log10(baseMean): 1.9868576<br />gene: FBgn0016080","log2FoldChange: -0.2136874488<br />-log10(padj): 4.295200e-01<br />log10(baseMean): 3.6925117<br />gene: FBgn0016081","log2FoldChange: 0.0783044569<br />-log10(padj): 6.187511e-02<br />log10(baseMean): 3.3108817<br />gene: FBgn0016119","log2FoldChange: -0.0614152369<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 3.3544942<br />gene: FBgn0016120","log2FoldChange: -0.0560130704<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.5080711<br />gene: FBgn0016122","log2FoldChange: 0.4267031125<br />-log10(padj): 5.293197e-01<br />log10(baseMean): 2.2074465<br />gene: FBgn0016123","log2FoldChange: -0.0198776628<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.5106757<br />gene: FBgn0016126","log2FoldChange: -0.0221355514<br />-log10(padj): 1.083072e-02<br />log10(baseMean): 2.5938006<br />gene: FBgn0016131","log2FoldChange: -0.1135917166<br />-log10(padj): 1.222563e-01<br />log10(baseMean): 2.8171088<br />gene: FBgn0016641","log2FoldChange: 0.3311018607<br />-log10(padj): 4.883024e-01<br />log10(baseMean): 2.6044813<br />gene: FBgn0016672","log2FoldChange: -0.0316415068<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 1.3864807<br />gene: FBgn0016684","log2FoldChange: 0.0528818625<br />-log10(padj): 6.600542e-02<br />log10(baseMean): 3.4397095<br />gene: FBgn0016685","log2FoldChange: 0.0495278961<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.3124922<br />gene: FBgn0016687","log2FoldChange: 0.1629274492<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 3.3342254<br />gene: FBgn0016691","log2FoldChange: -0.2705201715<br />-log10(padj): 4.918229e-01<br />log10(baseMean): 3.7940972<br />gene: FBgn0016693","log2FoldChange: -0.5532541957<br />-log10(padj): 1.103206e+00<br />log10(baseMean): 3.0819798<br />gene: FBgn0016694","log2FoldChange: 0.0568994789<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.0504049<br />gene: FBgn0016696","log2FoldChange: 0.3090457262<br />-log10(padj): 5.564255e-01<br />log10(baseMean): 3.0241884<br />gene: FBgn0016697","log2FoldChange: 0.1536582642<br />-log10(padj): 2.255230e-01<br />log10(baseMean): 2.6455232<br />gene: FBgn0016698","log2FoldChange: 0.0606725341<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.5992148<br />gene: FBgn0016700","log2FoldChange: -0.1132982269<br />-log10(padj): 1.406823e-01<br />log10(baseMean): 2.8371789<br />gene: FBgn0016701","log2FoldChange: -1.9736456418<br />-log10(padj): 1.906831e+01<br />log10(baseMean): 2.1786100<br />gene: FBgn0016715","log2FoldChange: -0.1968657309<br />-log10(padj): 2.311790e-01<br />log10(baseMean): 3.9223251<br />gene: FBgn0016726","log2FoldChange: -0.2963838652<br />-log10(padj): 8.599722e-01<br />log10(baseMean): 2.9615797<br />gene: FBgn0016754","log2FoldChange: 0.0432809877<br />-log10(padj): 2.763200e-02<br />log10(baseMean): 2.7692330<br />gene: FBgn0016756","log2FoldChange: 0.0891655772<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.4040654<br />gene: FBgn0016762","log2FoldChange: -0.3148529048<br />-log10(padj): 8.213913e-01<br />log10(baseMean): 2.9787994<br />gene: FBgn0016792","log2FoldChange: 0.0213553081<br />-log10(padj): 1.466187e-02<br />log10(baseMean): 3.4194344<br />gene: FBgn0016794","log2FoldChange: 0.2920055000<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 1.8471159<br />gene: FBgn0016797","log2FoldChange: -0.1796446557<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.7223309<br />gene: FBgn0016917","log2FoldChange: -0.3364051858<br />-log10(padj): 2.437044e-01<br />log10(baseMean): 1.8753378<br />gene: FBgn0016919","log2FoldChange: -0.4392371170<br />-log10(padj): 1.845010e+00<br />log10(baseMean): 2.9629489<br />gene: FBgn0016926","log2FoldChange: 0.0254508163<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.1243388<br />gene: FBgn0016930","log2FoldChange: -0.0658993063<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.7255475<br />gene: FBgn0016940","log2FoldChange: 0.2755195001<br />-log10(padj): 2.558107e-01<br />log10(baseMean): 2.0405956<br />gene: FBgn0016970","log2FoldChange: -0.1649946922<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 1.2101178<br />gene: FBgn0016974","log2FoldChange: -0.2178930536<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 4.0264729<br />gene: FBgn0016977","log2FoldChange: -0.2073878239<br />-log10(padj): 2.238635e-01<br />log10(baseMean): 3.0294757<br />gene: FBgn0016978","log2FoldChange: 0.0483377180<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 3.3664168<br />gene: FBgn0016983","log2FoldChange: -0.0268951014<br />-log10(padj): 2.570293e-02<br />log10(baseMean): 3.4094836<br />gene: FBgn0016984","log2FoldChange: 0.1168015002<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.0905014<br />gene: FBgn0017397","log2FoldChange: 0.4821337069<br />-log10(padj): 7.270587e-01<br />log10(baseMean): 1.9849941<br />gene: FBgn0017414","log2FoldChange: -0.1276518264<br />-log10(padj): 2.394155e-01<br />log10(baseMean): 2.9670295<br />gene: FBgn0017418","log2FoldChange: 0.0118899473<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.6237879<br />gene: FBgn0017429","log2FoldChange: -0.0143524941<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.2424176<br />gene: FBgn0017430","log2FoldChange: -0.1047007985<br />-log10(padj): 1.230588e-01<br />log10(baseMean): 3.2963309<br />gene: FBgn0017453","log2FoldChange: 0.6243514346<br />-log10(padj): 3.492311e-01<br />log10(baseMean): 1.3380309<br />gene: FBgn0017456","log2FoldChange: -0.2275906079<br />-log10(padj): 4.351803e-01<br />log10(baseMean): 2.8547171<br />gene: FBgn0017457","log2FoldChange: -0.2270054813<br />-log10(padj): 1.863749e-01<br />log10(baseMean): 1.9856665<br />gene: FBgn0017482","log2FoldChange: 0.0009807954<br />-log10(padj): 8.129480e-04<br />log10(baseMean): 4.4416599<br />gene: FBgn0017545","log2FoldChange: 0.2299657668<br />-log10(padj): 3.871890e-01<br />log10(baseMean): 2.8868889<br />gene: FBgn0017549","log2FoldChange: -0.2108957026<br />-log10(padj): 4.255595e-01<br />log10(baseMean): 3.3226726<br />gene: FBgn0017550","log2FoldChange: 0.4725967714<br />-log10(padj): 1.856296e+00<br />log10(baseMean): 2.4617135<br />gene: FBgn0017551","log2FoldChange: 0.0442192038<br />-log10(padj): 2.155213e-02<br />log10(baseMean): 2.7935458<br />gene: FBgn0017558","log2FoldChange: -0.1315764170<br />-log10(padj): 9.498057e-02<br />log10(baseMean): 3.4692989<br />gene: FBgn0017566","log2FoldChange: -0.0799287718<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.0314039<br />gene: FBgn0017567","log2FoldChange: -0.1311379482<br />-log10(padj): 2.640659e-01<br />log10(baseMean): 3.1806351<br />gene: FBgn0017572","log2FoldChange: 0.2869302869<br />-log10(padj): 8.797138e-01<br />log10(baseMean): 3.1347922<br />gene: FBgn0017577","log2FoldChange: -0.2746180483<br />-log10(padj): 1.397123e-01<br />log10(baseMean): 2.2266789<br />gene: FBgn0017578","log2FoldChange: -0.1076530997<br />-log10(padj): 6.642066e-02<br />log10(baseMean): 4.3736328<br />gene: FBgn0017579","log2FoldChange: -0.0799632781<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 4.3823334<br />gene: FBgn0017581","log2FoldChange: 0.0833586168<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.4134082<br />gene: FBgn0019624","log2FoldChange: 0.1104060009<br />-log10(padj): 1.081876e-01<br />log10(baseMean): 3.0895135<br />gene: FBgn0019637","log2FoldChange: 0.4714873157<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 1.2573722<br />gene: FBgn0019643","log2FoldChange: 0.1251036198<br />-log10(padj): 8.267977e-02<br />log10(baseMean): 3.3274619<br />gene: FBgn0019644","log2FoldChange: -0.1577148621<br />-log10(padj): 1.875434e-02<br />log10(baseMean): 0.8580417<br />gene: FBgn0019650","log2FoldChange: 0.1532385263<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.4041335<br />gene: FBgn0019660","log2FoldChange: 0.0505078846<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 2.1399935<br />gene: FBgn0019661","log2FoldChange: -0.1831940284<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 3.0936564<br />gene: FBgn0019662","log2FoldChange: 0.1108227200<br />-log10(padj): 1.522534e-01<br />log10(baseMean): 2.8878469<br />gene: FBgn0019686","log2FoldChange: 0.0005102460<br />-log10(padj): 6.626888e-04<br />log10(baseMean): 2.8046472<br />gene: FBgn0019830","log2FoldChange: 0.1367295132<br />-log10(padj): 2.909990e-01<br />log10(baseMean): 3.3168599<br />gene: FBgn0019886","log2FoldChange: -0.0634107853<br />-log10(padj): 6.742142e-02<br />log10(baseMean): 3.3595749<br />gene: FBgn0019890","log2FoldChange: 0.0470405261<br />-log10(padj): 3.728001e-02<br />log10(baseMean): 3.1021667<br />gene: FBgn0019925","log2FoldChange: -0.2189668756<br />-log10(padj): 1.024462e-01<br />log10(baseMean): 2.4651405<br />gene: FBgn0019932","log2FoldChange: 0.0004531067<br />-log10(padj): 8.760575e-04<br />log10(baseMean): 4.1285659<br />gene: FBgn0019936","log2FoldChange: -0.2304094160<br />-log10(padj): 4.041189e-01<br />log10(baseMean): 3.2478569<br />gene: FBgn0019938","log2FoldChange: -0.0170902603<br />-log10(padj): 9.053881e-03<br />log10(baseMean): 3.1858728<br />gene: FBgn0019947","log2FoldChange: -0.2012641284<br />-log10(padj): 5.580254e-01<br />log10(baseMean): 3.3593414<br />gene: FBgn0019948","log2FoldChange: 0.0912434214<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 2.5130290<br />gene: FBgn0019949","log2FoldChange: -0.3253575553<br />-log10(padj): 5.108891e-01<br />log10(baseMean): 3.1649946<br />gene: FBgn0019952","log2FoldChange: -0.3612770232<br />-log10(padj): 9.847395e-01<br />log10(baseMean): 3.0675224<br />gene: FBgn0019957","log2FoldChange: -0.0675989695<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.2390063<br />gene: FBgn0019960","log2FoldChange: 0.0106915442<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.3293471<br />gene: FBgn0019968","log2FoldChange: -0.0945919997<br />-log10(padj): 1.165047e-01<br />log10(baseMean): 3.0087401<br />gene: FBgn0019972","log2FoldChange: 0.0251902377<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.7639442<br />gene: FBgn0019982","log2FoldChange: -0.2701775916<br />-log10(padj): 1.101748e-01<br />log10(baseMean): 2.0667205<br />gene: FBgn0019985","log2FoldChange: -0.1963204068<br />-log10(padj): 3.669103e-01<br />log10(baseMean): 3.3158932<br />gene: FBgn0019990","log2FoldChange: 0.2890345107<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 2.4040497<br />gene: FBgn0020018","log2FoldChange: 0.0273882357<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.9895613<br />gene: FBgn0020224","log2FoldChange: -0.0265598537<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.9163321<br />gene: FBgn0020235","log2FoldChange: -0.1463159851<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 3.7091400<br />gene: FBgn0020236","log2FoldChange: 0.1038537462<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 4.1426223<br />gene: FBgn0020238","log2FoldChange: 0.6385266396<br />-log10(padj): 6.029373e+00<br />log10(baseMean): 3.3846820<br />gene: FBgn0020240","log2FoldChange: -0.0709355442<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.2391206<br />gene: FBgn0020245","log2FoldChange: 2.1229718480<br />-log10(padj): 8.335288e+00<br />log10(baseMean): 1.5517719<br />gene: FBgn0020248","log2FoldChange: 0.3903962371<br />-log10(padj): 1.375872e+00<br />log10(baseMean): 2.9977074<br />gene: FBgn0020249","log2FoldChange: -0.1570270870<br />-log10(padj): 1.467644e-01<br />log10(baseMean): 2.5229899<br />gene: FBgn0020251","log2FoldChange: 0.3306539544<br />-log10(padj): 8.238489e-01<br />log10(baseMean): 3.8125383<br />gene: FBgn0020255","log2FoldChange: -0.6796238324<br />-log10(padj): 7.514857e+00<br />log10(baseMean): 3.0334290<br />gene: FBgn0020257","log2FoldChange: 0.1032068499<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.8670538<br />gene: FBgn0020261","log2FoldChange: -0.1855281060<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 0.8024604<br />gene: FBgn0020269","log2FoldChange: 0.2594909146<br />-log10(padj): 1.067209e+00<br />log10(baseMean): 2.9189563<br />gene: FBgn0020270","log2FoldChange: 0.0140281899<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.7811522<br />gene: FBgn0020272","log2FoldChange: 0.0736352852<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.2290878<br />gene: FBgn0020278","log2FoldChange: -0.1375068132<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 4.0385671<br />gene: FBgn0020279","log2FoldChange: 1.0196403129<br />-log10(padj): 4.360557e-01<br />log10(baseMean): 1.0611370<br />gene: FBgn0020294","log2FoldChange: -0.4326001379<br />-log10(padj): 6.690143e-01<br />log10(baseMean): 2.2962878<br />gene: FBgn0020303","log2FoldChange: 0.0072517351<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.6530042<br />gene: FBgn0020304","log2FoldChange: -0.0217613709<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.7512193<br />gene: FBgn0020305","log2FoldChange: -0.1232653646<br />-log10(padj): 1.891520e-01<br />log10(baseMean): 3.8351522<br />gene: FBgn0020306","log2FoldChange: -1.1543860851<br />-log10(padj): 3.292970e-01<br />log10(baseMean): 0.8748422<br />gene: FBgn0020307","log2FoldChange: 0.2572853820<br />-log10(padj): 8.187828e-01<br />log10(baseMean): 3.9645587<br />gene: FBgn0020309","log2FoldChange: -0.0146563570<br />-log10(padj): 7.598195e-03<br />log10(baseMean): 3.1880535<br />gene: FBgn0020312","log2FoldChange: 0.1570849080<br />-log10(padj): 1.839264e-01<br />log10(baseMean): 3.5860681<br />gene: FBgn0020367","log2FoldChange: -0.0140492330<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 2.1946377<br />gene: FBgn0020368","log2FoldChange: 0.1812762644<br />-log10(padj): 2.368080e-01<br />log10(baseMean): 3.4161321<br />gene: FBgn0020369","log2FoldChange: -0.1508957498<br />-log10(padj): 2.399073e-01<br />log10(baseMean): 3.4122638<br />gene: FBgn0020370","log2FoldChange: -0.1712847913<br />-log10(padj): 6.232102e-02<br />log10(baseMean): 1.5860264<br />gene: FBgn0020372","log2FoldChange: -1.4896196331<br />-log10(padj): 3.367848e+00<br />log10(baseMean): 1.6107988<br />gene: FBgn0020376","log2FoldChange: -0.1104618804<br />-log10(padj): 7.959055e-02<br />log10(baseMean): 2.7031189<br />gene: FBgn0020379","log2FoldChange: 0.3039763085<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.1864246<br />gene: FBgn0020381","log2FoldChange: -0.3530246663<br />-log10(padj): 7.297988e-01<br />log10(baseMean): 2.3510794<br />gene: FBgn0020385","log2FoldChange: -0.3871034513<br />-log10(padj): 2.185871e+00<br />log10(baseMean): 3.4717365<br />gene: FBgn0020386","log2FoldChange: 0.0503632758<br />-log10(padj): 4.419180e-02<br />log10(baseMean): 3.1128516<br />gene: FBgn0020388","log2FoldChange: -0.0567934582<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.7337184<br />gene: FBgn0020389","log2FoldChange: 0.6438097758<br />-log10(padj): 3.205801e+00<br />log10(baseMean): 2.3465922<br />gene: FBgn0020391","log2FoldChange: 0.0729079133<br />-log10(padj): 6.536412e-02<br />log10(baseMean): 2.8730351<br />gene: FBgn0020392","log2FoldChange: -0.1167414926<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.5053593<br />gene: FBgn0020407","log2FoldChange: 0.0256657780<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 3.2157463<br />gene: FBgn0020412","log2FoldChange: -0.4285144444<br />-log10(padj): 2.059807e+00<br />log10(baseMean): 3.9675750<br />gene: FBgn0020414","log2FoldChange: -0.2051589599<br />-log10(padj): 3.789448e-01<br />log10(baseMean): 4.1207570<br />gene: FBgn0020415","log2FoldChange: -0.3505949964<br />-log10(padj): 1.388937e+00<br />log10(baseMean): 3.9403245<br />gene: FBgn0020416","log2FoldChange: -0.0869153022<br />-log10(padj): 6.787207e-02<br />log10(baseMean): 2.8993365<br />gene: FBgn0020440","log2FoldChange: -0.0825203520<br />-log10(padj): 1.401391e-01<br />log10(baseMean): 3.7146463<br />gene: FBgn0020443","log2FoldChange: 0.4383035761<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 1.1987652<br />gene: FBgn0020445","log2FoldChange: -0.7756597072<br />-log10(padj): 3.861566e+00<br />log10(baseMean): 2.5263463<br />gene: FBgn0020493","log2FoldChange: 0.2811652417<br />-log10(padj): 7.729714e-01<br />log10(baseMean): 3.3677343<br />gene: FBgn0020496","log2FoldChange: -0.0873581119<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.7466700<br />gene: FBgn0020497","log2FoldChange: -0.2001243513<br />-log10(padj): 1.669714e-01<br />log10(baseMean): 3.1152193<br />gene: FBgn0020503","log2FoldChange: -0.0273635781<br />-log10(padj): 2.063850e-02<br />log10(baseMean): 3.2393742<br />gene: FBgn0020510","log2FoldChange: 0.0894858955<br />-log10(padj): 7.875423e-02<br />log10(baseMean): 3.7532776<br />gene: FBgn0020513","log2FoldChange: -0.0381068511<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 0.8075723<br />gene: FBgn0020521","log2FoldChange: -0.2346119941<br />-log10(padj): 2.532958e-01<br />log10(baseMean): 3.1273753<br />gene: FBgn0020545","log2FoldChange: -0.0604874799<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 3.0765832<br />gene: FBgn0020611","log2FoldChange: -0.1298610641<br />-log10(padj): 1.863749e-01<br />log10(baseMean): 3.0740158<br />gene: FBgn0020616","log2FoldChange: -0.3721977461<br />-log10(padj): 7.755297e-01<br />log10(baseMean): 4.4488583<br />gene: FBgn0020618","log2FoldChange: -0.1620108189<br />-log10(padj): 4.295200e-01<br />log10(baseMean): 3.4042623<br />gene: FBgn0020620","log2FoldChange: 0.0364756686<br />-log10(padj): 3.434146e-02<br />log10(baseMean): 3.2354795<br />gene: FBgn0020621","log2FoldChange: -0.1944274478<br />-log10(padj): 2.556949e-01<br />log10(baseMean): 2.9449748<br />gene: FBgn0020622","log2FoldChange: -0.2125159056<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 1.1679571<br />gene: FBgn0020623","log2FoldChange: -0.0717541449<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 3.1851922<br />gene: FBgn0020626","log2FoldChange: 0.3658182540<br />-log10(padj): 1.630858e+00<br />log10(baseMean): 3.2719616<br />gene: FBgn0020633","log2FoldChange: -0.0108848606<br />-log10(padj): 4.852809e-03<br />log10(baseMean): 3.6041019<br />gene: FBgn0020647","log2FoldChange: 0.2552607977<br />-log10(padj): 5.085214e-01<br />log10(baseMean): 3.7034398<br />gene: FBgn0020653","log2FoldChange: -0.0682948019<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.0195427<br />gene: FBgn0020655","log2FoldChange: -0.2156181586<br />-log10(padj): 5.490181e-01<br />log10(baseMean): 3.3309841<br />gene: FBgn0020756","log2FoldChange: -0.3667564440<br />-log10(padj): 1.144949e+00<br />log10(baseMean): 3.0470153<br />gene: FBgn0020762","log2FoldChange: -0.3766853313<br />-log10(padj): 1.114114e+00<br />log10(baseMean): 2.9139015<br />gene: FBgn0020764","log2FoldChange: -0.1410690827<br />-log10(padj): 2.201481e-01<br />log10(baseMean): 2.7893553<br />gene: FBgn0020766","log2FoldChange: -0.1095162018<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.1020371<br />gene: FBgn0020767","log2FoldChange: -0.3127505049<br />-log10(padj): 1.284776e+00<br />log10(baseMean): 3.0381099<br />gene: FBgn0020887","log2FoldChange: -0.2463400533<br />-log10(padj): 4.300220e-01<br />log10(baseMean): 2.6568715<br />gene: FBgn0020909","log2FoldChange: -0.0751185105<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 4.5420690<br />gene: FBgn0020910","log2FoldChange: -0.0348488511<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.8049575<br />gene: FBgn0020930","log2FoldChange: 0.1636960368<br />-log10(padj): 1.977972e-01<br />log10(baseMean): 2.4787964<br />gene: FBgn0021750","log2FoldChange: -0.0796616248<br />-log10(padj): 1.101748e-01<br />log10(baseMean): 3.4287226<br />gene: FBgn0021760","log2FoldChange: -0.0325620081<br />-log10(padj): 2.131765e-02<br />log10(baseMean): 3.3396686<br />gene: FBgn0021761","log2FoldChange: 0.0348356813<br />-log10(padj): 2.185367e-02<br />log10(baseMean): 2.7996002<br />gene: FBgn0021764","log2FoldChange: 0.0357224969<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 3.1759158<br />gene: FBgn0021765","log2FoldChange: -0.2930468446<br />-log10(padj): 4.926677e-02<br />log10(baseMean): 1.0283406<br />gene: FBgn0021767","log2FoldChange: 0.0578966452<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.9077432<br />gene: FBgn0021768","log2FoldChange: 0.1378890540<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 0.9093889<br />gene: FBgn0021776","log2FoldChange: 0.4355931520<br />-log10(padj): 2.193800e+00<br />log10(baseMean): 3.3384125<br />gene: FBgn0021795","log2FoldChange: 0.1196629887<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 3.3772684<br />gene: FBgn0021796","log2FoldChange: 0.0475329768<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.6526463<br />gene: FBgn0021800","log2FoldChange: -0.1208891616<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8360208<br />gene: FBgn0021814","log2FoldChange: -0.0117533030<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.2273518<br />gene: FBgn0021818","log2FoldChange: 0.0780584101<br />-log10(padj): 7.084185e-02<br />log10(baseMean): 3.2126397<br />gene: FBgn0021825","log2FoldChange: -0.0677488381<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 2.7065464<br />gene: FBgn0021847","log2FoldChange: 0.2036135926<br />-log10(padj): 4.505560e-01<br />log10(baseMean): 2.8018901<br />gene: FBgn0021856","log2FoldChange: -0.0677686035<br />-log10(padj): 6.900827e-02<br />log10(baseMean): 4.1777472<br />gene: FBgn0021872","log2FoldChange: -0.0355644459<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.2575603<br />gene: FBgn0021873","log2FoldChange: -0.1222427503<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 2.7090065<br />gene: FBgn0021874","log2FoldChange: 0.1941725980<br />-log10(padj): 1.892046e-01<br />log10(baseMean): 2.4928277<br />gene: FBgn0021875","log2FoldChange: -0.0146800831<br />-log10(padj): 7.909689e-03<br />log10(baseMean): 3.3728215<br />gene: FBgn0021895","log2FoldChange: -0.1552029731<br />-log10(padj): 1.207488e-01<br />log10(baseMean): 3.3407140<br />gene: FBgn0021906","log2FoldChange: -0.1141184813<br />-log10(padj): 1.511863e-01<br />log10(baseMean): 3.1108554<br />gene: FBgn0021944","log2FoldChange: 0.1733857297<br />-log10(padj): 2.035081e-01<br />log10(baseMean): 3.1240490<br />gene: FBgn0021953","log2FoldChange: 0.0254350429<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.9687736<br />gene: FBgn0021967","log2FoldChange: 0.0080304662<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.2183635<br />gene: FBgn0021979","log2FoldChange: -0.1386218361<br />-log10(padj): 3.135429e-01<br />log10(baseMean): 3.0702197<br />gene: FBgn0021995","log2FoldChange: -0.1161261051<br />-log10(padj): 1.235517e-01<br />log10(baseMean): 3.8224248<br />gene: FBgn0022023","log2FoldChange: 0.0096319049<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.5276680<br />gene: FBgn0022027","log2FoldChange: 0.1599965877<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 3.0301869<br />gene: FBgn0022029","log2FoldChange: -0.0931948496<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 3.2349263<br />gene: FBgn0022063","log2FoldChange: -0.1301922217<br />-log10(padj): 2.140120e-01<br />log10(baseMean): 3.0086564<br />gene: FBgn0022069","log2FoldChange: -0.0966311571<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 2.5757352<br />gene: FBgn0022070","log2FoldChange: -0.2526387317<br />-log10(padj): 6.510071e-01<br />log10(baseMean): 3.6698861<br />gene: FBgn0022085","log2FoldChange: 0.2613808464<br />-log10(padj): 8.111198e-01<br />log10(baseMean): 3.0050010<br />gene: FBgn0022097","log2FoldChange: 0.0271526639<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 3.0366524<br />gene: FBgn0022131","log2FoldChange: 0.0120698940<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.4434553<br />gene: FBgn0022153","log2FoldChange: -0.8566089631<br />-log10(padj): 2.612745e+00<br />log10(baseMean): 3.1226440<br />gene: FBgn0022160","log2FoldChange: -0.1873157981<br />-log10(padj): 4.031715e-01<br />log10(baseMean): 3.4537894<br />gene: FBgn0022201","log2FoldChange: 0.2515430048<br />-log10(padj): 4.353194e-01<br />log10(baseMean): 3.4901852<br />gene: FBgn0022213","log2FoldChange: 0.0781323588<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.0986980<br />gene: FBgn0022224","log2FoldChange: -0.0438140092<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.8018774<br />gene: FBgn0022238","log2FoldChange: 0.2206689228<br />-log10(padj): 1.177783e-01<br />log10(baseMean): 1.8163324<br />gene: FBgn0022246","log2FoldChange: 0.0143025369<br />-log10(padj): 6.861557e-03<br />log10(baseMean): 3.4915962<br />gene: FBgn0022268","log2FoldChange: -0.2137127526<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 2.8159711<br />gene: FBgn0022288","log2FoldChange: -0.4713373521<br />-log10(padj): 4.074381e+00<br />log10(baseMean): 3.0198537<br />gene: FBgn0022338","log2FoldChange: 0.0231277777<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.0447307<br />gene: FBgn0022343","log2FoldChange: 0.0020863815<br />-log10(padj): 1.483411e-03<br />log10(baseMean): 2.6449717<br />gene: FBgn0022344","log2FoldChange: -0.2355606032<br />-log10(padj): 5.056095e-01<br />log10(baseMean): 3.4019218<br />gene: FBgn0022349","log2FoldChange: 0.9867682889<br />-log10(padj): 8.612358e-01<br />log10(baseMean): 1.3476779<br />gene: FBgn0022355","log2FoldChange: 0.3518854559<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 2.3849987<br />gene: FBgn0022359","log2FoldChange: -0.1107528836<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 1.9634802<br />gene: FBgn0022361","log2FoldChange: -0.5270000153<br />-log10(padj): 3.262004e+00<br />log10(baseMean): 2.7839596<br />gene: FBgn0022382","log2FoldChange: 0.0066798374<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.6916057<br />gene: FBgn0022699","log2FoldChange: 0.7276470250<br />-log10(padj): 1.561064e+00<br />log10(baseMean): 2.5264951<br />gene: FBgn0022702","log2FoldChange: 0.0367085870<br />-log10(padj): 3.771920e-02<br />log10(baseMean): 3.2548562<br />gene: FBgn0022708","log2FoldChange: 0.2918481386<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 2.0365743<br />gene: FBgn0022709","log2FoldChange: -0.1931936664<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 2.0212613<br />gene: FBgn0022710","log2FoldChange: 0.0036662685<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 3.2749659<br />gene: FBgn0022720","log2FoldChange: 0.0826822140<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 2.4297629<br />gene: FBgn0022724","log2FoldChange: -0.1944301680<br />-log10(padj): 5.075691e-01<br />log10(baseMean): 3.7322955<br />gene: FBgn0022764","log2FoldChange: -0.1864489585<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 3.3638975<br />gene: FBgn0022768","log2FoldChange: 0.7681518001<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 1.1992057<br />gene: FBgn0022770","log2FoldChange: 0.1196775214<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 2.6387260<br />gene: FBgn0022772","log2FoldChange: 0.2982511364<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.9515043<br />gene: FBgn0022774","log2FoldChange: 0.1012279685<br />-log10(padj): 9.789936e-02<br />log10(baseMean): 2.7875212<br />gene: FBgn0022786","log2FoldChange: 0.1192557618<br />-log10(padj): 2.684169e-01<br />log10(baseMean): 3.6693475<br />gene: FBgn0022787","log2FoldChange: -0.0028977264<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.7477933<br />gene: FBgn0022800","log2FoldChange: 0.2132764041<br />-log10(padj): 6.403508e-01<br />log10(baseMean): 4.1719074<br />gene: FBgn0022893","log2FoldChange: -0.0652811338<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.8167003<br />gene: FBgn0022935","log2FoldChange: 0.1185371761<br />-log10(padj): 1.080771e-01<br />log10(baseMean): 2.4302711<br />gene: FBgn0022936","log2FoldChange: 0.0817217192<br />-log10(padj): 1.094967e-01<br />log10(baseMean): 3.1680694<br />gene: FBgn0022942","log2FoldChange: 0.0932700997<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.4597468<br />gene: FBgn0022943","log2FoldChange: -0.0181894351<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.5886286<br />gene: FBgn0022959","log2FoldChange: 0.1183460419<br />-log10(padj): 2.200097e-01<br />log10(baseMean): 3.1380154<br />gene: FBgn0022960","log2FoldChange: 0.0613713630<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.6748957<br />gene: FBgn0022984","log2FoldChange: 0.1952919338<br />-log10(padj): 5.419624e-01<br />log10(baseMean): 3.1191083<br />gene: FBgn0022985","log2FoldChange: -0.0638238346<br />-log10(padj): 4.260051e-02<br />log10(baseMean): 2.5162457<br />gene: FBgn0022986","log2FoldChange: -0.0882513793<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 2.8998540<br />gene: FBgn0022987","log2FoldChange: 0.0485511036<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.6057428<br />gene: FBgn0023000","log2FoldChange: 0.0794899919<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.5278256<br />gene: FBgn0023023","log2FoldChange: 0.0394951081<br />-log10(padj): 4.103403e-02<br />log10(baseMean): 3.1927432<br />gene: FBgn0023081","log2FoldChange: -0.0357102863<br />-log10(padj): 2.570293e-02<br />log10(baseMean): 3.0584258<br />gene: FBgn0023083","log2FoldChange: 0.4735627260<br />-log10(padj): 8.513032e-02<br />log10(baseMean): 0.9216392<br />gene: FBgn0023090","log2FoldChange: 1.8587888074<br />-log10(padj): 1.415959e+00<br />log10(baseMean): 1.0402463<br />gene: FBgn0023091","log2FoldChange: -0.0431583869<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.3804516<br />gene: FBgn0023094","log2FoldChange: -0.0433166692<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.9499920<br />gene: FBgn0023095","log2FoldChange: 0.1403484424<br />-log10(padj): 3.861693e-02<br />log10(baseMean): 1.5044114<br />gene: FBgn0023096","log2FoldChange: -0.1496620015<br />-log10(padj): 1.931411e-01<br />log10(baseMean): 3.1245712<br />gene: FBgn0023097","log2FoldChange: -0.4540289062<br />-log10(padj): 1.834009e+00<br />log10(baseMean): 2.5568850<br />gene: FBgn0023129","log2FoldChange: -0.0067071399<br />-log10(padj): 4.054271e-03<br />log10(baseMean): 2.6004732<br />gene: FBgn0023130","log2FoldChange: -0.0055545757<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 3.9502978<br />gene: FBgn0023143","log2FoldChange: 0.1275036674<br />-log10(padj): 1.742844e-01<br />log10(baseMean): 2.9053044<br />gene: FBgn0023167","log2FoldChange: -0.3000494035<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 2.9258506<br />gene: FBgn0023169","log2FoldChange: 0.2388466876<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 3.9320805<br />gene: FBgn0023170","log2FoldChange: 0.1680119280<br />-log10(padj): 3.035299e-01<br />log10(baseMean): 2.6154788<br />gene: FBgn0023171","log2FoldChange: -0.0406569642<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 3.6313653<br />gene: FBgn0023172","log2FoldChange: 0.1606027373<br />-log10(padj): 2.407235e-01<br />log10(baseMean): 3.2246194<br />gene: FBgn0023174","log2FoldChange: 0.1936176324<br />-log10(padj): 5.537464e-01<br />log10(baseMean): 3.1107709<br />gene: FBgn0023175","log2FoldChange: 0.0780699345<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.2340005<br />gene: FBgn0023177","log2FoldChange: -0.3837739624<br />-log10(padj): 7.699618e-01<br />log10(baseMean): 2.2826679<br />gene: FBgn0023180","log2FoldChange: -0.3262619276<br />-log10(padj): 1.139055e+00<br />log10(baseMean): 2.6004766<br />gene: FBgn0023181","log2FoldChange: 0.0939031300<br />-log10(padj): 6.765428e-02<br />log10(baseMean): 2.8079966<br />gene: FBgn0023211","log2FoldChange: -0.1228560700<br />-log10(padj): 1.699820e-01<br />log10(baseMean): 3.1227573<br />gene: FBgn0023212","log2FoldChange: -0.2315825223<br />-log10(padj): 1.602229e-01<br />log10(baseMean): 3.9425759<br />gene: FBgn0023213","log2FoldChange: 0.3374077424<br />-log10(padj): 4.334978e-01<br />log10(baseMean): 2.7670672<br />gene: FBgn0023214","log2FoldChange: -0.1211778820<br />-log10(padj): 6.793828e-02<br />log10(baseMean): 2.9970524<br />gene: FBgn0023215","log2FoldChange: -0.0803711808<br />-log10(padj): 7.432824e-02<br />log10(baseMean): 2.5721773<br />gene: FBgn0023216","log2FoldChange: -0.1213380471<br />-log10(padj): 2.558107e-01<br />log10(baseMean): 3.1653234<br />gene: FBgn0023388","log2FoldChange: -0.0768748501<br />-log10(padj): 5.779384e-02<br />log10(baseMean): 2.5990200<br />gene: FBgn0023395","log2FoldChange: 0.0500997441<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.2451839<br />gene: FBgn0023407","log2FoldChange: 0.2249476144<br />-log10(padj): 3.885932e-01<br />log10(baseMean): 2.6744223<br />gene: FBgn0023416","log2FoldChange: 0.0051709162<br />-log10(padj): 3.983899e-03<br />log10(baseMean): 3.2143861<br />gene: FBgn0023423","log2FoldChange: -0.4239680140<br />-log10(padj): 1.997256e-01<br />log10(baseMean): 1.5293839<br />gene: FBgn0023441","log2FoldChange: -0.0839833616<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 3.2104473<br />gene: FBgn0023444","log2FoldChange: -0.0624209885<br />-log10(padj): 3.935620e-02<br />log10(baseMean): 3.7966646<br />gene: FBgn0023458","log2FoldChange: 0.4848428518<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 3.2565562<br />gene: FBgn0023477","log2FoldChange: -1.5253493010<br />-log10(padj): 2.462190e+01<br />log10(baseMean): 3.4902089<br />gene: FBgn0023479","log2FoldChange: -0.1648689671<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 2.3099458<br />gene: FBgn0023506","log2FoldChange: -0.3818286442<br />-log10(padj): 5.648955e-01<br />log10(baseMean): 2.9576507<br />gene: FBgn0023507","log2FoldChange: -0.0729237540<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 3.3103128<br />gene: FBgn0023508","log2FoldChange: 0.1006229772<br />-log10(padj): 8.541730e-02<br />log10(baseMean): 3.0824301<br />gene: FBgn0023509","log2FoldChange: -0.0109165287<br />-log10(padj): 6.267132e-03<br />log10(baseMean): 3.3412761<br />gene: FBgn0023510","log2FoldChange: 0.0826751538<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.2124414<br />gene: FBgn0023511","log2FoldChange: 0.2661219606<br />-log10(padj): 3.247082e-01<br />log10(baseMean): 3.1824778<br />gene: FBgn0023512","log2FoldChange: 0.0089867449<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.8810840<br />gene: FBgn0023513","log2FoldChange: 0.0302542465<br />-log10(padj): 2.579879e-02<br />log10(baseMean): 2.9894955<br />gene: FBgn0023514","log2FoldChange: 0.1475675726<br />-log10(padj): 2.545995e-01<br />log10(baseMean): 2.7806893<br />gene: FBgn0023515","log2FoldChange: -0.0652722490<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.7014694<br />gene: FBgn0023516","log2FoldChange: 0.2140564001<br />-log10(padj): 4.763013e-01<br />log10(baseMean): 2.8474426<br />gene: FBgn0023517","log2FoldChange: -0.1224259508<br />-log10(padj): 1.526116e-01<br />log10(baseMean): 3.4252094<br />gene: FBgn0023518","log2FoldChange: 0.0773670610<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.7692043<br />gene: FBgn0023519","log2FoldChange: 0.3206730202<br />-log10(padj): 5.666215e-01<br />log10(baseMean): 2.2078052<br />gene: FBgn0023520","log2FoldChange: 0.2668762924<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 2.1546144<br />gene: FBgn0023521","log2FoldChange: 0.1966830344<br />-log10(padj): 3.424323e-01<br />log10(baseMean): 2.9411420<br />gene: FBgn0023522","log2FoldChange: -0.1171806373<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 1.0033644<br />gene: FBgn0023523","log2FoldChange: 0.7924508754<br />-log10(padj): 5.264742e-01<br />log10(baseMean): 1.3107752<br />gene: FBgn0023524","log2FoldChange: 0.1576710080<br />-log10(padj): 7.166896e-02<br />log10(baseMean): 1.8619008<br />gene: FBgn0023525","log2FoldChange: -0.1174278791<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.5634027<br />gene: FBgn0023526","log2FoldChange: -0.1792532352<br />-log10(padj): 3.382998e-01<br />log10(baseMean): 2.7204902<br />gene: FBgn0023527","log2FoldChange: -0.0195198681<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 3.1025632<br />gene: FBgn0023528","log2FoldChange: -0.1278428726<br />-log10(padj): 1.431785e-01<br />log10(baseMean): 3.7810426<br />gene: FBgn0023529","log2FoldChange: 0.1728101149<br />-log10(padj): 1.669714e-01<br />log10(baseMean): 2.6524716<br />gene: FBgn0023530","log2FoldChange: 0.1339086682<br />-log10(padj): 1.307513e-01<br />log10(baseMean): 2.8529302<br />gene: FBgn0023531","log2FoldChange: 0.5174060991<br />-log10(padj): 8.483626e-01<br />log10(baseMean): 2.2337111<br />gene: FBgn0023535","log2FoldChange: 0.0483864583<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.9294032<br />gene: FBgn0023536","log2FoldChange: -0.3230477061<br />-log10(padj): 1.011330e+00<br />log10(baseMean): 3.2524792<br />gene: FBgn0023537","log2FoldChange: 0.0307193215<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.2373885<br />gene: FBgn0023540","log2FoldChange: -0.3960759988<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 1.3837639<br />gene: FBgn0023541","log2FoldChange: -0.0980520487<br />-log10(padj): 9.443517e-02<br />log10(baseMean): 2.8975970<br />gene: FBgn0023542","log2FoldChange: 0.3105848507<br />-log10(padj): 1.057636e+00<br />log10(baseMean): 3.0394064<br />gene: FBgn0023545","log2FoldChange: -1.5716365326<br />-log10(padj): 1.008535e+01<br />log10(baseMean): 2.3355341<br />gene: FBgn0023549","log2FoldChange: -0.5991438230<br />-log10(padj): 3.050935e+00<br />log10(baseMean): 3.3412383<br />gene: FBgn0024150","log2FoldChange: 0.4899969396<br />-log10(padj): 8.224991e-01<br />log10(baseMean): 2.6617759<br />gene: FBgn0024177","log2FoldChange: -0.0370085934<br />-log10(padj): 3.168799e-02<br />log10(baseMean): 3.5614266<br />gene: FBgn0024183","log2FoldChange: -0.1090655987<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.0106007<br />gene: FBgn0024187","log2FoldChange: 0.1065559523<br />-log10(padj): 9.220293e-02<br />log10(baseMean): 2.7284872<br />gene: FBgn0024188","log2FoldChange: 0.0391239053<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.4507060<br />gene: FBgn0024191","log2FoldChange: 0.1084307211<br />-log10(padj): 7.965921e-02<br />log10(baseMean): 2.7340337<br />gene: FBgn0024194","log2FoldChange: 0.0305741801<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.9679324<br />gene: FBgn0024196","log2FoldChange: -0.2136081099<br />-log10(padj): 2.685650e-01<br />log10(baseMean): 2.8209142<br />gene: FBgn0024222","log2FoldChange: 0.0008692568<br />-log10(padj): 1.068987e-03<br />log10(baseMean): 2.9140497<br />gene: FBgn0024227","log2FoldChange: -0.2108113413<br />-log10(padj): 1.977972e-01<br />log10(baseMean): 2.4109649<br />gene: FBgn0024230","log2FoldChange: 0.2490419684<br />-log10(padj): 2.347456e-01<br />log10(baseMean): 3.3034751<br />gene: FBgn0024234","log2FoldChange: -0.1521218370<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 3.5945259<br />gene: FBgn0024236","log2FoldChange: -0.2306498350<br />-log10(padj): 9.015875e-01<br />log10(baseMean): 3.5835365<br />gene: FBgn0024238","log2FoldChange: -0.0180258102<br />-log10(padj): 7.564141e-03<br />log10(baseMean): 2.4006974<br />gene: FBgn0024245","log2FoldChange: -0.3244556792<br />-log10(padj): 5.751573e-01<br />log10(baseMean): 2.9466941<br />gene: FBgn0024248","log2FoldChange: -0.7141810421<br />-log10(padj): 3.427043e+00<br />log10(baseMean): 2.5929571<br />gene: FBgn0024250","log2FoldChange: -0.0943951776<br />-log10(padj): 7.571023e-02<br />log10(baseMean): 3.1009645<br />gene: FBgn0024252","log2FoldChange: -0.3284383216<br />-log10(padj): 8.007301e-01<br />log10(baseMean): 3.1947051<br />gene: FBgn0024273","log2FoldChange: 0.2546063598<br />-log10(padj): 1.036394e+00<br />log10(baseMean): 3.8151531<br />gene: FBgn0024277","log2FoldChange: -0.0012263593<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.3274581<br />gene: FBgn0024285","log2FoldChange: -4.5858334065<br />-log10(padj): 1.983438e+01<br />log10(baseMean): 1.7697427<br />gene: FBgn0024288","log2FoldChange: -0.2500511566<br />-log10(padj): 6.730876e-01<br />log10(baseMean): 2.9792063<br />gene: FBgn0024291","log2FoldChange: -0.3268163551<br />-log10(padj): 5.911432e-01<br />log10(baseMean): 4.3857881<br />gene: FBgn0024308","log2FoldChange: 0.0727790924<br />-log10(padj): 5.083022e-02<br />log10(baseMean): 3.1791067<br />gene: FBgn0024314","log2FoldChange: -2.2866984636<br />-log10(padj): 1.960852e+01<br />log10(baseMean): 1.9641843<br />gene: FBgn0024315","log2FoldChange: -0.2871375235<br />-log10(padj): 5.553243e-01<br />log10(baseMean): 3.3144591<br />gene: FBgn0024320","log2FoldChange: -0.0996114844<br />-log10(padj): 8.126759e-02<br />log10(baseMean): 2.9570840<br />gene: FBgn0024321","log2FoldChange: -0.0366275900<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 3.1039201<br />gene: FBgn0024326","log2FoldChange: 0.0766847188<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.4773397<br />gene: FBgn0024329","log2FoldChange: -0.1259802395<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 2.5077801<br />gene: FBgn0024330","log2FoldChange: 0.1598402176<br />-log10(padj): 1.236393e-01<br />log10(baseMean): 3.1204220<br />gene: FBgn0024332","log2FoldChange: 0.0399878938<br />-log10(padj): 2.733646e-02<br />log10(baseMean): 3.5792075<br />gene: FBgn0024352","log2FoldChange: 0.1474089353<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.3477130<br />gene: FBgn0024360","log2FoldChange: -0.1130777914<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 1.3384391<br />gene: FBgn0024361","log2FoldChange: 0.0241310466<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.7633098<br />gene: FBgn0024362","log2FoldChange: -0.0769413104<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.7489479<br />gene: FBgn0024364","log2FoldChange: -0.0034912980<br />-log10(padj): 2.659248e-03<br />log10(baseMean): 3.0252350<br />gene: FBgn0024365","log2FoldChange: 0.1156213756<br />-log10(padj): 2.809186e-02<br />log10(baseMean): 1.5349837<br />gene: FBgn0024366","log2FoldChange: -0.2410749928<br />-log10(padj): 8.612358e-01<br />log10(baseMean): 2.8451940<br />gene: FBgn0024371","log2FoldChange: -0.1829156969<br />-log10(padj): 4.139797e-01<br />log10(baseMean): 3.2345845<br />gene: FBgn0024432","log2FoldChange: 0.6208264324<br />-log10(padj): 4.503592e+00<br />log10(baseMean): 2.7158710<br />gene: FBgn0024491","log2FoldChange: 0.1174475407<br />-log10(padj): 8.273506e-02<br />log10(baseMean): 3.4292876<br />gene: FBgn0024509","log2FoldChange: 0.2400757812<br />-log10(padj): 3.779447e-01<br />log10(baseMean): 2.3565628<br />gene: FBgn0024510","log2FoldChange: 0.0775989970<br />-log10(padj): 5.257367e-02<br />log10(baseMean): 2.2326456<br />gene: FBgn0024542","log2FoldChange: -0.0878850235<br />-log10(padj): 8.004060e-02<br />log10(baseMean): 3.4516361<br />gene: FBgn0024555","log2FoldChange: 0.0745449255<br />-log10(padj): 4.397230e-02<br />log10(baseMean): 3.3072899<br />gene: FBgn0024556","log2FoldChange: 0.0333782128<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.9246683<br />gene: FBgn0024558","log2FoldChange: 0.1234495008<br />-log10(padj): 1.980409e-01<br />log10(baseMean): 2.9194116<br />gene: FBgn0024689","log2FoldChange: -0.2752245420<br />-log10(padj): 4.735582e-01<br />log10(baseMean): 3.2453196<br />gene: FBgn0024698","log2FoldChange: -0.1989973418<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8925046<br />gene: FBgn0024728","log2FoldChange: 0.1857435431<br />-log10(padj): 1.986805e-01<br />log10(baseMean): 2.6605472<br />gene: FBgn0024732","log2FoldChange: 0.1586141818<br />-log10(padj): 9.874229e-02<br />log10(baseMean): 4.1835186<br />gene: FBgn0024733","log2FoldChange: -0.2414646508<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 3.4207978<br />gene: FBgn0024734","log2FoldChange: -0.0679396492<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.8216104<br />gene: FBgn0024753","log2FoldChange: -0.0573118306<br />-log10(padj): 3.287411e-02<br />log10(baseMean): 3.2781145<br />gene: FBgn0024754","log2FoldChange: -0.2083629796<br />-log10(padj): 3.149084e-01<br />log10(baseMean): 3.1557630<br />gene: FBgn0024806","log2FoldChange: 0.0584727416<br />-log10(padj): 3.287411e-02<br />log10(baseMean): 3.2467264<br />gene: FBgn0024807","log2FoldChange: -0.1089295918<br />-log10(padj): 3.030893e-02<br />log10(baseMean): 2.4580875<br />gene: FBgn0024811","log2FoldChange: -0.0244818287<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 3.2412579<br />gene: FBgn0024814","log2FoldChange: -0.0726866764<br />-log10(padj): 7.066721e-02<br />log10(baseMean): 3.1255797<br />gene: FBgn0024832","log2FoldChange: -0.1423811806<br />-log10(padj): 1.782943e-01<br />log10(baseMean): 3.1343495<br />gene: FBgn0024833","log2FoldChange: 1.5034563246<br />-log10(padj): 1.648017e+00<br />log10(baseMean): 1.2163414<br />gene: FBgn0024836","log2FoldChange: 0.3704157544<br />-log10(padj): 1.758016e+00<br />log10(baseMean): 2.8384196<br />gene: FBgn0024841","log2FoldChange: -0.1085730789<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 2.8085542<br />gene: FBgn0024846","log2FoldChange: -0.0662891379<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 2.4282340<br />gene: FBgn0024887","log2FoldChange: 0.0691422715<br />-log10(padj): 6.482097e-02<br />log10(baseMean): 2.8207488<br />gene: FBgn0024889","log2FoldChange: 0.3813341581<br />-log10(padj): 1.529819e+00<br />log10(baseMean): 3.1759617<br />gene: FBgn0024891","log2FoldChange: -0.1035992671<br />-log10(padj): 4.021223e-02<br />log10(baseMean): 2.0727133<br />gene: FBgn0024897","log2FoldChange: -0.1588411685<br />-log10(padj): 1.526703e-01<br />log10(baseMean): 2.4612877<br />gene: FBgn0024909","log2FoldChange: 0.2075860480<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.0527551<br />gene: FBgn0024912","log2FoldChange: -0.2264582741<br />-log10(padj): 4.487936e-01<br />log10(baseMean): 3.2137055<br />gene: FBgn0024920","log2FoldChange: -0.0918629478<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 3.3080135<br />gene: FBgn0024921","log2FoldChange: -0.5463146439<br />-log10(padj): 9.921493e-01<br />log10(baseMean): 4.6942578<br />gene: FBgn0024939","log2FoldChange: 0.0285777911<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.2806624<br />gene: FBgn0024943","log2FoldChange: -0.0709175798<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.6719809<br />gene: FBgn0024945","log2FoldChange: 0.1075805354<br />-log10(padj): 9.840526e-02<br />log10(baseMean): 3.5319865<br />gene: FBgn0024947","log2FoldChange: -0.0131413523<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.6599261<br />gene: FBgn0024956","log2FoldChange: 0.0837426712<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.0944596<br />gene: FBgn0024957","log2FoldChange: 0.0483449094<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 2.9775679<br />gene: FBgn0024958","log2FoldChange: 0.2738192995<br />-log10(padj): 3.702691e-01<br />log10(baseMean): 2.9137397<br />gene: FBgn0024973","log2FoldChange: 0.1052232977<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.3165850<br />gene: FBgn0024975","log2FoldChange: 1.0678024135<br />-log10(padj): 1.561096e+00<br />log10(baseMean): 1.4539868<br />gene: FBgn0024977","log2FoldChange: -0.1910214932<br />-log10(padj): 2.201481e-01<br />log10(baseMean): 2.7558910<br />gene: FBgn0024980","log2FoldChange: 0.0681224739<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 2.3242216<br />gene: FBgn0024983","log2FoldChange: 1.4829847487<br />-log10(padj): 1.145801e+01<br />log10(baseMean): 2.0817950<br />gene: FBgn0024984","log2FoldChange: -0.1095678005<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 2.8825778<br />gene: FBgn0024985","log2FoldChange: -0.1758083737<br />-log10(padj): 2.120283e-01<br />log10(baseMean): 2.4052696<br />gene: FBgn0024986","log2FoldChange: -0.0415509861<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 3.0911203<br />gene: FBgn0024987","log2FoldChange: 0.1411899088<br />-log10(padj): 1.335171e-01<br />log10(baseMean): 2.6696491<br />gene: FBgn0024991","log2FoldChange: 0.2755930808<br />-log10(padj): 6.139946e-01<br />log10(baseMean): 3.2678203<br />gene: FBgn0024992","log2FoldChange: 0.1998017838<br />-log10(padj): 1.711819e-01<br />log10(baseMean): 2.1571326<br />gene: FBgn0024993","log2FoldChange: -0.1903664476<br />-log10(padj): 1.948001e-01<br />log10(baseMean): 2.7897535<br />gene: FBgn0024994","log2FoldChange: 0.1735884584<br />-log10(padj): 1.836853e-01<br />log10(baseMean): 2.8144828<br />gene: FBgn0024996","log2FoldChange: 0.1204327968<br />-log10(padj): 7.424202e-02<br />log10(baseMean): 2.5869441<br />gene: FBgn0024997","log2FoldChange: -0.0150068697<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.4404283<br />gene: FBgn0024998","log2FoldChange: -0.0542730729<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 2.5482896<br />gene: FBgn0025109","log2FoldChange: -0.1226947411<br />-log10(padj): 1.441906e-01<br />log10(baseMean): 3.3473682<br />gene: FBgn0025117","log2FoldChange: -0.1246134023<br />-log10(padj): 2.147990e-01<br />log10(baseMean): 2.9453529<br />gene: FBgn0025140","log2FoldChange: 0.2617899192<br />-log10(padj): 4.674523e-01<br />log10(baseMean): 2.4557653<br />gene: FBgn0025185","log2FoldChange: 0.0035473900<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.6278358<br />gene: FBgn0025186","log2FoldChange: -0.1458341818<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 4.3121491<br />gene: FBgn0025286","log2FoldChange: -0.1663931887<br />-log10(padj): 1.052752e-01<br />log10(baseMean): 3.0135275<br />gene: FBgn0025335","log2FoldChange: 0.0049136337<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.7339978<br />gene: FBgn0025336","log2FoldChange: -0.2436768965<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 3.3794568<br />gene: FBgn0025352","log2FoldChange: -0.2246451270<br />-log10(padj): 2.764490e-01<br />log10(baseMean): 2.4419013<br />gene: FBgn0025355","log2FoldChange: -0.3490510971<br />-log10(padj): 8.669735e-01<br />log10(baseMean): 3.8212877<br />gene: FBgn0025366","log2FoldChange: 0.0209683507<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.8415469<br />gene: FBgn0025373","log2FoldChange: -0.0201644180<br />-log10(padj): 1.124980e-02<br />log10(baseMean): 3.8371888<br />gene: FBgn0025381","log2FoldChange: 0.0782702850<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 2.2880513<br />gene: FBgn0025382","log2FoldChange: -0.8410456565<br />-log10(padj): 1.277061e+00<br />log10(baseMean): 1.9202581<br />gene: FBgn0025383","log2FoldChange: -0.1151477380<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.0613153<br />gene: FBgn0025387","log2FoldChange: -0.0265019724<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.6247916<br />gene: FBgn0025388","log2FoldChange: 0.6427891578<br />-log10(padj): 1.089969e+00<br />log10(baseMean): 2.2015525<br />gene: FBgn0025390","log2FoldChange: -0.0656953186<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 3.2462426<br />gene: FBgn0025394","log2FoldChange: -0.1532075998<br />-log10(padj): 1.515658e-01<br />log10(baseMean): 3.4535210<br />gene: FBgn0025455","log2FoldChange: 0.8062543821<br />-log10(padj): 7.454429e-01<br />log10(baseMean): 1.5604490<br />gene: FBgn0025456","log2FoldChange: 0.0246199272<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 3.0998684<br />gene: FBgn0025457","log2FoldChange: -0.1221204512<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 3.1089896<br />gene: FBgn0025458","log2FoldChange: -0.0082009513<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 3.3763320<br />gene: FBgn0025463","log2FoldChange: 0.1532598133<br />-log10(padj): 1.177172e-01<br />log10(baseMean): 2.4191403<br />gene: FBgn0025469","log2FoldChange: 0.2208973397<br />-log10(padj): 5.778945e-01<br />log10(baseMean): 2.7453411<br />gene: FBgn0025519","log2FoldChange: 0.7177736336<br />-log10(padj): 1.058512e+00<br />log10(baseMean): 1.7720398<br />gene: FBgn0025525","log2FoldChange: -0.0615163139<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 2.5885208<br />gene: FBgn0025549","log2FoldChange: -0.0476191547<br />-log10(padj): 1.931985e-02<br />log10(baseMean): 2.3663378<br />gene: FBgn0025558","log2FoldChange: -0.2320430054<br />-log10(padj): 3.611022e-01<br />log10(baseMean): 3.2609706<br />gene: FBgn0025571","log2FoldChange: 0.4647131846<br />-log10(padj): 1.521890e+00<br />log10(baseMean): 2.6098717<br />gene: FBgn0025574","log2FoldChange: -0.0781330574<br />-log10(padj): 6.990324e-02<br />log10(baseMean): 3.6652619<br />gene: FBgn0025582","log2FoldChange: -0.0156956505<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.8424423<br />gene: FBgn0025592","log2FoldChange: 1.8466158141<br />-log10(padj): 1.440397e+00<br />log10(baseMean): 1.0333184<br />gene: FBgn0025593","log2FoldChange: -0.0664929508<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 3.2001366<br />gene: FBgn0025608","log2FoldChange: -0.0794100861<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.1043671<br />gene: FBgn0025615","log2FoldChange: -0.7809741485<br />-log10(padj): 2.942722e-01<br />log10(baseMean): 1.1933163<br />gene: FBgn0025616","log2FoldChange: 0.0226845522<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.7154666<br />gene: FBgn0025621","log2FoldChange: -0.0511642587<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.9578115<br />gene: FBgn0025624","log2FoldChange: -0.2613819511<br />-log10(padj): 2.809169e-01<br />log10(baseMean): 2.5271430<br />gene: FBgn0025625","log2FoldChange: 0.1425408803<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 3.2370937<br />gene: FBgn0025626","log2FoldChange: -0.0758585687<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 1.3216910<br />gene: FBgn0025627","log2FoldChange: -0.5033652824<br />-log10(padj): 3.164854e+00<br />log10(baseMean): 3.5485725<br />gene: FBgn0025628","log2FoldChange: -0.0480525035<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.8219303<br />gene: FBgn0025629","log2FoldChange: 0.2086879515<br />-log10(padj): 2.303742e-01<br />log10(baseMean): 2.5439719<br />gene: FBgn0025630","log2FoldChange: -0.1461208517<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.9420882<br />gene: FBgn0025631","log2FoldChange: 0.2145244175<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 1.9736180<br />gene: FBgn0025632","log2FoldChange: 0.0383037095<br />-log10(padj): 3.075722e-02<br />log10(baseMean): 2.9307231<br />gene: FBgn0025633","log2FoldChange: -0.1728553541<br />-log10(padj): 1.223780e-01<br />log10(baseMean): 2.3220229<br />gene: FBgn0025634","log2FoldChange: 0.1292627364<br />-log10(padj): 8.308833e-02<br />log10(baseMean): 2.0979017<br />gene: FBgn0025635","log2FoldChange: 0.0163436838<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.0917684<br />gene: FBgn0025637","log2FoldChange: 0.2553083943<br />-log10(padj): 7.234091e-01<br />log10(baseMean): 2.7565573<br />gene: FBgn0025638","log2FoldChange: -0.0774251167<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 3.0325717<br />gene: FBgn0025639","log2FoldChange: 0.0383457851<br />-log10(padj): 2.211415e-02<br />log10(baseMean): 2.5543782<br />gene: FBgn0025640","log2FoldChange: 0.3956956507<br />-log10(padj): 7.704394e-01<br />log10(baseMean): 2.9193422<br />gene: FBgn0025641","log2FoldChange: 0.1687438066<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 1.9354110<br />gene: FBgn0025642","log2FoldChange: 0.3716790816<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 1.1772333<br />gene: FBgn0025643","log2FoldChange: 0.0611153502<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 3.3216291<br />gene: FBgn0025674","log2FoldChange: 0.0684283881<br />-log10(padj): 2.499648e-02<br />log10(baseMean): 2.5630634<br />gene: FBgn0025676","log2FoldChange: 0.5478547650<br />-log10(padj): 5.342133e+00<br />log10(baseMean): 4.0115787<br />gene: FBgn0025678","log2FoldChange: -0.4945096125<br />-log10(padj): 2.441565e+00<br />log10(baseMean): 3.5430882<br />gene: FBgn0025681","log2FoldChange: -0.3303091530<br />-log10(padj): 1.324846e+00<br />log10(baseMean): 3.9477960<br />gene: FBgn0025682","log2FoldChange: -0.2971522569<br />-log10(padj): 2.249640e+00<br />log10(baseMean): 3.6444810<br />gene: FBgn0025683","log2FoldChange: -0.0793241799<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.9047928<br />gene: FBgn0025684","log2FoldChange: -0.3735235755<br />-log10(padj): 1.284760e+00<br />log10(baseMean): 3.6877382<br />gene: FBgn0025687","log2FoldChange: 0.3097824702<br />-log10(padj): 1.977972e-01<br />log10(baseMean): 1.8110015<br />gene: FBgn0025692","log2FoldChange: -0.3861613462<br />-log10(padj): 1.036394e+00<br />log10(baseMean): 2.9317509<br />gene: FBgn0025693","log2FoldChange: -0.3441938417<br />-log10(padj): 3.135429e-01<br />log10(baseMean): 1.8661453<br />gene: FBgn0025697","log2FoldChange: -0.1833914029<br />-log10(padj): 5.854518e-01<br />log10(baseMean): 3.6222938<br />gene: FBgn0025698","log2FoldChange: 0.2682203909<br />-log10(padj): 7.093263e-01<br />log10(baseMean): 3.0429197<br />gene: FBgn0025700","log2FoldChange: -0.2214016103<br />-log10(padj): 6.772263e-01<br />log10(baseMean): 3.5938514<br />gene: FBgn0025701","log2FoldChange: 0.4343939458<br />-log10(padj): 7.086281e-01<br />log10(baseMean): 2.0301052<br />gene: FBgn0025702","log2FoldChange: -0.0450554921<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.2163684<br />gene: FBgn0025712","log2FoldChange: -0.0016832539<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 2.9572913<br />gene: FBgn0025716","log2FoldChange: -0.1094691624<br />-log10(padj): 1.385050e-01<br />log10(baseMean): 2.8333713<br />gene: FBgn0025720","log2FoldChange: -0.0074105783<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 3.6207320<br />gene: FBgn0025724","log2FoldChange: -0.0573181806<br />-log10(padj): 4.021223e-02<br />log10(baseMean): 3.7765950<br />gene: FBgn0025725","log2FoldChange: 0.2394925970<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 3.5399326<br />gene: FBgn0025726","log2FoldChange: 0.3944543771<br />-log10(padj): 1.359839e+00<br />log10(baseMean): 2.7058308<br />gene: FBgn0025739","log2FoldChange: -0.5714822120<br />-log10(padj): 9.881310e-01<br />log10(baseMean): 2.9250410<br />gene: FBgn0025740","log2FoldChange: -0.0337789413<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.4224067<br />gene: FBgn0025741","log2FoldChange: 0.1032278413<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 3.0593698<br />gene: FBgn0025742","log2FoldChange: 0.0290094825<br />-log10(padj): 1.952727e-02<br />log10(baseMean): 2.9622470<br />gene: FBgn0025743","log2FoldChange: -0.0819632967<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.8472335<br />gene: FBgn0025777","log2FoldChange: 0.1600403708<br />-log10(padj): 1.414879e-01<br />log10(baseMean): 2.5376008<br />gene: FBgn0025781","log2FoldChange: -0.0907591717<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 2.9636532<br />gene: FBgn0025790","log2FoldChange: -0.1652529048<br />-log10(padj): 2.640659e-01<br />log10(baseMean): 3.2380723<br />gene: FBgn0025800","log2FoldChange: -0.0879193974<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 3.1623504<br />gene: FBgn0025802","log2FoldChange: 0.0411171775<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 3.4550841<br />gene: FBgn0025803","log2FoldChange: -0.0611673757<br />-log10(padj): 2.322730e-02<br />log10(baseMean): 2.6570623<br />gene: FBgn0025806","log2FoldChange: -0.1583643861<br />-log10(padj): 9.200579e-02<br />log10(baseMean): 2.0986376<br />gene: FBgn0025807","log2FoldChange: 0.2405521737<br />-log10(padj): 2.347456e-01<br />log10(baseMean): 2.1644295<br />gene: FBgn0025808","log2FoldChange: 0.2646786628<br />-log10(padj): 2.608363e-01<br />log10(baseMean): 2.2683168<br />gene: FBgn0025809","log2FoldChange: 0.0415841923<br />-log10(padj): 1.486559e-02<br />log10(baseMean): 3.0073642<br />gene: FBgn0025814","log2FoldChange: 0.1514476371<br />-log10(padj): 1.564430e-01<br />log10(baseMean): 3.3252746<br />gene: FBgn0025815","log2FoldChange: 0.4695215323<br />-log10(padj): 9.843728e-01<br />log10(baseMean): 2.2849819<br />gene: FBgn0025820","log2FoldChange: 0.1065518378<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.8835951<br />gene: FBgn0025825","log2FoldChange: -0.0615757349<br />-log10(padj): 4.958099e-02<br />log10(baseMean): 2.6466161<br />gene: FBgn0025830","log2FoldChange: -0.1251742535<br />-log10(padj): 1.082707e-01<br />log10(baseMean): 2.6531555<br />gene: FBgn0025832","log2FoldChange: -1.1252273598<br />-log10(padj): 3.009787e+00<br />log10(baseMean): 1.9718530<br />gene: FBgn0025836","log2FoldChange: 0.0593370134<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 0.8926034<br />gene: FBgn0025838","log2FoldChange: 0.2507113986<br />-log10(padj): 4.231477e-01<br />log10(baseMean): 2.6411463<br />gene: FBgn0025839","log2FoldChange: 0.0651083135<br />-log10(padj): 4.021223e-02<br />log10(baseMean): 3.6439138<br />gene: FBgn0025864","log2FoldChange: 0.3333968486<br />-log10(padj): 1.568056e+00<br />log10(baseMean): 3.6989179<br />gene: FBgn0025865","log2FoldChange: -0.0210642609<br />-log10(padj): 1.388958e-02<br />log10(baseMean): 3.0730634<br />gene: FBgn0025866","log2FoldChange: 0.0961308410<br />-log10(padj): 6.900827e-02<br />log10(baseMean): 2.4622707<br />gene: FBgn0025874","log2FoldChange: -0.5249588371<br />-log10(padj): 8.188638e-02<br />log10(baseMean): 0.8453333<br />gene: FBgn0025879","log2FoldChange: 0.3254900455<br />-log10(padj): 7.234091e-01<br />log10(baseMean): 4.2622829<br />gene: FBgn0025885","log2FoldChange: 0.0867883132<br />-log10(padj): 2.814148e-02<br />log10(baseMean): 3.0277073<br />gene: FBgn0025936","log2FoldChange: 0.1512711091<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.1696182<br />gene: FBgn0026015","log2FoldChange: 0.0627305778<br />-log10(padj): 4.633512e-02<br />log10(baseMean): 2.5090316<br />gene: FBgn0026056","log2FoldChange: 0.0664439295<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.0550762<br />gene: FBgn0026059","log2FoldChange: 0.1027198700<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.5988583<br />gene: FBgn0026060","log2FoldChange: -0.1799056846<br />-log10(padj): 1.938633e-01<br />log10(baseMean): 2.7424444<br />gene: FBgn0026076","log2FoldChange: 0.0775908226<br />-log10(padj): 5.645306e-02<br />log10(baseMean): 2.8278395<br />gene: FBgn0026079","log2FoldChange: -0.0673951425<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.6785934<br />gene: FBgn0026080","log2FoldChange: 0.2631137569<br />-log10(padj): 4.592152e-01<br />log10(baseMean): 3.6389748<br />gene: FBgn0026083","log2FoldChange: 0.4340975757<br />-log10(padj): 1.065231e+00<br />log10(baseMean): 2.9273608<br />gene: FBgn0026084","log2FoldChange: 0.0270254400<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.2331443<br />gene: FBgn0026085","log2FoldChange: -0.1297202965<br />-log10(padj): 9.795881e-02<br />log10(baseMean): 2.8827350<br />gene: FBgn0026086","log2FoldChange: 0.2828656998<br />-log10(padj): 6.092959e-01<br />log10(baseMean): 2.6518257<br />gene: FBgn0026088","log2FoldChange: -0.0193096387<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.6230274<br />gene: FBgn0026089","log2FoldChange: -0.2331751881<br />-log10(padj): 4.918229e-01<br />log10(baseMean): 2.8508072<br />gene: FBgn0026090","log2FoldChange: -0.3946835286<br />-log10(padj): 1.030985e+00<br />log10(baseMean): 2.4990378<br />gene: FBgn0026143","log2FoldChange: -0.5547732921<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 1.0821149<br />gene: FBgn0026144","log2FoldChange: -0.0940333048<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 2.8558076<br />gene: FBgn0026147","log2FoldChange: 0.2990848798<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 1.7346927<br />gene: FBgn0026148","log2FoldChange: 0.2285519077<br />-log10(padj): 3.329052e-01<br />log10(baseMean): 2.4345315<br />gene: FBgn0026149","log2FoldChange: 0.3911838420<br />-log10(padj): 2.669658e+00<br />log10(baseMean): 2.9903072<br />gene: FBgn0026150","log2FoldChange: -0.2742697241<br />-log10(padj): 1.863749e-01<br />log10(baseMean): 3.2360908<br />gene: FBgn0026160","log2FoldChange: -0.0455099211<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.6297292<br />gene: FBgn0026170","log2FoldChange: 0.5500543646<br />-log10(padj): 2.579744e-01<br />log10(baseMean): 1.3369571<br />gene: FBgn0026174","log2FoldChange: 0.2372292428<br />-log10(padj): 1.788166e-01<br />log10(baseMean): 2.4047226<br />gene: FBgn0026175","log2FoldChange: -0.2069035746<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 1.1029600<br />gene: FBgn0026176","log2FoldChange: 0.6848852043<br />-log10(padj): 2.777385e+00<br />log10(baseMean): 2.6693475<br />gene: FBgn0026179","log2FoldChange: -0.0670762991<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.5712467<br />gene: FBgn0026181","log2FoldChange: 0.3948779359<br />-log10(padj): 2.315537e+00<br />log10(baseMean): 4.0593862<br />gene: FBgn0026189","log2FoldChange: 0.1199820890<br />-log10(padj): 1.096547e-01<br />log10(baseMean): 3.1537265<br />gene: FBgn0026192","log2FoldChange: -0.0867390963<br />-log10(padj): 7.933913e-02<br />log10(baseMean): 3.2850751<br />gene: FBgn0026196","log2FoldChange: 0.0115335826<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 1.7786837<br />gene: FBgn0026197","log2FoldChange: -0.2213495952<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.1253971<br />gene: FBgn0026199","log2FoldChange: 0.0783379591<br />-log10(padj): 6.037484e-02<br />log10(baseMean): 3.3273253<br />gene: FBgn0026206","log2FoldChange: 0.0755710483<br />-log10(padj): 7.166896e-02<br />log10(baseMean): 2.7374479<br />gene: FBgn0026207","log2FoldChange: 0.0815798742<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 3.2406613<br />gene: FBgn0026208","log2FoldChange: -0.0278294758<br />-log10(padj): 7.564141e-03<br />log10(baseMean): 2.8115380<br />gene: FBgn0026238","log2FoldChange: -0.0596732674<br />-log10(padj): 2.122202e-02<br />log10(baseMean): 1.8582205<br />gene: FBgn0026239","log2FoldChange: -0.0078312258<br />-log10(padj): 4.158319e-03<br />log10(baseMean): 3.5729114<br />gene: FBgn0026250","log2FoldChange: -0.0724912974<br />-log10(padj): 1.129479e-01<br />log10(baseMean): 3.9223676<br />gene: FBgn0026252","log2FoldChange: 0.7532329806<br />-log10(padj): 2.974653e-01<br />log10(baseMean): 1.0943053<br />gene: FBgn0026255","log2FoldChange: 0.1704781425<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.0867322<br />gene: FBgn0026257","log2FoldChange: -0.0632155525<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.8216717<br />gene: FBgn0026259","log2FoldChange: 0.0985109163<br />-log10(padj): 6.609605e-02<br />log10(baseMean): 2.5381241<br />gene: FBgn0026261","log2FoldChange: -0.2348098144<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.0222648<br />gene: FBgn0026262","log2FoldChange: 0.2219023562<br />-log10(padj): 2.588560e-01<br />log10(baseMean): 2.2505178<br />gene: FBgn0026263","log2FoldChange: -0.0106592641<br />-log10(padj): 5.265523e-03<br />log10(baseMean): 2.6076551<br />gene: FBgn0026309","log2FoldChange: -0.0041694320<br />-log10(padj): 3.581171e-03<br />log10(baseMean): 3.3434666<br />gene: FBgn0026313","log2FoldChange: -0.2117505809<br />-log10(padj): 2.560635e-01<br />log10(baseMean): 3.0717475<br />gene: FBgn0026315","log2FoldChange: -0.0181291567<br />-log10(padj): 8.278629e-03<br />log10(baseMean): 2.7338380<br />gene: FBgn0026316","log2FoldChange: 0.1104427140<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.9083141<br />gene: FBgn0026317","log2FoldChange: 0.1041386469<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.2501439<br />gene: FBgn0026318","log2FoldChange: 0.4288367643<br />-log10(padj): 1.416180e+00<br />log10(baseMean): 2.7496326<br />gene: FBgn0026319","log2FoldChange: -0.2484045278<br />-log10(padj): 6.066753e-01<br />log10(baseMean): 3.1837309<br />gene: FBgn0026323","log2FoldChange: 0.0193796288<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.3684839<br />gene: FBgn0026324","log2FoldChange: 0.0086521874<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.7313719<br />gene: FBgn0026326","log2FoldChange: 0.2281048879<br />-log10(padj): 3.790452e-01<br />log10(baseMean): 2.4595568<br />gene: FBgn0026361","log2FoldChange: -0.2117326958<br />-log10(padj): 6.756424e-01<br />log10(baseMean): 3.1583265<br />gene: FBgn0026369","log2FoldChange: -0.1270725629<br />-log10(padj): 1.251486e-01<br />log10(baseMean): 3.4263715<br />gene: FBgn0026370","log2FoldChange: 0.0472053985<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 2.5365864<br />gene: FBgn0026371","log2FoldChange: -0.2883521985<br />-log10(padj): 5.380087e-01<br />log10(baseMean): 4.7958115<br />gene: FBgn0026372","log2FoldChange: 0.1026264809<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 2.6653478<br />gene: FBgn0026373","log2FoldChange: 0.1349234710<br />-log10(padj): 1.258130e-01<br />log10(baseMean): 2.4509333<br />gene: FBgn0026374","log2FoldChange: 0.0430552359<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 3.5076930<br />gene: FBgn0026375","log2FoldChange: -1.0061396294<br />-log10(padj): 5.556612e+00<br />log10(baseMean): 3.1462105<br />gene: FBgn0026376","log2FoldChange: 0.2212107839<br />-log10(padj): 4.266784e-01<br />log10(baseMean): 2.6831451<br />gene: FBgn0026378","log2FoldChange: 0.1310498218<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 2.4837293<br />gene: FBgn0026379","log2FoldChange: 0.3434276278<br />-log10(padj): 1.437407e+00<br />log10(baseMean): 3.0715435<br />gene: FBgn0026380","log2FoldChange: 0.1925394020<br />-log10(padj): 1.748230e-01<br />log10(baseMean): 2.1466509<br />gene: FBgn0026393","log2FoldChange: 0.0756639712<br />-log10(padj): 8.199836e-02<br />log10(baseMean): 2.8287025<br />gene: FBgn0026400","log2FoldChange: 0.0999410411<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.4186404<br />gene: FBgn0026401","log2FoldChange: -0.0962441268<br />-log10(padj): 8.517467e-02<br />log10(baseMean): 2.4352642<br />gene: FBgn0026402","log2FoldChange: 0.0432262530<br />-log10(padj): 3.197684e-02<br />log10(baseMean): 2.7264937<br />gene: FBgn0026404","log2FoldChange: -0.2839495955<br />-log10(padj): 6.090994e-01<br />log10(baseMean): 3.9419706<br />gene: FBgn0026409","log2FoldChange: 1.1561010036<br />-log10(padj): 3.298813e+00<br />log10(baseMean): 1.6716893<br />gene: FBgn0026415","log2FoldChange: 0.3700579236<br />-log10(padj): 4.203212e-01<br />log10(baseMean): 2.3511533<br />gene: FBgn0026417","log2FoldChange: -0.2158125547<br />-log10(padj): 5.355160e-01<br />log10(baseMean): 3.9409981<br />gene: FBgn0026418","log2FoldChange: -0.1072098917<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.3883815<br />gene: FBgn0026427","log2FoldChange: -0.0324182967<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.7946815<br />gene: FBgn0026428","log2FoldChange: 0.3086351763<br />-log10(padj): 8.212087e-01<br />log10(baseMean): 3.0156772<br />gene: FBgn0026430","log2FoldChange: 0.1681927437<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.3893340<br />gene: FBgn0026431","log2FoldChange: 0.0914223527<br />-log10(padj): 5.961065e-02<br />log10(baseMean): 2.6510131<br />gene: FBgn0026432","log2FoldChange: 0.3045263052<br />-log10(padj): 5.942977e-01<br />log10(baseMean): 2.5543376<br />gene: FBgn0026433","log2FoldChange: -0.4256261796<br />-log10(padj): 1.415959e+00<br />log10(baseMean): 2.4416860<br />gene: FBgn0026439","log2FoldChange: -0.1124587292<br />-log10(padj): 1.326801e-01<br />log10(baseMean): 3.0167652<br />gene: FBgn0026441","log2FoldChange: 0.1124412793<br />-log10(padj): 1.192651e-01<br />log10(baseMean): 3.2820390<br />gene: FBgn0026479","log2FoldChange: -0.0703200127<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.6540775<br />gene: FBgn0026533","log2FoldChange: -2.3624586337<br />-log10(padj): 2.231268e+01<br />log10(baseMean): 4.6425579<br />gene: FBgn0026562","log2FoldChange: 0.1795201652<br />-log10(padj): 3.019219e-01<br />log10(baseMean): 2.8815949<br />gene: FBgn0026566","log2FoldChange: 0.1611330970<br />-log10(padj): 3.347556e-01<br />log10(baseMean): 3.1080147<br />gene: FBgn0026567","log2FoldChange: -0.0038971869<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.7640563<br />gene: FBgn0026570","log2FoldChange: -0.1065867605<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.0173862<br />gene: FBgn0026573","log2FoldChange: -0.0466020457<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 3.8142870<br />gene: FBgn0026575","log2FoldChange: -0.4280466955<br />-log10(padj): 1.863883e+00<br />log10(baseMean): 2.8947812<br />gene: FBgn0026576","log2FoldChange: -0.0769365368<br />-log10(padj): 5.315955e-02<br />log10(baseMean): 3.6289835<br />gene: FBgn0026577","log2FoldChange: -0.3446836477<br />-log10(padj): 7.947596e-01<br />log10(baseMean): 2.5072476<br />gene: FBgn0026582","log2FoldChange: 0.5341501722<br />-log10(padj): 5.315683e+00<br />log10(baseMean): 3.3057607<br />gene: FBgn0026585","log2FoldChange: -0.9074075413<br />-log10(padj): 6.261658e-01<br />log10(baseMean): 1.2480407<br />gene: FBgn0026592","log2FoldChange: -0.0310400155<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 2.0031675<br />gene: FBgn0026593","log2FoldChange: -0.0261702859<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 3.2070608<br />gene: FBgn0026597","log2FoldChange: 0.3452083186<br />-log10(padj): 1.130951e+00<br />log10(baseMean): 2.5973274<br />gene: FBgn0026598","log2FoldChange: 0.2261696834<br />-log10(padj): 1.672272e-01<br />log10(baseMean): 2.3265425<br />gene: FBgn0026602","log2FoldChange: -0.2464870452<br />-log10(padj): 5.014606e-01<br />log10(baseMean): 3.6790725<br />gene: FBgn0026611","log2FoldChange: 0.1368420480<br />-log10(padj): 1.399139e-01<br />log10(baseMean): 2.5158117<br />gene: FBgn0026616","log2FoldChange: 0.0183822072<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.4680995<br />gene: FBgn0026619","log2FoldChange: -0.0140957473<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 3.4792074<br />gene: FBgn0026620","log2FoldChange: -0.1349531151<br />-log10(padj): 1.274255e-01<br />log10(baseMean): 3.0188699<br />gene: FBgn0026630","log2FoldChange: -0.0432482292<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 2.5229758<br />gene: FBgn0026634","log2FoldChange: 0.1871837708<br />-log10(padj): 1.983976e-01<br />log10(baseMean): 2.2775401<br />gene: FBgn0026666","log2FoldChange: 0.0169353625<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.8968678<br />gene: FBgn0026679","log2FoldChange: -0.0432928002<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.5768838<br />gene: FBgn0026702","log2FoldChange: 0.0422662329<br />-log10(padj): 1.902713e-02<br />log10(baseMean): 3.3168618<br />gene: FBgn0026713","log2FoldChange: 0.3426892833<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 1.8390285<br />gene: FBgn0026718","log2FoldChange: -0.6764094865<br />-log10(padj): 2.480568e+00<br />log10(baseMean): 3.5849453<br />gene: FBgn0026721","log2FoldChange: 0.1147412722<br />-log10(padj): 1.487293e-01<br />log10(baseMean): 2.7637302<br />gene: FBgn0026722","log2FoldChange: 0.1788791175<br />-log10(padj): 1.425243e-01<br />log10(baseMean): 2.3682022<br />gene: FBgn0026737","log2FoldChange: -0.0141864967<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.4525656<br />gene: FBgn0026738","log2FoldChange: -0.1448984294<br />-log10(padj): 1.129479e-01<br />log10(baseMean): 2.3332446<br />gene: FBgn0026741","log2FoldChange: -0.0113382367<br />-log10(padj): 5.431473e-03<br />log10(baseMean): 2.5164460<br />gene: FBgn0026749","log2FoldChange: 0.0309843248<br />-log10(padj): 2.221399e-02<br />log10(baseMean): 2.8879916<br />gene: FBgn0026751","log2FoldChange: 0.1826196120<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 3.2773086<br />gene: FBgn0026753","log2FoldChange: 0.0858915412<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 3.3089325<br />gene: FBgn0026758","log2FoldChange: -1.2691201927<br />-log10(padj): 1.460841e+00<br />log10(baseMean): 1.3064260<br />gene: FBgn0026760","log2FoldChange: -0.2229262066<br />-log10(padj): 6.154252e-01<br />log10(baseMean): 3.0923248<br />gene: FBgn0026761","log2FoldChange: -0.1069251744<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 2.8945573<br />gene: FBgn0026777","log2FoldChange: -0.3298128457<br />-log10(padj): 7.918610e-01<br />log10(baseMean): 2.4094652<br />gene: FBgn0026787","log2FoldChange: -0.1896234231<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 3.4156361<br />gene: FBgn0026869","log2FoldChange: -0.3578339398<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 2.2750045<br />gene: FBgn0026872","log2FoldChange: 0.2554216344<br />-log10(padj): 6.395845e-01<br />log10(baseMean): 2.6686531<br />gene: FBgn0026873","log2FoldChange: -0.4172325409<br />-log10(padj): 2.819451e-01<br />log10(baseMean): 1.5934212<br />gene: FBgn0026874","log2FoldChange: 0.1568547938<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 2.4106298<br />gene: FBgn0026876","log2FoldChange: 0.0429344311<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 0.8741631<br />gene: FBgn0026878","log2FoldChange: 0.0180610523<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.4793944<br />gene: FBgn0026879","log2FoldChange: -0.0131903519<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.4110703<br />gene: FBgn0027052","log2FoldChange: 0.1956952825<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.8923882<br />gene: FBgn0027053","log2FoldChange: 0.0512850084<br />-log10(padj): 3.790338e-02<br />log10(baseMean): 2.7645205<br />gene: FBgn0027054","log2FoldChange: -0.0429382395<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.9159915<br />gene: FBgn0027055","log2FoldChange: -0.0030613684<br />-log10(padj): 2.725201e-03<br />log10(baseMean): 2.7400754<br />gene: FBgn0027057","log2FoldChange: 0.0252046502<br />-log10(padj): 1.375785e-02<br />log10(baseMean): 3.6016903<br />gene: FBgn0027066","log2FoldChange: 0.6401076037<br />-log10(padj): 4.393303e-01<br />log10(baseMean): 1.4895045<br />gene: FBgn0027070","log2FoldChange: 0.1595211320<br />-log10(padj): 2.222556e-01<br />log10(baseMean): 3.5066729<br />gene: FBgn0027079","log2FoldChange: -0.0806303130<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.1737679<br />gene: FBgn0027080","log2FoldChange: -0.0067532002<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 3.6959469<br />gene: FBgn0027081","log2FoldChange: 0.0182916959<br />-log10(padj): 8.177369e-03<br />log10(baseMean): 2.3200698<br />gene: FBgn0027082","log2FoldChange: -0.0726398601<br />-log10(padj): 4.202573e-02<br />log10(baseMean): 2.3466623<br />gene: FBgn0027083","log2FoldChange: 0.0194684627<br />-log10(padj): 1.032306e-02<br />log10(baseMean): 3.2963075<br />gene: FBgn0027084","log2FoldChange: 0.0552886562<br />-log10(padj): 4.537254e-02<br />log10(baseMean): 2.6236092<br />gene: FBgn0027085","log2FoldChange: -0.2578061962<br />-log10(padj): 5.566581e-01<br />log10(baseMean): 3.4784765<br />gene: FBgn0027086","log2FoldChange: 0.1303051901<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.4180747<br />gene: FBgn0027087","log2FoldChange: 0.1906052675<br />-log10(padj): 4.287800e-01<br />log10(baseMean): 3.4168175<br />gene: FBgn0027088","log2FoldChange: 0.0139643353<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 3.0959769<br />gene: FBgn0027090","log2FoldChange: -0.6593365510<br />-log10(padj): 7.042842e+00<br />log10(baseMean): 3.0739813<br />gene: FBgn0027091","log2FoldChange: -0.0323197043<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 3.0405491<br />gene: FBgn0027093","log2FoldChange: 0.0790068155<br />-log10(padj): 8.619087e-02<br />log10(baseMean): 3.7948616<br />gene: FBgn0027094","log2FoldChange: 0.2149898109<br />-log10(padj): 4.291021e-01<br />log10(baseMean): 3.2673883<br />gene: FBgn0027095","log2FoldChange: -0.1165994718<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 2.7406254<br />gene: FBgn0027101","log2FoldChange: 0.1416586376<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.4488018<br />gene: FBgn0027103","log2FoldChange: -0.3273701100<br />-log10(padj): 6.117078e-01<br />log10(baseMean): 2.6609415<br />gene: FBgn0027108","log2FoldChange: -1.0436921317<br />-log10(padj): 7.932671e-01<br />log10(baseMean): 1.3347302<br />gene: FBgn0027111","log2FoldChange: 0.1591354223<br />-log10(padj): 2.238635e-01<br />log10(baseMean): 2.5259289<br />gene: FBgn0027259","log2FoldChange: -1.1920220933<br />-log10(padj): 3.291697e+01<br />log10(baseMean): 3.4705439<br />gene: FBgn0027279","log2FoldChange: -0.0501764759<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 3.7462565<br />gene: FBgn0027280","log2FoldChange: -0.1630155453<br />-log10(padj): 3.329052e-01<br />log10(baseMean): 3.1313426<br />gene: FBgn0027287","log2FoldChange: -0.0327380530<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.3364972<br />gene: FBgn0027291","log2FoldChange: -0.1617739727<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.3443516<br />gene: FBgn0027296","log2FoldChange: 0.0853909407<br />-log10(padj): 7.046529e-02<br />log10(baseMean): 3.8031692<br />gene: FBgn0027329","log2FoldChange: -0.0434805947<br />-log10(padj): 3.467316e-02<br />log10(baseMean): 2.8780617<br />gene: FBgn0027330","log2FoldChange: -0.2915462777<br />-log10(padj): 6.232611e-01<br />log10(baseMean): 2.6896013<br />gene: FBgn0027334","log2FoldChange: 0.0074049520<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 4.0183480<br />gene: FBgn0027335","log2FoldChange: -0.0837210144<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 3.5352576<br />gene: FBgn0027338","log2FoldChange: -0.3542735825<br />-log10(padj): 7.932671e-01<br />log10(baseMean): 3.1479026<br />gene: FBgn0027339","log2FoldChange: -0.0394441270<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.5132130<br />gene: FBgn0027342","log2FoldChange: -1.1842211760<br />-log10(padj): 5.563605e+00<br />log10(baseMean): 2.2379854<br />gene: FBgn0027348","log2FoldChange: -0.0924903616<br />-log10(padj): 7.677997e-02<br />log10(baseMean): 2.7938834<br />gene: FBgn0027356","log2FoldChange: 0.0535124212<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.6354029<br />gene: FBgn0027359","log2FoldChange: -0.0960619284<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 3.2617202<br />gene: FBgn0027363","log2FoldChange: -0.5469187777<br />-log10(padj): 8.153982e-01<br />log10(baseMean): 2.9937817<br />gene: FBgn0027375","log2FoldChange: -0.0662751533<br />-log10(padj): 6.037484e-02<br />log10(baseMean): 2.6276322<br />gene: FBgn0027378","log2FoldChange: -0.0125031020<br />-log10(padj): 7.932954e-03<br />log10(baseMean): 2.9292222<br />gene: FBgn0027453","log2FoldChange: -0.1725455323<br />-log10(padj): 3.280476e-01<br />log10(baseMean): 2.7072990<br />gene: FBgn0027490","log2FoldChange: 0.4576165767<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 1.2868051<br />gene: FBgn0027491","log2FoldChange: -0.1017031149<br />-log10(padj): 1.165047e-01<br />log10(baseMean): 3.7251107<br />gene: FBgn0027492","log2FoldChange: 0.1344943133<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 3.7628621<br />gene: FBgn0027493","log2FoldChange: -0.0525737803<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 2.9234532<br />gene: FBgn0027495","log2FoldChange: -0.0087834210<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.9870584<br />gene: FBgn0027496","log2FoldChange: 0.1345801656<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.1956993<br />gene: FBgn0027497","log2FoldChange: 0.2951968250<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.3495793<br />gene: FBgn0027498","log2FoldChange: -0.1257182781<br />-log10(padj): 9.410274e-02<br />log10(baseMean): 2.9549615<br />gene: FBgn0027499","log2FoldChange: 0.0628014406<br />-log10(padj): 5.895435e-02<br />log10(baseMean): 2.8487810<br />gene: FBgn0027500","log2FoldChange: -0.0179919558<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.9104449<br />gene: FBgn0027503","log2FoldChange: 0.0400445744<br />-log10(padj): 3.177149e-02<br />log10(baseMean): 3.1900288<br />gene: FBgn0027504","log2FoldChange: 0.0520436725<br />-log10(padj): 4.017965e-02<br />log10(baseMean): 2.6084396<br />gene: FBgn0027505","log2FoldChange: -0.2943348316<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 2.7147568<br />gene: FBgn0027506","log2FoldChange: -0.1794921106<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 2.8460639<br />gene: FBgn0027507","log2FoldChange: 0.0130708428<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.8739093<br />gene: FBgn0027508","log2FoldChange: 0.0845163007<br />-log10(padj): 9.597665e-02<br />log10(baseMean): 3.2277880<br />gene: FBgn0027509","log2FoldChange: 0.0227978812<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.8485735<br />gene: FBgn0027512","log2FoldChange: -0.2073420960<br />-log10(padj): 3.339066e-01<br />log10(baseMean): 2.4199545<br />gene: FBgn0027513","log2FoldChange: 0.3219031972<br />-log10(padj): 5.500723e-01<br />log10(baseMean): 2.4506495<br />gene: FBgn0027514","log2FoldChange: 0.9008616887<br />-log10(padj): 1.031943e+01<br />log10(baseMean): 2.6496291<br />gene: FBgn0027515","log2FoldChange: -0.1904176901<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.9458655<br />gene: FBgn0027518","log2FoldChange: 0.1967713372<br />-log10(padj): 6.639638e-02<br />log10(baseMean): 1.4836100<br />gene: FBgn0027521","log2FoldChange: 0.1849405904<br />-log10(padj): 1.782943e-01<br />log10(baseMean): 2.4076611<br />gene: FBgn0027524","log2FoldChange: -0.3658450601<br />-log10(padj): 1.568056e+00<br />log10(baseMean): 3.5199206<br />gene: FBgn0027525","log2FoldChange: 0.3152506557<br />-log10(padj): 4.285157e-01<br />log10(baseMean): 2.4875248<br />gene: FBgn0027526","log2FoldChange: 0.2498936252<br />-log10(padj): 8.666742e-01<br />log10(baseMean): 3.0875209<br />gene: FBgn0027529","log2FoldChange: -0.1356965840<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 2.9752820<br />gene: FBgn0027532","log2FoldChange: -0.1183703810<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 3.0853956<br />gene: FBgn0027534","log2FoldChange: -0.0733628580<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.9538434<br />gene: FBgn0027535","log2FoldChange: -0.0729482431<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 3.5747051<br />gene: FBgn0027537","log2FoldChange: -0.1063126679<br />-log10(padj): 6.243927e-02<br />log10(baseMean): 2.2459053<br />gene: FBgn0027538","log2FoldChange: 0.1341935923<br />-log10(padj): 1.245799e-01<br />log10(baseMean): 2.6817260<br />gene: FBgn0027539","log2FoldChange: -0.0247405997<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 1.0287540<br />gene: FBgn0027544","log2FoldChange: -0.0226629587<br />-log10(padj): 1.790419e-02<br />log10(baseMean): 2.9818381<br />gene: FBgn0027547","log2FoldChange: -0.1192904691<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 3.1357213<br />gene: FBgn0027548","log2FoldChange: -0.1974675104<br />-log10(padj): 4.505560e-01<br />log10(baseMean): 3.0580555<br />gene: FBgn0027549","log2FoldChange: -0.3882182740<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 2.3484895<br />gene: FBgn0027550","log2FoldChange: -0.4993089378<br />-log10(padj): 1.815871e+00<br />log10(baseMean): 3.0075390<br />gene: FBgn0027552","log2FoldChange: -0.1473403095<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 2.6367991<br />gene: FBgn0027553","log2FoldChange: -0.0442391358<br />-log10(padj): 3.415356e-02<br />log10(baseMean): 3.0275246<br />gene: FBgn0027554","log2FoldChange: 0.1183090251<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 2.6169606<br />gene: FBgn0027556","log2FoldChange: -0.0841775194<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.0671373<br />gene: FBgn0027558","log2FoldChange: -0.1139704463<br />-log10(padj): 9.728847e-02<br />log10(baseMean): 2.5503317<br />gene: FBgn0027559","log2FoldChange: 0.5079726574<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 0.8738875<br />gene: FBgn0027560","log2FoldChange: -0.4428025748<br />-log10(padj): 1.507634e+00<br />log10(baseMean): 3.3275452<br />gene: FBgn0027561","log2FoldChange: -0.1105282819<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 1.1373569<br />gene: FBgn0027563","log2FoldChange: 0.1812748767<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 2.5650861<br />gene: FBgn0027564","log2FoldChange: -0.1092605333<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 2.5307424<br />gene: FBgn0027565","log2FoldChange: 0.0293014640<br />-log10(padj): 1.891881e-02<br />log10(baseMean): 3.4512325<br />gene: FBgn0027567","log2FoldChange: -0.0256949161<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.2737455<br />gene: FBgn0027568","log2FoldChange: -0.1824474964<br />-log10(padj): 4.437963e-01<br />log10(baseMean): 3.2798210<br />gene: FBgn0027569","log2FoldChange: -0.8971272273<br />-log10(padj): 8.099063e-01<br />log10(baseMean): 1.4045940<br />gene: FBgn0027570","log2FoldChange: 0.2599415714<br />-log10(padj): 8.280480e-01<br />log10(baseMean): 4.8441582<br />gene: FBgn0027571","log2FoldChange: 0.0251331627<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.9163299<br />gene: FBgn0027572","log2FoldChange: -0.1595095745<br />-log10(padj): 1.798294e-01<br />log10(baseMean): 2.6271732<br />gene: FBgn0027574","log2FoldChange: -0.0680801520<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.4765549<br />gene: FBgn0027575","log2FoldChange: -0.5072079135<br />-log10(padj): 4.151904e+00<br />log10(baseMean): 3.1375765<br />gene: FBgn0027578","log2FoldChange: -0.2670583209<br />-log10(padj): 8.385468e-01<br />log10(baseMean): 3.0907117<br />gene: FBgn0027579","log2FoldChange: -0.8448775519<br />-log10(padj): 5.414132e+00<br />log10(baseMean): 3.1172905<br />gene: FBgn0027580","log2FoldChange: 0.0528119508<br />-log10(padj): 5.660673e-02<br />log10(baseMean): 3.1642223<br />gene: FBgn0027581","log2FoldChange: -0.1231737241<br />-log10(padj): 2.352411e-01<br />log10(baseMean): 3.3293909<br />gene: FBgn0027582","log2FoldChange: 0.1212931152<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 2.9403104<br />gene: FBgn0027583","log2FoldChange: -0.7935564445<br />-log10(padj): 7.318301e-01<br />log10(baseMean): 1.6965423<br />gene: FBgn0027585","log2FoldChange: -0.2312671881<br />-log10(padj): 3.548792e-01<br />log10(baseMean): 3.0267758<br />gene: FBgn0027587","log2FoldChange: -0.1814796185<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 3.7041360<br />gene: FBgn0027588","log2FoldChange: -0.5525935265<br />-log10(padj): 3.525957e+00<br />log10(baseMean): 2.6663097<br />gene: FBgn0027590","log2FoldChange: -0.1780215010<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 3.1100163<br />gene: FBgn0027592","log2FoldChange: -0.5249659450<br />-log10(padj): 2.443435e+00<br />log10(baseMean): 3.8457493<br />gene: FBgn0027594","log2FoldChange: -0.8349625872<br />-log10(padj): 5.055793e+00<br />log10(baseMean): 2.8770967<br />gene: FBgn0027596","log2FoldChange: 0.6485491090<br />-log10(padj): 5.563605e+00<br />log10(baseMean): 2.6224628<br />gene: FBgn0027597","log2FoldChange: 0.2533948959<br />-log10(padj): 8.318124e-01<br />log10(baseMean): 3.8505569<br />gene: FBgn0027598","log2FoldChange: 0.1027155023<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.5868389<br />gene: FBgn0027599","log2FoldChange: -0.3348613998<br />-log10(padj): 9.684093e-01<br />log10(baseMean): 2.6586469<br />gene: FBgn0027601","log2FoldChange: -0.0042688613<br />-log10(padj): 3.057405e-03<br />log10(baseMean): 2.7600431<br />gene: FBgn0027602","log2FoldChange: -0.1246802488<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.2763726<br />gene: FBgn0027603","log2FoldChange: -0.5808524835<br />-log10(padj): 1.026584e+00<br />log10(baseMean): 2.4516206<br />gene: FBgn0027604","log2FoldChange: -0.0908855865<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 3.5564661<br />gene: FBgn0027605","log2FoldChange: -0.2285034708<br />-log10(padj): 8.154356e-01<br />log10(baseMean): 3.3310419<br />gene: FBgn0027607","log2FoldChange: -0.1160042245<br />-log10(padj): 1.331016e-01<br />log10(baseMean): 2.8465400<br />gene: FBgn0027609","log2FoldChange: -0.2271994602<br />-log10(padj): 8.110079e-01<br />log10(baseMean): 3.2182338<br />gene: FBgn0027610","log2FoldChange: 0.0634239868<br />-log10(padj): 7.702384e-02<br />log10(baseMean): 3.4631858<br />gene: FBgn0027611","log2FoldChange: 0.0678800048<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 2.4150800<br />gene: FBgn0027615","log2FoldChange: 0.0374258054<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.2470183<br />gene: FBgn0027616","log2FoldChange: 0.0722855742<br />-log10(padj): 4.487485e-02<br />log10(baseMean): 2.3487520<br />gene: FBgn0027617","log2FoldChange: -0.0112251759<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 3.2769572<br />gene: FBgn0027619","log2FoldChange: -0.3069785033<br />-log10(padj): 1.165532e+00<br />log10(baseMean): 3.3110250<br />gene: FBgn0027620","log2FoldChange: -0.3717776838<br />-log10(padj): 1.110429e+00<br />log10(baseMean): 2.9052807<br />gene: FBgn0027621","log2FoldChange: -0.2155942844<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.5380107<br />gene: FBgn0027654","log2FoldChange: -0.0728471937<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.4715123<br />gene: FBgn0027655","log2FoldChange: -0.4704727144<br />-log10(padj): 1.969127e+00<br />log10(baseMean): 2.5998476<br />gene: FBgn0027657","log2FoldChange: -0.1615221236<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 1.9960723<br />gene: FBgn0027660","log2FoldChange: -0.0944883812<br />-log10(padj): 9.260568e-02<br />log10(baseMean): 3.3165285<br />gene: FBgn0027779","log2FoldChange: 0.0115827146<br />-log10(padj): 5.423471e-03<br />log10(baseMean): 2.5872714<br />gene: FBgn0027780","log2FoldChange: -0.1422524354<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.1974238<br />gene: FBgn0027783","log2FoldChange: 0.1495713220<br />-log10(padj): 9.544956e-02<br />log10(baseMean): 2.1556774<br />gene: FBgn0027784","log2FoldChange: 0.1188015225<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 3.0411092<br />gene: FBgn0027785","log2FoldChange: 0.0999903960<br />-log10(padj): 1.136128e-01<br />log10(baseMean): 3.0705069<br />gene: FBgn0027786","log2FoldChange: -0.1359886201<br />-log10(padj): 7.495853e-02<br />log10(baseMean): 2.9029455<br />gene: FBgn0027791","log2FoldChange: 0.0878693938<br />-log10(padj): 1.685188e-02<br />log10(baseMean): 1.4311871<br />gene: FBgn0027793","log2FoldChange: 0.0103269057<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 2.8907445<br />gene: FBgn0027794","log2FoldChange: 0.1597430546<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 1.8642776<br />gene: FBgn0027795","log2FoldChange: -0.0640425994<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 3.9712700<br />gene: FBgn0027835","log2FoldChange: 0.5796844822<br />-log10(padj): 3.036947e+00<br />log10(baseMean): 3.5189816<br />gene: FBgn0027836","log2FoldChange: 0.2350610113<br />-log10(padj): 2.978285e-01<br />log10(baseMean): 2.3702770<br />gene: FBgn0027841","log2FoldChange: -0.2258657447<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 3.0560834<br />gene: FBgn0027842","log2FoldChange: 0.5941991082<br />-log10(padj): 3.120745e+00<br />log10(baseMean): 2.8709700<br />gene: FBgn0027843","log2FoldChange: -0.3488430523<br />-log10(padj): 2.390894e-01<br />log10(baseMean): 2.0124078<br />gene: FBgn0027864","log2FoldChange: 0.6331449353<br />-log10(padj): 1.340977e+00<br />log10(baseMean): 2.6099881<br />gene: FBgn0027865","log2FoldChange: -0.1040548122<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 3.2834172<br />gene: FBgn0027866","log2FoldChange: -0.1340842273<br />-log10(padj): 9.544956e-02<br />log10(baseMean): 3.2495236<br />gene: FBgn0027868","log2FoldChange: 0.2653519443<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 2.1773275<br />gene: FBgn0027872","log2FoldChange: 0.0171692300<br />-log10(padj): 1.079808e-02<br />log10(baseMean): 2.8323662<br />gene: FBgn0027873","log2FoldChange: 0.2874315764<br />-log10(padj): 7.632928e-01<br />log10(baseMean): 3.3643088<br />gene: FBgn0027885","log2FoldChange: 0.2142847489<br />-log10(padj): 7.399142e-01<br />log10(baseMean): 3.0909926<br />gene: FBgn0027889","log2FoldChange: -0.0660698374<br />-log10(padj): 5.118578e-02<br />log10(baseMean): 2.7178816<br />gene: FBgn0027903","log2FoldChange: 0.2404916877<br />-log10(padj): 3.810681e-01<br />log10(baseMean): 2.3035130<br />gene: FBgn0027914","log2FoldChange: 0.4657139716<br />-log10(padj): 1.049229e+00<br />log10(baseMean): 2.6944421<br />gene: FBgn0027929","log2FoldChange: 0.1126777131<br />-log10(padj): 1.866767e-01<br />log10(baseMean): 3.0842980<br />gene: FBgn0027930","log2FoldChange: 0.2732676811<br />-log10(padj): 1.127608e+00<br />log10(baseMean): 4.1455598<br />gene: FBgn0027932","log2FoldChange: 0.1832217982<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.8443373<br />gene: FBgn0027936","log2FoldChange: 0.5543352735<br />-log10(padj): 1.208505e+00<br />log10(baseMean): 2.3116495<br />gene: FBgn0027945","log2FoldChange: 0.2601486728<br />-log10(padj): 5.660703e-01<br />log10(baseMean): 3.8965997<br />gene: FBgn0027948","log2FoldChange: 0.6462232079<br />-log10(padj): 7.850214e+00<br />log10(baseMean): 3.1608229<br />gene: FBgn0027949","log2FoldChange: 0.1768691933<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 2.6148706<br />gene: FBgn0027950","log2FoldChange: -0.1270723338<br />-log10(padj): 1.807875e-01<br />log10(baseMean): 3.3472933<br />gene: FBgn0027951","log2FoldChange: -0.0434332835<br />-log10(padj): 2.710448e-02<br />log10(baseMean): 2.9351064<br />gene: FBgn0028274","log2FoldChange: -0.0499156400<br />-log10(padj): 3.566099e-02<br />log10(baseMean): 2.6743876<br />gene: FBgn0028292","log2FoldChange: -0.3263275513<br />-log10(padj): 9.091652e-01<br />log10(baseMean): 3.4290508<br />gene: FBgn0028325","log2FoldChange: 0.6074742268<br />-log10(padj): 6.437222e+00<br />log10(baseMean): 3.5766130<br />gene: FBgn0028327","log2FoldChange: -0.3629399002<br />-log10(padj): 1.309617e+00<br />log10(baseMean): 3.6507662<br />gene: FBgn0028331","log2FoldChange: 0.0907253834<br />-log10(padj): 7.570770e-02<br />log10(baseMean): 3.2009224<br />gene: FBgn0028336","log2FoldChange: -0.1152597109<br />-log10(padj): 9.428386e-02<br />log10(baseMean): 3.0122313<br />gene: FBgn0028341","log2FoldChange: 0.1488567031<br />-log10(padj): 1.638394e-01<br />log10(baseMean): 3.1775578<br />gene: FBgn0028342","log2FoldChange: 0.1170463943<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 3.2919400<br />gene: FBgn0028343","log2FoldChange: 0.3098767842<br />-log10(padj): 8.224991e-01<br />log10(baseMean): 2.8251178<br />gene: FBgn0028360","log2FoldChange: 0.1631547491<br />-log10(padj): 1.836853e-01<br />log10(baseMean): 2.4313980<br />gene: FBgn0028372","log2FoldChange: 1.2966293221<br />-log10(padj): 2.358199e+00<br />log10(baseMean): 1.6238788<br />gene: FBgn0028373","log2FoldChange: 0.2229618307<br />-log10(padj): 3.279484e-01<br />log10(baseMean): 2.8868510<br />gene: FBgn0028375","log2FoldChange: -0.0183449848<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 0.9579660<br />gene: FBgn0028379","log2FoldChange: -0.1431887444<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 2.6953060<br />gene: FBgn0028380","log2FoldChange: 0.0409430631<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.4088585<br />gene: FBgn0028382","log2FoldChange: -0.3904547326<br />-log10(padj): 8.534688e-01<br />log10(baseMean): 3.8052616<br />gene: FBgn0028386","log2FoldChange: -0.0914487183<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.5926092<br />gene: FBgn0028387","log2FoldChange: -0.6735163959<br />-log10(padj): 2.332214e+00<br />log10(baseMean): 2.1759024<br />gene: FBgn0028394","log2FoldChange: -0.7454091241<br />-log10(padj): 2.611743e+00<br />log10(baseMean): 2.5489840<br />gene: FBgn0028397","log2FoldChange: -0.4159133208<br />-log10(padj): 4.117913e-01<br />log10(baseMean): 2.3364043<br />gene: FBgn0028398","log2FoldChange: -0.2464818703<br />-log10(padj): 4.212496e-01<br />log10(baseMean): 3.4791393<br />gene: FBgn0028399","log2FoldChange: -0.0787900726<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.9150423<br />gene: FBgn0028401","log2FoldChange: 0.0344472143<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.5225582<br />gene: FBgn0028402","log2FoldChange: -0.0302933804<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.3159532<br />gene: FBgn0028406","log2FoldChange: -0.3499313644<br />-log10(padj): 1.751247e-01<br />log10(baseMean): 1.5939366<br />gene: FBgn0028408","log2FoldChange: 0.1859576229<br />-log10(padj): 1.816269e-01<br />log10(baseMean): 2.6503381<br />gene: FBgn0028411","log2FoldChange: 0.7443176929<br />-log10(padj): 2.024417e-01<br />log10(baseMean): 0.9351053<br />gene: FBgn0028418","log2FoldChange: -0.0250912505<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.2633985<br />gene: FBgn0028419","log2FoldChange: -0.3456562783<br />-log10(padj): 8.721040e-01<br />log10(baseMean): 3.1495735<br />gene: FBgn0028420","log2FoldChange: 0.0478727571<br />-log10(padj): 4.218202e-02<br />log10(baseMean): 2.8412516<br />gene: FBgn0028421","log2FoldChange: 0.1693332650<br />-log10(padj): 1.823087e-01<br />log10(baseMean): 3.0379929<br />gene: FBgn0028424","log2FoldChange: -0.2231845319<br />-log10(padj): 8.453524e-01<br />log10(baseMean): 3.5528332<br />gene: FBgn0028425","log2FoldChange: -0.2025677487<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 2.7880492<br />gene: FBgn0028426","log2FoldChange: 0.1547603178<br />-log10(padj): 3.007806e-01<br />log10(baseMean): 2.9436182<br />gene: FBgn0028427","log2FoldChange: 0.4606420843<br />-log10(padj): 8.930645e-01<br />log10(baseMean): 1.9808909<br />gene: FBgn0028428","log2FoldChange: -0.1011056096<br />-log10(padj): 7.229956e-02<br />log10(baseMean): 2.7653698<br />gene: FBgn0028429","log2FoldChange: -0.2482805765<br />-log10(padj): 4.949434e-02<br />log10(baseMean): 3.2382467<br />gene: FBgn0028430","log2FoldChange: -1.3481269437<br />-log10(padj): 7.281408e+00<br />log10(baseMean): 1.9671890<br />gene: FBgn0028433","log2FoldChange: 0.9843183550<br />-log10(padj): 3.343682e+00<br />log10(baseMean): 1.9225462<br />gene: FBgn0028434","log2FoldChange: 0.0207660289<br />-log10(padj): 9.236129e-03<br />log10(baseMean): 2.3296230<br />gene: FBgn0028436","log2FoldChange: -0.1508969230<br />-log10(padj): 2.869994e-01<br />log10(baseMean): 3.0480623<br />gene: FBgn0028467","log2FoldChange: -0.0478549941<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 3.0114663<br />gene: FBgn0028468","log2FoldChange: -0.0586843164<br />-log10(padj): 6.024283e-02<br />log10(baseMean): 3.1309544<br />gene: FBgn0028470","log2FoldChange: -0.0100579074<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 2.8700811<br />gene: FBgn0028471","log2FoldChange: -0.5292372673<br />-log10(padj): 2.587527e+00<br />log10(baseMean): 4.3476308<br />gene: FBgn0028473","log2FoldChange: -0.0498746360<br />-log10(padj): 3.802180e-02<br />log10(baseMean): 3.2231150<br />gene: FBgn0028474","log2FoldChange: 0.0684425658<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 3.1775083<br />gene: FBgn0028475","log2FoldChange: -0.0156435546<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.9989689<br />gene: FBgn0028476","log2FoldChange: -0.2194338190<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 3.4673830<br />gene: FBgn0028479","log2FoldChange: -0.1502853527<br />-log10(padj): 1.634192e-01<br />log10(baseMean): 3.2773134<br />gene: FBgn0028480","log2FoldChange: -0.5253704389<br />-log10(padj): 4.125754e-01<br />log10(baseMean): 1.8794704<br />gene: FBgn0028482","log2FoldChange: 0.0149502438<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.1145996<br />gene: FBgn0028484","log2FoldChange: -0.2698582353<br />-log10(padj): 5.848240e-01<br />log10(baseMean): 2.9837583<br />gene: FBgn0028485","log2FoldChange: -0.2342693060<br />-log10(padj): 5.242480e-01<br />log10(baseMean): 2.5845978<br />gene: FBgn0028487","log2FoldChange: -0.4057105503<br />-log10(padj): 1.313096e+00<br />log10(baseMean): 3.9148108<br />gene: FBgn0028490","log2FoldChange: 0.6527114680<br />-log10(padj): 1.515411e+00<br />log10(baseMean): 2.2276375<br />gene: FBgn0028491","log2FoldChange: -0.1540798062<br />-log10(padj): 2.311790e-01<br />log10(baseMean): 2.9546724<br />gene: FBgn0028494","log2FoldChange: -0.0916954950<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 3.1734480<br />gene: FBgn0028497","log2FoldChange: 0.2641372323<br />-log10(padj): 2.435905e-01<br />log10(baseMean): 2.3026248<br />gene: FBgn0028499","log2FoldChange: -0.1964381860<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 3.1168737<br />gene: FBgn0028500","log2FoldChange: -0.0407720566<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.1959608<br />gene: FBgn0028504","log2FoldChange: -0.0036176644<br />-log10(padj): 1.734822e-03<br />log10(baseMean): 2.6600848<br />gene: FBgn0028506","log2FoldChange: 0.2039779055<br />-log10(padj): 2.764490e-01<br />log10(baseMean): 2.7682559<br />gene: FBgn0028507","log2FoldChange: -0.1352504449<br />-log10(padj): 1.868422e-01<br />log10(baseMean): 3.0412799<br />gene: FBgn0028509","log2FoldChange: -0.0528229259<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 1.6996846<br />gene: FBgn0028513","log2FoldChange: -1.0511661425<br />-log10(padj): 6.416928e+00<br />log10(baseMean): 2.7774518<br />gene: FBgn0028514","log2FoldChange: -0.0997577979<br />-log10(padj): 1.588341e-01<br />log10(baseMean): 3.1545122<br />gene: FBgn0028515","log2FoldChange: 0.5391593086<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 1.0622632<br />gene: FBgn0028519","log2FoldChange: 0.1457108204<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.8083048<br />gene: FBgn0028523","log2FoldChange: 0.2339835401<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 2.4033080<br />gene: FBgn0028530","log2FoldChange: 0.0467661232<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.4339227<br />gene: FBgn0028535","log2FoldChange: -0.0348968420<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 3.2365393<br />gene: FBgn0028538","log2FoldChange: -0.0693440435<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 3.3532815<br />gene: FBgn0028539","log2FoldChange: -0.8348143282<br />-log10(padj): 2.545535e+00<br />log10(baseMean): 2.0611713<br />gene: FBgn0028540","log2FoldChange: 0.0013975987<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 3.2373289<br />gene: FBgn0028541","log2FoldChange: -0.9561724551<br />-log10(padj): 3.694157e+00<br />log10(baseMean): 3.0659576<br />gene: FBgn0028542","log2FoldChange: -0.5397093747<br />-log10(padj): 1.606061e+00<br />log10(baseMean): 2.8399360<br />gene: FBgn0028543","log2FoldChange: 0.1257376245<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 2.8977655<br />gene: FBgn0028546","log2FoldChange: 0.0146843636<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.2061382<br />gene: FBgn0028550","log2FoldChange: -0.0212992410<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.6951136<br />gene: FBgn0028552","log2FoldChange: 0.0730814608<br />-log10(padj): 6.981422e-02<br />log10(baseMean): 3.1129901<br />gene: FBgn0028554","log2FoldChange: -0.4799392417<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 0.8783796<br />gene: FBgn0028562","log2FoldChange: -0.6089747811<br />-log10(padj): 5.392197e+00<br />log10(baseMean): 3.0230912<br />gene: FBgn0028563","log2FoldChange: -0.1859802322<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.5631074<br />gene: FBgn0028572","log2FoldChange: 0.6047312789<br />-log10(padj): 7.347053e-01<br />log10(baseMean): 1.9972851<br />gene: FBgn0028573","log2FoldChange: -0.1394262881<br />-log10(padj): 1.345004e-01<br />log10(baseMean): 3.4210295<br />gene: FBgn0028577","log2FoldChange: 0.0119914752<br />-log10(padj): 7.820026e-03<br />log10(baseMean): 2.7726911<br />gene: FBgn0028579","log2FoldChange: -0.0828141762<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 3.3268400<br />gene: FBgn0028582","log2FoldChange: 0.6301947042<br />-log10(padj): 1.207488e-01<br />log10(baseMean): 0.8802450<br />gene: FBgn0028622","log2FoldChange: -0.3093412973<br />-log10(padj): 1.254010e+00<br />log10(baseMean): 2.8335127<br />gene: FBgn0028646","log2FoldChange: 0.1377410267<br />-log10(padj): 1.038634e-01<br />log10(baseMean): 2.3374650<br />gene: FBgn0028647","log2FoldChange: 0.0321324107<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.4415046<br />gene: FBgn0028648","log2FoldChange: 0.2538839557<br />-log10(padj): 1.019524e-01<br />log10(baseMean): 1.5180596<br />gene: FBgn0028658","log2FoldChange: -0.0120892431<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 3.3675052<br />gene: FBgn0028662","log2FoldChange: -0.0343470318<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.9471916<br />gene: FBgn0028663","log2FoldChange: -0.3754988659<br />-log10(padj): 3.304748e-01<br />log10(baseMean): 2.1130033<br />gene: FBgn0028664","log2FoldChange: 0.0112885845<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 3.3729417<br />gene: FBgn0028665","log2FoldChange: 0.2493900333<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 3.5827304<br />gene: FBgn0028670","log2FoldChange: -0.2223737521<br />-log10(padj): 4.709759e-01<br />log10(baseMean): 3.4257804<br />gene: FBgn0028671","log2FoldChange: 0.1462792444<br />-log10(padj): 9.580390e-02<br />log10(baseMean): 2.2879731<br />gene: FBgn0028675","log2FoldChange: -0.0671588179<br />-log10(padj): 2.009957e-02<br />log10(baseMean): 2.2352604<br />gene: FBgn0028683","log2FoldChange: 0.2930866028<br />-log10(padj): 1.066951e+00<br />log10(baseMean): 3.0088851<br />gene: FBgn0028684","log2FoldChange: 0.0841477175<br />-log10(padj): 6.926412e-02<br />log10(baseMean): 3.2849429<br />gene: FBgn0028685","log2FoldChange: 0.2169003017<br />-log10(padj): 4.181491e-01<br />log10(baseMean): 3.3654682<br />gene: FBgn0028686","log2FoldChange: 0.1003872442<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 3.4520716<br />gene: FBgn0028687","log2FoldChange: 0.0852173365<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 3.1545120<br />gene: FBgn0028688","log2FoldChange: 0.0461586612<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 3.3615984<br />gene: FBgn0028689","log2FoldChange: 0.2356790201<br />-log10(padj): 8.949731e-01<br />log10(baseMean): 3.2434893<br />gene: FBgn0028690","log2FoldChange: 0.2084766891<br />-log10(padj): 5.584344e-01<br />log10(baseMean): 2.9843371<br />gene: FBgn0028691","log2FoldChange: 0.2284815084<br />-log10(padj): 5.490360e-01<br />log10(baseMean): 3.5846970<br />gene: FBgn0028692","log2FoldChange: 0.1429583567<br />-log10(padj): 1.805616e-01<br />log10(baseMean): 2.9741569<br />gene: FBgn0028693","log2FoldChange: 0.2817954268<br />-log10(padj): 9.402181e-01<br />log10(baseMean): 3.2054396<br />gene: FBgn0028694","log2FoldChange: 0.1664580954<br />-log10(padj): 1.827227e-01<br />log10(baseMean): 3.4615586<br />gene: FBgn0028695","log2FoldChange: -0.1252877366<br />-log10(padj): 1.397724e-01<br />log10(baseMean): 4.1757130<br />gene: FBgn0028696","log2FoldChange: -0.1442668810<br />-log10(padj): 1.223780e-01<br />log10(baseMean): 2.5200629<br />gene: FBgn0028700","log2FoldChange: -0.2551603294<br />-log10(padj): 5.943739e-01<br />log10(baseMean): 2.7475247<br />gene: FBgn0028703","log2FoldChange: 0.2102796616<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 1.4577298<br />gene: FBgn0028704","log2FoldChange: -0.1640204444<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.7126779<br />gene: FBgn0028707","log2FoldChange: -0.0577890161<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.3700072<br />gene: FBgn0028708","log2FoldChange: -0.2571458540<br />-log10(padj): 8.213913e-01<br />log10(baseMean): 3.1499141<br />gene: FBgn0028717","log2FoldChange: 0.0345638251<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 3.5792851<br />gene: FBgn0028734","log2FoldChange: -0.0664716840<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 3.9331748<br />gene: FBgn0028737","log2FoldChange: 0.0514144999<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.8967830<br />gene: FBgn0028738","log2FoldChange: 0.4300507186<br />-log10(padj): 5.354932e-01<br />log10(baseMean): 1.8974982<br />gene: FBgn0028740","log2FoldChange: -0.1095437159<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.3968590<br />gene: FBgn0028743","log2FoldChange: -0.2282923104<br />-log10(padj): 8.110079e-01<br />log10(baseMean): 3.0709127<br />gene: FBgn0028744","log2FoldChange: 0.1143585377<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 2.0258647<br />gene: FBgn0028746","log2FoldChange: 0.1023856457<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 2.6724753<br />gene: FBgn0028836","log2FoldChange: 0.0558606139<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 2.6721732<br />gene: FBgn0028837","log2FoldChange: 0.4296971646<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 1.1545770<br />gene: FBgn0028859","log2FoldChange: 0.2440346870<br />-log10(padj): 1.465198e-01<br />log10(baseMean): 2.1738212<br />gene: FBgn0028863","log2FoldChange: 0.0858049021<br />-log10(padj): 4.675633e-02<br />log10(baseMean): 2.0753632<br />gene: FBgn0028886","log2FoldChange: -0.2012408578<br />-log10(padj): 2.146110e-01<br />log10(baseMean): 2.8228172<br />gene: FBgn0028894","log2FoldChange: -0.1014978393<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 1.8493100<br />gene: FBgn0028895","log2FoldChange: 0.0442916946<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.0051050<br />gene: FBgn0028897","log2FoldChange: 0.8553456589<br />-log10(padj): 3.418896e-01<br />log10(baseMean): 1.2351143<br />gene: FBgn0028899","log2FoldChange: 1.2494827693<br />-log10(padj): 4.763013e-01<br />log10(baseMean): 0.8445246<br />gene: FBgn0028903","log2FoldChange: -0.3026398729<br />-log10(padj): 4.548636e-02<br />log10(baseMean): 0.8903606<br />gene: FBgn0028905","log2FoldChange: -0.3958759447<br />-log10(padj): 1.333676e-01<br />log10(baseMean): 1.5436298<br />gene: FBgn0028906","log2FoldChange: -0.3140853006<br />-log10(padj): 8.588928e-01<br />log10(baseMean): 2.9390931<br />gene: FBgn0028916","log2FoldChange: 0.1847665334<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 2.2993274<br />gene: FBgn0028919","log2FoldChange: -0.1152594542<br />-log10(padj): 7.401100e-02<br />log10(baseMean): 2.1738406<br />gene: FBgn0028925","log2FoldChange: -0.0951884898<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.4822944<br />gene: FBgn0028926","log2FoldChange: 0.0916956739<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.0513970<br />gene: FBgn0028931","log2FoldChange: 0.1247629838<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.9195771<br />gene: FBgn0028932","log2FoldChange: -0.4335109003<br />-log10(padj): 7.436854e-01<br />log10(baseMean): 3.0103272<br />gene: FBgn0028936","log2FoldChange: -2.4044816248<br />-log10(padj): 3.594672e+00<br />log10(baseMean): 1.3016316<br />gene: FBgn0028939","log2FoldChange: -0.3487399003<br />-log10(padj): 7.919026e-01<br />log10(baseMean): 2.6532636<br />gene: FBgn0028940","log2FoldChange: 0.9812990162<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 1.0939059<br />gene: FBgn0028941","log2FoldChange: 0.2106738555<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 1.8958968<br />gene: FBgn0028952","log2FoldChange: -0.1590142858<br />-log10(padj): 2.167831e-01<br />log10(baseMean): 3.2403227<br />gene: FBgn0028953","log2FoldChange: 0.3502454541<br />-log10(padj): 1.074248e+00<br />log10(baseMean): 3.2393434<br />gene: FBgn0028954","log2FoldChange: -0.7062714172<br />-log10(padj): 1.148396e+00<br />log10(baseMean): 1.7037407<br />gene: FBgn0028955","log2FoldChange: -0.1947520342<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 2.6090519<br />gene: FBgn0028956","log2FoldChange: -0.1390504633<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 2.6580148<br />gene: FBgn0028962","log2FoldChange: -0.0410549796<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.7762992<br />gene: FBgn0028965","log2FoldChange: -0.0782086370<br />-log10(padj): 6.938076e-02<br />log10(baseMean): 3.5259942<br />gene: FBgn0028968","log2FoldChange: 0.0028670683<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 3.6608657<br />gene: FBgn0028969","log2FoldChange: -0.4318098328<br />-log10(padj): 6.627975e-01<br />log10(baseMean): 2.6633141<br />gene: FBgn0028970","log2FoldChange: -0.0925427476<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.8422751<br />gene: FBgn0028974","log2FoldChange: 0.2587782227<br />-log10(padj): 3.989651e-01<br />log10(baseMean): 2.7243383<br />gene: FBgn0028978","log2FoldChange: -0.1265127028<br />-log10(padj): 1.322521e-01<br />log10(baseMean): 2.6142408<br />gene: FBgn0028979","log2FoldChange: 0.0800427032<br />-log10(padj): 4.732637e-02<br />log10(baseMean): 2.3551303<br />gene: FBgn0028980","log2FoldChange: -0.0872507152<br />-log10(padj): 8.188638e-02<br />log10(baseMean): 3.5740167<br />gene: FBgn0028982","log2FoldChange: 0.0971920886<br />-log10(padj): 8.310444e-02<br />log10(baseMean): 2.5352330<br />gene: FBgn0028983","log2FoldChange: -0.0085446765<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.7924674<br />gene: FBgn0028984","log2FoldChange: 0.2305841836<br />-log10(padj): 7.704394e-01<br />log10(baseMean): 2.9214849<br />gene: FBgn0028985","log2FoldChange: 0.5547703723<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 1.2564243<br />gene: FBgn0028988","log2FoldChange: -0.7151992363<br />-log10(padj): 4.535543e+00<br />log10(baseMean): 3.1336401<br />gene: FBgn0028990","log2FoldChange: 0.1011197135<br />-log10(padj): 5.763448e-02<br />log10(baseMean): 2.5303848<br />gene: FBgn0028991","log2FoldChange: 0.1272472263<br />-log10(padj): 1.034432e-01<br />log10(baseMean): 2.7999814<br />gene: FBgn0028992","log2FoldChange: -0.1848165986<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 1.6221597<br />gene: FBgn0028996","log2FoldChange: 0.1354346796<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 2.3352776<br />gene: FBgn0028997","log2FoldChange: 1.4377905091<br />-log10(padj): 8.146922e+00<br />log10(baseMean): 3.2007803<br />gene: FBgn0029002","log2FoldChange: -0.2448829608<br />-log10(padj): 1.183927e+00<br />log10(baseMean): 3.3125366<br />gene: FBgn0029006","log2FoldChange: -0.1952129010<br />-log10(padj): 1.551123e-01<br />log10(baseMean): 3.0557223<br />gene: FBgn0029067","log2FoldChange: -0.0294722598<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.6699310<br />gene: FBgn0029079","log2FoldChange: 0.0913178678<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.0562632<br />gene: FBgn0029088","log2FoldChange: -1.4699863364<br />-log10(padj): 3.962396e+00<br />log10(baseMean): 1.6388553<br />gene: FBgn0029090","log2FoldChange: -0.4498710637<br />-log10(padj): 2.077428e+00<br />log10(baseMean): 3.8940522<br />gene: FBgn0029092","log2FoldChange: -0.0927232699<br />-log10(padj): 6.890100e-02<br />log10(baseMean): 4.1050496<br />gene: FBgn0029093","log2FoldChange: -0.1264672761<br />-log10(padj): 1.803835e-01<br />log10(baseMean): 2.8458043<br />gene: FBgn0029094","log2FoldChange: -0.4524607733<br />-log10(padj): 1.792584e+00<br />log10(baseMean): 3.6191079<br />gene: FBgn0029095","log2FoldChange: 0.1142287846<br />-log10(padj): 2.396890e-01<br />log10(baseMean): 3.3795156<br />gene: FBgn0029113","log2FoldChange: -0.4392171589<br />-log10(padj): 1.988693e+00<br />log10(baseMean): 3.1429805<br />gene: FBgn0029114","log2FoldChange: -0.0147048330<br />-log10(padj): 5.544660e-03<br />log10(baseMean): 2.2030785<br />gene: FBgn0029117","log2FoldChange: 0.2546220596<br />-log10(padj): 8.051506e-01<br />log10(baseMean): 2.7440768<br />gene: FBgn0029118","log2FoldChange: 0.2261804457<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 2.2768172<br />gene: FBgn0029121","log2FoldChange: -0.6662919076<br />-log10(padj): 1.291793e-01<br />log10(baseMean): 0.8278152<br />gene: FBgn0029128","log2FoldChange: -0.2590615731<br />-log10(padj): 5.710417e-01<br />log10(baseMean): 3.1285616<br />gene: FBgn0029131","log2FoldChange: 0.0198498091<br />-log10(padj): 1.300138e-02<br />log10(baseMean): 3.4170612<br />gene: FBgn0029133","log2FoldChange: 0.1983401829<br />-log10(padj): 3.960721e-01<br />log10(baseMean): 3.2493386<br />gene: FBgn0029134","log2FoldChange: 0.1283099003<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 1.4521984<br />gene: FBgn0029137","log2FoldChange: 0.5320774029<br />-log10(padj): 1.982457e+00<br />log10(baseMean): 2.9981794<br />gene: FBgn0029147","log2FoldChange: 0.1353820686<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 2.7902169<br />gene: FBgn0029148","log2FoldChange: 0.0268840970<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.8117495<br />gene: FBgn0029152","log2FoldChange: -0.2225028455<br />-log10(padj): 9.130654e-01<br />log10(baseMean): 3.5925357<br />gene: FBgn0029155","log2FoldChange: -0.0291988793<br />-log10(padj): 1.650947e-02<br />log10(baseMean): 3.0640269<br />gene: FBgn0029157","log2FoldChange: -0.0374017242<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.3229574<br />gene: FBgn0029158","log2FoldChange: 0.0618907697<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.4330031<br />gene: FBgn0029173","log2FoldChange: 0.0920168259<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 3.3292712<br />gene: FBgn0029174","log2FoldChange: -0.0856381446<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 4.4329325<br />gene: FBgn0029176","log2FoldChange: 0.0649909261<br />-log10(padj): 5.118578e-02<br />log10(baseMean): 2.6200317<br />gene: FBgn0029502","log2FoldChange: -0.1339832121<br />-log10(padj): 1.327424e-01<br />log10(baseMean): 2.5153886<br />gene: FBgn0029503","log2FoldChange: -0.4032883887<br />-log10(padj): 1.139055e+00<br />log10(baseMean): 3.0345788<br />gene: FBgn0029504","log2FoldChange: 0.3999797750<br />-log10(padj): 1.054532e+00<br />log10(baseMean): 4.0677085<br />gene: FBgn0029506","log2FoldChange: 0.0321891680<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 3.1948951<br />gene: FBgn0029507","log2FoldChange: 0.1878985033<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.9623363<br />gene: FBgn0029508","log2FoldChange: 0.0363843660<br />-log10(padj): 2.903413e-02<br />log10(baseMean): 3.0065682<br />gene: FBgn0029512","log2FoldChange: 0.2634556589<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 2.2850764<br />gene: FBgn0029514","log2FoldChange: 0.5592483358<br />-log10(padj): 9.064644e-01<br />log10(baseMean): 2.0017366<br />gene: FBgn0029521","log2FoldChange: 0.4491290889<br />-log10(padj): 3.548454e-01<br />log10(baseMean): 1.5924331<br />gene: FBgn0029522","log2FoldChange: 0.3882626412<br />-log10(padj): 1.271027e-01<br />log10(baseMean): 1.2656632<br />gene: FBgn0029523","log2FoldChange: 0.8594514968<br />-log10(padj): 3.646662e-01<br />log10(baseMean): 1.1724124<br />gene: FBgn0029524","log2FoldChange: 0.0209808000<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 2.5452375<br />gene: FBgn0029525","log2FoldChange: 0.6550638298<br />-log10(padj): 7.904199e-01<br />log10(baseMean): 1.6962585<br />gene: FBgn0029526","log2FoldChange: 0.2634192441<br />-log10(padj): 2.215655e-01<br />log10(baseMean): 1.9725945<br />gene: FBgn0029529","log2FoldChange: -0.0606521539<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.7600675<br />gene: FBgn0029538","log2FoldChange: 0.5447638122<br />-log10(padj): 1.627297e-01<br />log10(baseMean): 1.0965446<br />gene: FBgn0029568","log2FoldChange: 0.3298377335<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.2819805<br />gene: FBgn0029573","log2FoldChange: 0.1925254331<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 1.2716642<br />gene: FBgn0029580","log2FoldChange: 0.0085694449<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 0.9719220<br />gene: FBgn0029587","log2FoldChange: 0.4638135125<br />-log10(padj): 2.368448e-01<br />log10(baseMean): 1.4354633<br />gene: FBgn0029588","log2FoldChange: 0.3424420744<br />-log10(padj): 9.951660e-01<br />log10(baseMean): 2.6018193<br />gene: FBgn0029594","log2FoldChange: 0.5054470952<br />-log10(padj): 5.375839e-01<br />log10(baseMean): 1.7175035<br />gene: FBgn0029596","log2FoldChange: -0.5415631121<br />-log10(padj): 2.518764e+00<br />log10(baseMean): 2.8038897<br />gene: FBgn0029608","log2FoldChange: -0.0173620937<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.7857491<br />gene: FBgn0029629","log2FoldChange: 0.2596646315<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 0.8324715<br />gene: FBgn0029643","log2FoldChange: 0.1117800697<br />-log10(padj): 3.572320e-02<br />log10(baseMean): 1.6063060<br />gene: FBgn0029648","log2FoldChange: 0.2176470864<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 1.2081000<br />gene: FBgn0029658","log2FoldChange: 0.8530804070<br />-log10(padj): 1.120929e+00<br />log10(baseMean): 1.7546550<br />gene: FBgn0029659","log2FoldChange: 0.0077042874<br />-log10(padj): 1.611385e-03<br />log10(baseMean): 1.9006754<br />gene: FBgn0029660","log2FoldChange: 0.2284391149<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 1.8312275<br />gene: FBgn0029661","log2FoldChange: 0.2033387236<br />-log10(padj): 1.598990e-01<br />log10(baseMean): 2.6356872<br />gene: FBgn0029662","log2FoldChange: 0.2612261029<br />-log10(padj): 1.026860e-01<br />log10(baseMean): 1.7795618<br />gene: FBgn0029663","log2FoldChange: 0.5454006232<br />-log10(padj): 3.172190e+00<br />log10(baseMean): 2.7827542<br />gene: FBgn0029664","log2FoldChange: 0.1557039338<br />-log10(padj): 9.410274e-02<br />log10(baseMean): 2.1218836<br />gene: FBgn0029665","log2FoldChange: 0.0665937675<br />-log10(padj): 3.989442e-02<br />log10(baseMean): 2.6586493<br />gene: FBgn0029666","log2FoldChange: 0.4511307080<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 1.3565804<br />gene: FBgn0029667","log2FoldChange: 0.2111967913<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 1.2355094<br />gene: FBgn0029669","log2FoldChange: -0.1541798535<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.5010191<br />gene: FBgn0029672","log2FoldChange: 0.4540709241<br />-log10(padj): 2.944630e-01<br />log10(baseMean): 2.3072420<br />gene: FBgn0029676","log2FoldChange: -0.2635278049<br />-log10(padj): 7.348363e-01<br />log10(baseMean): 3.1368918<br />gene: FBgn0029685","log2FoldChange: 0.1320649547<br />-log10(padj): 5.444957e-02<br />log10(baseMean): 2.0299186<br />gene: FBgn0029686","log2FoldChange: 0.0461391469<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 3.3508816<br />gene: FBgn0029687","log2FoldChange: -0.1219538867<br />-log10(padj): 2.035081e-01<br />log10(baseMean): 3.5513930<br />gene: FBgn0029688","log2FoldChange: -0.0326807197<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.5116111<br />gene: FBgn0029689","log2FoldChange: 0.2820362471<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 2.8090837<br />gene: FBgn0029693","log2FoldChange: 0.7549753749<br />-log10(padj): 1.517235e-01<br />log10(baseMean): 0.9225916<br />gene: FBgn0029697","log2FoldChange: 0.0159490414<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.7621684<br />gene: FBgn0029704","log2FoldChange: -0.2013541133<br />-log10(padj): 3.982645e-01<br />log10(baseMean): 3.0035745<br />gene: FBgn0029706","log2FoldChange: 0.0734981711<br />-log10(padj): 5.435965e-02<br />log10(baseMean): 3.2321107<br />gene: FBgn0029709","log2FoldChange: -0.1344475333<br />-log10(padj): 3.216101e-02<br />log10(baseMean): 1.4746583<br />gene: FBgn0029710","log2FoldChange: -0.0118146361<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 2.2458419<br />gene: FBgn0029711","log2FoldChange: 0.2096805312<br />-log10(padj): 4.949434e-02<br />log10(baseMean): 1.3673220<br />gene: FBgn0029712","log2FoldChange: 0.3193539103<br />-log10(padj): 1.192265e+00<br />log10(baseMean): 2.7245146<br />gene: FBgn0029713","log2FoldChange: -0.0417645686<br />-log10(padj): 1.984290e-02<br />log10(baseMean): 2.3618508<br />gene: FBgn0029714","log2FoldChange: -0.1001857077<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.8586287<br />gene: FBgn0029715","log2FoldChange: 0.0090105022<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.4169717<br />gene: FBgn0029718","log2FoldChange: 0.4048338476<br />-log10(padj): 1.414067e-01<br />log10(baseMean): 1.3995800<br />gene: FBgn0029719","log2FoldChange: -0.2640998307<br />-log10(padj): 6.143645e-02<br />log10(baseMean): 1.1392379<br />gene: FBgn0029722","log2FoldChange: 0.0935672834<br />-log10(padj): 6.787207e-02<br />log10(baseMean): 3.0232465<br />gene: FBgn0029736","log2FoldChange: 0.0200334573<br />-log10(padj): 6.149316e-03<br />log10(baseMean): 3.0966705<br />gene: FBgn0029737","log2FoldChange: -0.1053016110<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 3.4549929<br />gene: FBgn0029738","log2FoldChange: -0.2571241811<br />-log10(padj): 3.778552e-02<br />log10(baseMean): 1.0289685<br />gene: FBgn0029747","log2FoldChange: -0.4887620542<br />-log10(padj): 1.181581e-01<br />log10(baseMean): 1.0788442<br />gene: FBgn0029750","log2FoldChange: -0.0597048995<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.6453026<br />gene: FBgn0029755","log2FoldChange: -0.1074525862<br />-log10(padj): 6.688615e-02<br />log10(baseMean): 2.1896158<br />gene: FBgn0029756","log2FoldChange: 1.0099340279<br />-log10(padj): 9.005737e-01<br />log10(baseMean): 1.3016288<br />gene: FBgn0029761","log2FoldChange: -0.3062129715<br />-log10(padj): 6.539725e-02<br />log10(baseMean): 1.3560257<br />gene: FBgn0029762","log2FoldChange: -0.0028769892<br />-log10(padj): 1.883530e-03<br />log10(baseMean): 3.0641807<br />gene: FBgn0029763","log2FoldChange: 0.0782704309<br />-log10(padj): 7.698958e-02<br />log10(baseMean): 3.6081074<br />gene: FBgn0029764","log2FoldChange: 0.4907275568<br />-log10(padj): 1.537132e-01<br />log10(baseMean): 1.1248564<br />gene: FBgn0029765","log2FoldChange: -0.5598836533<br />-log10(padj): 7.861074e-01<br />log10(baseMean): 4.2843862<br />gene: FBgn0029766","log2FoldChange: 0.1648836594<br />-log10(padj): 2.165823e-01<br />log10(baseMean): 2.6971344<br />gene: FBgn0029771","log2FoldChange: 0.2804756929<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 2.7414529<br />gene: FBgn0029778","log2FoldChange: -0.1283459195<br />-log10(padj): 6.605863e-02<br />log10(baseMean): 1.9700671<br />gene: FBgn0029783","log2FoldChange: -0.0319424746<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 4.3727242<br />gene: FBgn0029785","log2FoldChange: -0.0029942889<br />-log10(padj): 1.734822e-03<br />log10(baseMean): 2.5794413<br />gene: FBgn0029789","log2FoldChange: -0.4039510403<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.1766435<br />gene: FBgn0029791","log2FoldChange: -0.3226343700<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 1.1608446<br />gene: FBgn0029795","log2FoldChange: -0.3234752789<br />-log10(padj): 1.164929e+00<br />log10(baseMean): 2.7784458<br />gene: FBgn0029798","log2FoldChange: 0.1628148340<br />-log10(padj): 7.348271e-02<br />log10(baseMean): 1.8358268<br />gene: FBgn0029799","log2FoldChange: -0.4031360611<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 1.9752402<br />gene: FBgn0029800","log2FoldChange: -0.8099251417<br />-log10(padj): 8.452491e+00<br />log10(baseMean): 3.1738445<br />gene: FBgn0029801","log2FoldChange: -0.1115566470<br />-log10(padj): 4.419180e-02<br />log10(baseMean): 1.8868156<br />gene: FBgn0029803","log2FoldChange: 0.3044379099<br />-log10(padj): 3.647743e-02<br />log10(baseMean): 0.8460890<br />gene: FBgn0029804","log2FoldChange: -0.1594343437<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 0.7985886<br />gene: FBgn0029807","log2FoldChange: 0.0005350492<br />-log10(padj): 4.855687e-04<br />log10(baseMean): 2.2627026<br />gene: FBgn0029808","log2FoldChange: 0.0776238921<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 1.7845800<br />gene: FBgn0029813","log2FoldChange: 0.0695534393<br />-log10(padj): 5.965048e-02<br />log10(baseMean): 3.1087274<br />gene: FBgn0029818","log2FoldChange: 0.0337463204<br />-log10(padj): 1.897739e-02<br />log10(baseMean): 2.8231421<br />gene: FBgn0029819","log2FoldChange: -0.2474046790<br />-log10(padj): 4.767426e-01<br />log10(baseMean): 3.0949633<br />gene: FBgn0029820","log2FoldChange: -0.0728031829<br />-log10(padj): 2.955797e-02<br />log10(baseMean): 1.9428366<br />gene: FBgn0029821","log2FoldChange: -0.0619547662<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.8078598<br />gene: FBgn0029822","log2FoldChange: 0.4248878632<br />-log10(padj): 1.049229e+00<br />log10(baseMean): 3.3695717<br />gene: FBgn0029823","log2FoldChange: 0.1531213799<br />-log10(padj): 9.517988e-02<br />log10(baseMean): 2.4605748<br />gene: FBgn0029824","log2FoldChange: 0.5838862578<br />-log10(padj): 9.004293e-01<br />log10(baseMean): 1.9404265<br />gene: FBgn0029825","log2FoldChange: 0.8103918500<br />-log10(padj): 3.104823e-01<br />log10(baseMean): 0.9951702<br />gene: FBgn0029826","log2FoldChange: -0.0266621700<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 0.8985211<br />gene: FBgn0029830","log2FoldChange: 0.0439572892<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.6962515<br />gene: FBgn0029833","log2FoldChange: 0.1294091829<br />-log10(padj): 2.763200e-02<br />log10(baseMean): 1.2850452<br />gene: FBgn0029834","log2FoldChange: -0.2169848742<br />-log10(padj): 2.368448e-01<br />log10(baseMean): 3.0247943<br />gene: FBgn0029840","log2FoldChange: 0.1675929126<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 0.7962147<br />gene: FBgn0029846","log2FoldChange: 1.3388535559<br />-log10(padj): 1.209635e+00<br />log10(baseMean): 1.1275891<br />gene: FBgn0029848","log2FoldChange: -0.2795541166<br />-log10(padj): 4.094269e-01<br />log10(baseMean): 2.6585127<br />gene: FBgn0029849","log2FoldChange: 0.2387493800<br />-log10(padj): 4.385364e-01<br />log10(baseMean): 3.1556600<br />gene: FBgn0029850","log2FoldChange: -0.0127617570<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.4611002<br />gene: FBgn0029851","log2FoldChange: 0.1497172009<br />-log10(padj): 2.325270e-01<br />log10(baseMean): 2.8993617<br />gene: FBgn0029853","log2FoldChange: 0.3016080425<br />-log10(padj): 8.006545e-01<br />log10(baseMean): 2.5934609<br />gene: FBgn0029854","log2FoldChange: 0.2525273896<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 3.2070149<br />gene: FBgn0029856","log2FoldChange: -0.1171674867<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 2.7895681<br />gene: FBgn0029857","log2FoldChange: 0.1201531423<br />-log10(padj): 5.636143e-02<br />log10(baseMean): 2.2759289<br />gene: FBgn0029858","log2FoldChange: 0.4519224054<br />-log10(padj): 1.777528e+00<br />log10(baseMean): 2.8225660<br />gene: FBgn0029861","log2FoldChange: -0.1690449619<br />-log10(padj): 3.635924e-02<br />log10(baseMean): 1.2815248<br />gene: FBgn0029866","log2FoldChange: -0.1346786601<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 3.1985215<br />gene: FBgn0029867","log2FoldChange: -0.0920329763<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 3.0393742<br />gene: FBgn0029868","log2FoldChange: -0.2718181582<br />-log10(padj): 9.750975e-01<br />log10(baseMean): 3.3765055<br />gene: FBgn0029870","log2FoldChange: 0.0421935641<br />-log10(padj): 2.319476e-02<br />log10(baseMean): 2.9665668<br />gene: FBgn0029873","log2FoldChange: 0.1348146094<br />-log10(padj): 8.308833e-02<br />log10(baseMean): 2.5602909<br />gene: FBgn0029874","log2FoldChange: -0.1604044794<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 1.2142759<br />gene: FBgn0029877","log2FoldChange: 0.1260673369<br />-log10(padj): 1.515658e-01<br />log10(baseMean): 3.2863169<br />gene: FBgn0029878","log2FoldChange: 0.1734795451<br />-log10(padj): 4.021223e-02<br />log10(baseMean): 1.3663055<br />gene: FBgn0029879","log2FoldChange: 0.2902791219<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 1.1717127<br />gene: FBgn0029880","log2FoldChange: -0.5056350733<br />-log10(padj): 2.059361e+00<br />log10(baseMean): 3.6283888<br />gene: FBgn0029881","log2FoldChange: 0.0280105774<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.0363722<br />gene: FBgn0029882","log2FoldChange: -0.1583879048<br />-log10(padj): 2.348156e-01<br />log10(baseMean): 2.5810871<br />gene: FBgn0029885","log2FoldChange: 0.0068586131<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.1438824<br />gene: FBgn0029887","log2FoldChange: -0.0162021600<br />-log10(padj): 8.465413e-03<br />log10(baseMean): 2.7743961<br />gene: FBgn0029888","log2FoldChange: 0.0822349081<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.8979137<br />gene: FBgn0029890","log2FoldChange: -0.2335200059<br />-log10(padj): 4.145580e-01<br />log10(baseMean): 3.0812917<br />gene: FBgn0029891","log2FoldChange: 0.4049145401<br />-log10(padj): 3.818032e-01<br />log10(baseMean): 1.9186082<br />gene: FBgn0029892","log2FoldChange: -0.3147815095<br />-log10(padj): 8.453524e-01<br />log10(baseMean): 3.3387170<br />gene: FBgn0029893","log2FoldChange: -0.2267842882<br />-log10(padj): 1.728416e-01<br />log10(baseMean): 2.4676190<br />gene: FBgn0029894","log2FoldChange: 0.6280411257<br />-log10(padj): 1.049229e+00<br />log10(baseMean): 2.0744093<br />gene: FBgn0029895","log2FoldChange: -2.4449449388<br />-log10(padj): 5.430982e+01<br />log10(baseMean): 2.6900873<br />gene: FBgn0029896","log2FoldChange: -0.1667602543<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 4.5376151<br />gene: FBgn0029897","log2FoldChange: -0.2013957590<br />-log10(padj): 2.005583e-02<br />log10(baseMean): 0.8153651<br />gene: FBgn0029898","log2FoldChange: 0.0413551960<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 3.5223155<br />gene: FBgn0029899","log2FoldChange: 0.0949201508<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 3.6065881<br />gene: FBgn0029903","log2FoldChange: -0.0869347621<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 2.8398463<br />gene: FBgn0029905","log2FoldChange: 0.9539153679<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 0.8836544<br />gene: FBgn0029909","log2FoldChange: -0.0914833911<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.8653370<br />gene: FBgn0029911","log2FoldChange: -0.1599195311<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 2.8927560<br />gene: FBgn0029912","log2FoldChange: 0.2311928378<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.0096515<br />gene: FBgn0029913","log2FoldChange: 0.0258819866<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 2.5396238<br />gene: FBgn0029914","log2FoldChange: 0.4219663971<br />-log10(padj): 1.153382e+00<br />log10(baseMean): 2.5573119<br />gene: FBgn0029915","log2FoldChange: 0.2757241981<br />-log10(padj): 2.253295e-01<br />log10(baseMean): 2.1201869<br />gene: FBgn0029924","log2FoldChange: 0.1326341302<br />-log10(padj): 1.201659e-01<br />log10(baseMean): 2.3767471<br />gene: FBgn0029925","log2FoldChange: 0.2696140185<br />-log10(padj): 1.026860e-01<br />log10(baseMean): 1.4881405<br />gene: FBgn0029928","log2FoldChange: -0.0692929190<br />-log10(padj): 4.686228e-02<br />log10(baseMean): 2.5419968<br />gene: FBgn0029929","log2FoldChange: -0.2789166238<br />-log10(padj): 4.081606e-01<br />log10(baseMean): 2.2605320<br />gene: FBgn0029935","log2FoldChange: 0.0717035190<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.3334820<br />gene: FBgn0029936","log2FoldChange: -0.0720554315<br />-log10(padj): 5.286181e-02<br />log10(baseMean): 2.8938559<br />gene: FBgn0029937","log2FoldChange: -0.4621751866<br />-log10(padj): 7.995419e-02<br />log10(baseMean): 0.8691695<br />gene: FBgn0029939","log2FoldChange: 0.1541571416<br />-log10(padj): 1.661605e-01<br />log10(baseMean): 3.4288817<br />gene: FBgn0029941","log2FoldChange: 0.0467525164<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.2965992<br />gene: FBgn0029942","log2FoldChange: -0.0581415269<br />-log10(padj): 3.727518e-02<br />log10(baseMean): 2.4972074<br />gene: FBgn0029943","log2FoldChange: 0.3168956805<br />-log10(padj): 8.007301e-01<br />log10(baseMean): 2.7344531<br />gene: FBgn0029944","log2FoldChange: 0.7022722171<br />-log10(padj): 3.575793e-01<br />log10(baseMean): 1.2621137<br />gene: FBgn0029945","log2FoldChange: -0.5679184634<br />-log10(padj): 1.084626e-01<br />log10(baseMean): 1.2872952<br />gene: FBgn0029950","log2FoldChange: -0.7236331235<br />-log10(padj): 1.986805e-01<br />log10(baseMean): 1.2661006<br />gene: FBgn0029951","log2FoldChange: 0.1628735694<br />-log10(padj): 1.140984e-01<br />log10(baseMean): 2.8777826<br />gene: FBgn0029957","log2FoldChange: 0.0518606386<br />-log10(padj): 2.985434e-02<br />log10(baseMean): 2.5270256<br />gene: FBgn0029958","log2FoldChange: -0.1591831404<br />-log10(padj): 2.259036e-01<br />log10(baseMean): 2.5409062<br />gene: FBgn0029959","log2FoldChange: -0.3387670033<br />-log10(padj): 3.756638e-01<br />log10(baseMean): 1.9681430<br />gene: FBgn0029968","log2FoldChange: -0.2205672481<br />-log10(padj): 4.125754e-01<br />log10(baseMean): 2.6685422<br />gene: FBgn0029969","log2FoldChange: 0.6176583440<br />-log10(padj): 2.171234e+00<br />log10(baseMean): 2.1267620<br />gene: FBgn0029970","log2FoldChange: -0.0730478430<br />-log10(padj): 6.237762e-02<br />log10(baseMean): 2.9312241<br />gene: FBgn0029971","log2FoldChange: 0.2650157602<br />-log10(padj): 1.092943e-01<br />log10(baseMean): 1.5268645<br />gene: FBgn0029974","log2FoldChange: 0.3591896509<br />-log10(padj): 1.330054e+00<br />log10(baseMean): 3.4361294<br />gene: FBgn0029975","log2FoldChange: -0.2211266855<br />-log10(padj): 1.000222e+00<br />log10(baseMean): 3.3640984<br />gene: FBgn0029976","log2FoldChange: 0.3294449901<br />-log10(padj): 1.467644e-01<br />log10(baseMean): 1.6123388<br />gene: FBgn0029977","log2FoldChange: -0.1240457062<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 3.3824912<br />gene: FBgn0029979","log2FoldChange: -0.1984509441<br />-log10(padj): 1.648969e-01<br />log10(baseMean): 2.4909767<br />gene: FBgn0029980","log2FoldChange: 0.7779643173<br />-log10(padj): 5.725357e-01<br />log10(baseMean): 1.3036744<br />gene: FBgn0029986","log2FoldChange: -0.1048440010<br />-log10(padj): 1.393747e-02<br />log10(baseMean): 0.9254919<br />gene: FBgn0029989","log2FoldChange: -0.5094936906<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 1.2685166<br />gene: FBgn0029990","log2FoldChange: 0.0921131132<br />-log10(padj): 1.065672e-01<br />log10(baseMean): 2.9623080<br />gene: FBgn0029992","log2FoldChange: 0.0563828650<br />-log10(padj): 8.393752e-03<br />log10(baseMean): 1.1967986<br />gene: FBgn0029994","log2FoldChange: -0.3396932249<br />-log10(padj): 6.355729e-01<br />log10(baseMean): 3.1190753<br />gene: FBgn0029996","log2FoldChange: -0.1163549482<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 2.3296136<br />gene: FBgn0029997","log2FoldChange: 0.2062490854<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 2.7541655<br />gene: FBgn0029999","log2FoldChange: -0.1963258752<br />-log10(padj): 2.580649e-01<br />log10(baseMean): 2.5848662<br />gene: FBgn0030000","log2FoldChange: 0.6571870085<br />-log10(padj): 6.143040e-01<br />log10(baseMean): 1.6354307<br />gene: FBgn0030001","log2FoldChange: -0.0251354300<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.7704469<br />gene: FBgn0030003","log2FoldChange: 0.1747360379<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 1.9101786<br />gene: FBgn0030004","log2FoldChange: 0.0101011095<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.0707055<br />gene: FBgn0030006","log2FoldChange: 0.0721822847<br />-log10(padj): 4.985557e-02<br />log10(baseMean): 2.8982464<br />gene: FBgn0030007","log2FoldChange: 0.0804035791<br />-log10(padj): 4.923107e-02<br />log10(baseMean): 2.3235056<br />gene: FBgn0030008","log2FoldChange: 0.0638266716<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.8618837<br />gene: FBgn0030010","log2FoldChange: 0.6749306298<br />-log10(padj): 4.125902e-01<br />log10(baseMean): 1.3749541<br />gene: FBgn0030011","log2FoldChange: 0.3053461471<br />-log10(padj): 3.202619e-01<br />log10(baseMean): 2.3645447<br />gene: FBgn0030012","log2FoldChange: -0.1242339759<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 2.6237484<br />gene: FBgn0030013","log2FoldChange: 0.0192905204<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 2.0058177<br />gene: FBgn0030017","log2FoldChange: -0.0999093468<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 3.1655714<br />gene: FBgn0030018","log2FoldChange: 0.1986005984<br />-log10(padj): 1.742225e-01<br />log10(baseMean): 2.4453550<br />gene: FBgn0030025","log2FoldChange: 0.5661914402<br />-log10(padj): 1.013921e+00<br />log10(baseMean): 2.3266844<br />gene: FBgn0030026","log2FoldChange: 0.2451672106<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 1.6103798<br />gene: FBgn0030028","log2FoldChange: 0.7114380777<br />-log10(padj): 1.382248e-01<br />log10(baseMean): 0.7945815<br />gene: FBgn0030029","log2FoldChange: -0.0894721755<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 2.1726820<br />gene: FBgn0030030","log2FoldChange: -0.1821886715<br />-log10(padj): 2.136474e-01<br />log10(baseMean): 3.1187683<br />gene: FBgn0030034","log2FoldChange: 0.1773274192<br />-log10(padj): 1.858855e-01<br />log10(baseMean): 3.1602357<br />gene: FBgn0030035","log2FoldChange: 0.7277903603<br />-log10(padj): 9.906922e+00<br />log10(baseMean): 2.8535708<br />gene: FBgn0030037","log2FoldChange: 0.1671727113<br />-log10(padj): 2.260508e-01<br />log10(baseMean): 3.2819374<br />gene: FBgn0030038","log2FoldChange: -0.2139677318<br />-log10(padj): 1.553941e-01<br />log10(baseMean): 2.2830836<br />gene: FBgn0030039","log2FoldChange: 0.3807059540<br />-log10(padj): 9.062172e-01<br />log10(baseMean): 3.3318771<br />gene: FBgn0030040","log2FoldChange: -2.8432190516<br />-log10(padj): 1.360174e+01<br />log10(baseMean): 1.9388743<br />gene: FBgn0030041","log2FoldChange: -0.1108491190<br />-log10(padj): 6.470652e-02<br />log10(baseMean): 2.8152000<br />gene: FBgn0030048","log2FoldChange: -0.1397907510<br />-log10(padj): 1.031211e-01<br />log10(baseMean): 3.4064758<br />gene: FBgn0030049","log2FoldChange: 1.1355478582<br />-log10(padj): 1.889861e+00<br />log10(baseMean): 1.5172017<br />gene: FBgn0030051","log2FoldChange: -0.3318325186<br />-log10(padj): 8.150812e-01<br />log10(baseMean): 3.8456754<br />gene: FBgn0030052","log2FoldChange: 0.3383382522<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 2.6985715<br />gene: FBgn0030053","log2FoldChange: -0.3214805068<br />-log10(padj): 7.348363e-01<br />log10(baseMean): 3.3995154<br />gene: FBgn0030054","log2FoldChange: -0.1931213622<br />-log10(padj): 2.395816e-01<br />log10(baseMean): 2.5193492<br />gene: FBgn0030055","log2FoldChange: 0.2270866859<br />-log10(padj): 6.871396e-01<br />log10(baseMean): 3.1529669<br />gene: FBgn0030056","log2FoldChange: 0.3226371003<br />-log10(padj): 1.003309e+00<br />log10(baseMean): 2.5447647<br />gene: FBgn0030057","log2FoldChange: 0.0465297015<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.7971478<br />gene: FBgn0030060","log2FoldChange: -0.1221570518<br />-log10(padj): 1.836853e-01<br />log10(baseMean): 2.8306819<br />gene: FBgn0030061","log2FoldChange: -0.2022931697<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 2.3748504<br />gene: FBgn0030063","log2FoldChange: 0.0084415647<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 3.2068223<br />gene: FBgn0030065","log2FoldChange: -0.2689610006<br />-log10(padj): 3.982645e-01<br />log10(baseMean): 2.3570707<br />gene: FBgn0030066","log2FoldChange: -0.2243223920<br />-log10(padj): 4.069892e-01<br />log10(baseMean): 3.2882346<br />gene: FBgn0030067","log2FoldChange: 0.2427316025<br />-log10(padj): 9.339008e-02<br />log10(baseMean): 1.6571286<br />gene: FBgn0030071","log2FoldChange: -0.1804892447<br />-log10(padj): 2.072149e-01<br />log10(baseMean): 3.0727672<br />gene: FBgn0030073","log2FoldChange: -0.3397728008<br />-log10(padj): 3.154508e-01<br />log10(baseMean): 2.5839392<br />gene: FBgn0030077","log2FoldChange: 0.3828729136<br />-log10(padj): 2.523253e-02<br />log10(baseMean): 1.8413946<br />gene: FBgn0030078","log2FoldChange: 0.3415125508<br />-log10(padj): 5.803467e-01<br />log10(baseMean): 3.4079158<br />gene: FBgn0030079","log2FoldChange: -0.1070419734<br />-log10(padj): 8.837435e-02<br />log10(baseMean): 2.3944445<br />gene: FBgn0030081","log2FoldChange: 0.0093148868<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 2.7074459<br />gene: FBgn0030082","log2FoldChange: 0.1062544140<br />-log10(padj): 7.066721e-02<br />log10(baseMean): 3.8335232<br />gene: FBgn0030086","log2FoldChange: 0.0830955661<br />-log10(padj): 6.485116e-02<br />log10(baseMean): 3.3982059<br />gene: FBgn0030087","log2FoldChange: 0.0785752013<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 2.2123456<br />gene: FBgn0030088","log2FoldChange: 0.0008854095<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.3754436<br />gene: FBgn0030089","log2FoldChange: -0.7642654990<br />-log10(padj): 1.993047e+00<br />log10(baseMean): 2.1996423<br />gene: FBgn0030090","log2FoldChange: 0.0488870915<br />-log10(padj): 4.415545e-02<br />log10(baseMean): 2.9816187<br />gene: FBgn0030091","log2FoldChange: 0.0780248177<br />-log10(padj): 4.130831e-02<br />log10(baseMean): 2.2299337<br />gene: FBgn0030092","log2FoldChange: 0.0098524631<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 3.2651224<br />gene: FBgn0030093","log2FoldChange: -0.0652828538<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.6983098<br />gene: FBgn0030096","log2FoldChange: 0.0953870272<br />-log10(padj): 6.938076e-02<br />log10(baseMean): 2.5281492<br />gene: FBgn0030099","log2FoldChange: 0.1733148506<br />-log10(padj): 7.750817e-02<br />log10(baseMean): 1.7226819<br />gene: FBgn0030100","log2FoldChange: 0.3001345809<br />-log10(padj): 1.938542e-01<br />log10(baseMean): 1.8233455<br />gene: FBgn0030101","log2FoldChange: -0.0407440715<br />-log10(padj): 3.436600e-02<br />log10(baseMean): 3.3877317<br />gene: FBgn0030109","log2FoldChange: -0.2854241832<br />-log10(padj): 3.168145e-01<br />log10(baseMean): 3.0098790<br />gene: FBgn0030114","log2FoldChange: -0.8572757930<br />-log10(padj): 4.687799e-01<br />log10(baseMean): 1.2886053<br />gene: FBgn0030120","log2FoldChange: -0.1040294118<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.6248903<br />gene: FBgn0030121","log2FoldChange: 0.0411331503<br />-log10(padj): 1.891881e-02<br />log10(baseMean): 2.6105880<br />gene: FBgn0030122","log2FoldChange: -0.1511029239<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 4.2704431<br />gene: FBgn0030136","log2FoldChange: -0.2089809575<br />-log10(padj): 9.859279e-02<br />log10(baseMean): 1.8974978<br />gene: FBgn0030137","log2FoldChange: 0.1110831511<br />-log10(padj): 1.000366e-01<br />log10(baseMean): 2.8519415<br />gene: FBgn0030141","log2FoldChange: 0.2039789389<br />-log10(padj): 5.230630e-01<br />log10(baseMean): 2.7168767<br />gene: FBgn0030142","log2FoldChange: -0.0781556691<br />-log10(padj): 4.732637e-02<br />log10(baseMean): 2.8022124<br />gene: FBgn0030144","log2FoldChange: 0.6620973742<br />-log10(padj): 3.818161e+00<br />log10(baseMean): 2.6668973<br />gene: FBgn0030145","log2FoldChange: 0.2058386822<br />-log10(padj): 4.452796e-01<br />log10(baseMean): 3.1636094<br />gene: FBgn0030151","log2FoldChange: 0.9166955902<br />-log10(padj): 1.048490e+00<br />log10(baseMean): 1.3938445<br />gene: FBgn0030160","log2FoldChange: -0.2373270551<br />-log10(padj): 2.388550e-01<br />log10(baseMean): 2.3722005<br />gene: FBgn0030170","log2FoldChange: -0.2100092542<br />-log10(padj): 7.847011e-02<br />log10(baseMean): 1.7306384<br />gene: FBgn0030171","log2FoldChange: -0.1763320835<br />-log10(padj): 3.240448e-01<br />log10(baseMean): 2.7363528<br />gene: FBgn0030177","log2FoldChange: -0.0565747066<br />-log10(padj): 2.200484e-02<br />log10(baseMean): 1.9755251<br />gene: FBgn0030178","log2FoldChange: -0.0568290720<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.9766595<br />gene: FBgn0030183","log2FoldChange: 0.3566599241<br />-log10(padj): 8.401327e-01<br />log10(baseMean): 2.5488962<br />gene: FBgn0030189","log2FoldChange: -0.3625054192<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 1.2001584<br />gene: FBgn0030195","log2FoldChange: 0.3125838976<br />-log10(padj): 6.820856e-01<br />log10(baseMean): 2.3929985<br />gene: FBgn0030196","log2FoldChange: 0.0630291122<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 4.1166186<br />gene: FBgn0030205","log2FoldChange: 0.0059834734<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.5559461<br />gene: FBgn0030206","log2FoldChange: 0.0724472440<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 3.0355234<br />gene: FBgn0030208","log2FoldChange: 0.0886695610<br />-log10(padj): 4.371691e-02<br />log10(baseMean): 2.0425684<br />gene: FBgn0030217","log2FoldChange: 0.0446225135<br />-log10(padj): 1.767870e-02<br />log10(baseMean): 2.0752155<br />gene: FBgn0030218","log2FoldChange: -0.0601107592<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 2.5839388<br />gene: FBgn0030228","log2FoldChange: -0.1360288677<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 2.8918023<br />gene: FBgn0030230","log2FoldChange: 0.2702346218<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 2.2146995<br />gene: FBgn0030234","log2FoldChange: -0.0336282996<br />-log10(padj): 1.254009e-02<br />log10(baseMean): 3.5189565<br />gene: FBgn0030235","log2FoldChange: -0.9439590962<br />-log10(padj): 1.338429e+01<br />log10(baseMean): 3.3030289<br />gene: FBgn0030237","log2FoldChange: 0.1280311876<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.4729939<br />gene: FBgn0030239","log2FoldChange: -0.2523750718<br />-log10(padj): 4.158692e-01<br />log10(baseMean): 2.8750930<br />gene: FBgn0030240","log2FoldChange: 0.3505248747<br />-log10(padj): 8.111198e-01<br />log10(baseMean): 3.0774258<br />gene: FBgn0030241","log2FoldChange: 0.1101683910<br />-log10(padj): 7.472126e-02<br />log10(baseMean): 2.2554118<br />gene: FBgn0030242","log2FoldChange: -0.2776932249<br />-log10(padj): 3.713583e-01<br />log10(baseMean): 3.2475058<br />gene: FBgn0030243","log2FoldChange: 1.6141837091<br />-log10(padj): 7.434783e-01<br />log10(baseMean): 0.8341926<br />gene: FBgn0030244","log2FoldChange: -0.2169882481<br />-log10(padj): 4.374410e-01<br />log10(baseMean): 3.3484785<br />gene: FBgn0030245","log2FoldChange: 0.0648489295<br />-log10(padj): 6.149538e-02<br />log10(baseMean): 3.1667164<br />gene: FBgn0030246","log2FoldChange: -1.3220553525<br />-log10(padj): 3.661991e-01<br />log10(baseMean): 0.9188304<br />gene: FBgn0030249","log2FoldChange: -0.2151303953<br />-log10(padj): 5.407902e-02<br />log10(baseMean): 1.3716301<br />gene: FBgn0030252","log2FoldChange: -1.1175279122<br />-log10(padj): 8.159360e-01<br />log10(baseMean): 1.2307542<br />gene: FBgn0030262","log2FoldChange: 0.1459028896<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 3.2285694<br />gene: FBgn0030263","log2FoldChange: -0.0379863199<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.3707021<br />gene: FBgn0030266","log2FoldChange: 0.1474924897<br />-log10(padj): 1.786471e-01<br />log10(baseMean): 3.5479858<br />gene: FBgn0030268","log2FoldChange: -0.0639033763<br />-log10(padj): 6.121591e-02<br />log10(baseMean): 2.7355891<br />gene: FBgn0030269","log2FoldChange: -0.5681459101<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 0.8924146<br />gene: FBgn0030270","log2FoldChange: -0.4756945418<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 1.4965017<br />gene: FBgn0030271","log2FoldChange: 0.5280317191<br />-log10(padj): 1.097356e-01<br />log10(baseMean): 0.8813826<br />gene: FBgn0030272","log2FoldChange: -0.0293404424<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.7317819<br />gene: FBgn0030274","log2FoldChange: -0.1121049799<br />-log10(padj): 1.622799e-01<br />log10(baseMean): 3.2171498<br />gene: FBgn0030276","log2FoldChange: -0.0864645017<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 3.2842848<br />gene: FBgn0030286","log2FoldChange: -0.0047613974<br />-log10(padj): 2.818228e-03<br />log10(baseMean): 3.0227474<br />gene: FBgn0030289","log2FoldChange: -0.2968726232<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.0151111<br />gene: FBgn0030291","log2FoldChange: -0.0322677209<br />-log10(padj): 1.685188e-02<br />log10(baseMean): 2.3339716<br />gene: FBgn0030292","log2FoldChange: -0.0379806232<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 3.2028478<br />gene: FBgn0030293","log2FoldChange: 0.0194006521<br />-log10(padj): 5.773460e-03<br />log10(baseMean): 2.4196974<br />gene: FBgn0030294","log2FoldChange: -0.1111103921<br />-log10(padj): 1.001750e-01<br />log10(baseMean): 2.9329815<br />gene: FBgn0030299","log2FoldChange: -0.3901348853<br />-log10(padj): 6.233606e-01<br />log10(baseMean): 2.0424606<br />gene: FBgn0030300","log2FoldChange: -0.0545339516<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.9799405<br />gene: FBgn0030301","log2FoldChange: 0.4790284989<br />-log10(padj): 2.312321e+00<br />log10(baseMean): 2.7155433<br />gene: FBgn0030305","log2FoldChange: -0.0533377808<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.0184656<br />gene: FBgn0030306","log2FoldChange: -0.1478485435<br />-log10(padj): 1.511863e-01<br />log10(baseMean): 3.8676593<br />gene: FBgn0030309","log2FoldChange: 0.1674627079<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 3.3345243<br />gene: FBgn0030310","log2FoldChange: -0.0468046554<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.8490775<br />gene: FBgn0030311","log2FoldChange: 0.0479209715<br />-log10(padj): 2.774356e-02<br />log10(baseMean): 2.7705540<br />gene: FBgn0030314","log2FoldChange: 0.1287297614<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.9692483<br />gene: FBgn0030316","log2FoldChange: -0.0402893665<br />-log10(padj): 1.124980e-02<br />log10(baseMean): 1.7204883<br />gene: FBgn0030317","log2FoldChange: 0.3988328209<br />-log10(padj): 2.389925e+00<br />log10(baseMean): 3.3549397<br />gene: FBgn0030318","log2FoldChange: 0.2364457316<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 0.9769862<br />gene: FBgn0030319","log2FoldChange: 0.1625789815<br />-log10(padj): 2.777246e-01<br />log10(baseMean): 2.8808689<br />gene: FBgn0030320","log2FoldChange: 0.0190239677<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 3.4714503<br />gene: FBgn0030321","log2FoldChange: 0.5139366860<br />-log10(padj): 1.493031e+00<br />log10(baseMean): 2.6315294<br />gene: FBgn0030322","log2FoldChange: -0.0059766038<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.1915882<br />gene: FBgn0030323","log2FoldChange: -1.2395582579<br />-log10(padj): 4.777831e+00<br />log10(baseMean): 2.1480381<br />gene: FBgn0030326","log2FoldChange: -0.0827965187<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.6026161<br />gene: FBgn0030327","log2FoldChange: 0.0369310289<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.3090308<br />gene: FBgn0030328","log2FoldChange: -0.0548049010<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.9235877<br />gene: FBgn0030329","log2FoldChange: 0.0722319892<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 2.5562229<br />gene: FBgn0030330","log2FoldChange: -0.0309557345<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.0245047<br />gene: FBgn0030331","log2FoldChange: 0.0028189925<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 2.7184922<br />gene: FBgn0030332","log2FoldChange: -0.0049803311<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.9766814<br />gene: FBgn0030334","log2FoldChange: 0.0912316161<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.1350534<br />gene: FBgn0030336","log2FoldChange: -0.0686367568<br />-log10(padj): 3.566099e-02<br />log10(baseMean): 3.0226197<br />gene: FBgn0030340","log2FoldChange: 0.1719931590<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.0670746<br />gene: FBgn0030341","log2FoldChange: -0.0991976919<br />-log10(padj): 7.472126e-02<br />log10(baseMean): 2.7599999<br />gene: FBgn0030342","log2FoldChange: 0.4943934372<br />-log10(padj): 1.711693e+00<br />log10(baseMean): 3.2045806<br />gene: FBgn0030343","log2FoldChange: 0.2548462396<br />-log10(padj): 6.984918e-01<br />log10(baseMean): 3.1654232<br />gene: FBgn0030344","log2FoldChange: 0.1534612776<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.3983589<br />gene: FBgn0030345","log2FoldChange: 0.1249325365<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.2115247<br />gene: FBgn0030346","log2FoldChange: -0.3128937692<br />-log10(padj): 4.925073e-01<br />log10(baseMean): 2.4369154<br />gene: FBgn0030347","log2FoldChange: 0.7890751636<br />-log10(padj): 2.481823e-01<br />log10(baseMean): 0.9777284<br />gene: FBgn0030348","log2FoldChange: -0.5866617643<br />-log10(padj): 5.946438e+00<br />log10(baseMean): 3.2678452<br />gene: FBgn0030349","log2FoldChange: 0.1192013752<br />-log10(padj): 1.238255e-01<br />log10(baseMean): 2.9182171<br />gene: FBgn0030350","log2FoldChange: 0.4055907771<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 2.1393950<br />gene: FBgn0030351","log2FoldChange: 0.0318252243<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.4136419<br />gene: FBgn0030352","log2FoldChange: -0.1487207236<br />-log10(padj): 2.672544e-01<br />log10(baseMean): 3.2814170<br />gene: FBgn0030354","log2FoldChange: -0.1072796921<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 1.9381623<br />gene: FBgn0030357","log2FoldChange: -0.2273195763<br />-log10(padj): 2.459786e-01<br />log10(baseMean): 2.3378648<br />gene: FBgn0030358","log2FoldChange: 0.3408282545<br />-log10(padj): 1.265198e-01<br />log10(baseMean): 1.4175325<br />gene: FBgn0030360","log2FoldChange: 0.6273640960<br />-log10(padj): 2.043501e+00<br />log10(baseMean): 2.1211850<br />gene: FBgn0030361","log2FoldChange: -0.4030358541<br />-log10(padj): 1.641330e+00<br />log10(baseMean): 3.8199812<br />gene: FBgn0030362","log2FoldChange: 0.3217685321<br />-log10(padj): 1.124926e+00<br />log10(baseMean): 2.7400983<br />gene: FBgn0030364","log2FoldChange: 0.0469361015<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.8927068<br />gene: FBgn0030365","log2FoldChange: 0.0019857951<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 3.6072693<br />gene: FBgn0030366","log2FoldChange: 0.1418134724<br />-log10(padj): 6.540085e-02<br />log10(baseMean): 1.8304738<br />gene: FBgn0030367","log2FoldChange: 0.1171379217<br />-log10(padj): 2.944976e-02<br />log10(baseMean): 1.7593588<br />gene: FBgn0030369","log2FoldChange: 0.1594562344<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.4380575<br />gene: FBgn0030373","log2FoldChange: -0.0503163335<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.0993440<br />gene: FBgn0030377","log2FoldChange: 0.2251527535<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.5510612<br />gene: FBgn0030391","log2FoldChange: -0.6943554999<br />-log10(padj): 1.394616e-01<br />log10(baseMean): 0.8662583<br />gene: FBgn0030394","log2FoldChange: 0.5708351702<br />-log10(padj): 4.064506e-01<br />log10(baseMean): 1.5030826<br />gene: FBgn0030396","log2FoldChange: 0.2044462006<br />-log10(padj): 2.281480e-02<br />log10(baseMean): 0.9104959<br />gene: FBgn0030399","log2FoldChange: -0.0029403186<br />-log10(padj): 1.734822e-03<br />log10(baseMean): 3.1308416<br />gene: FBgn0030400","log2FoldChange: -0.2614101780<br />-log10(padj): 3.294375e-01<br />log10(baseMean): 3.0288839<br />gene: FBgn0030403","log2FoldChange: 0.0125191163<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 2.3083598<br />gene: FBgn0030406","log2FoldChange: -0.1463328464<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 1.6915795<br />gene: FBgn0030407","log2FoldChange: 0.1910439152<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 2.0215006<br />gene: FBgn0030410","log2FoldChange: 0.2005984076<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 2.0224413<br />gene: FBgn0030411","log2FoldChange: -0.3173529058<br />-log10(padj): 1.548827e+00<br />log10(baseMean): 3.1167944<br />gene: FBgn0030412","log2FoldChange: -0.0903486809<br />-log10(padj): 2.007602e-02<br />log10(baseMean): 2.0373777<br />gene: FBgn0030417","log2FoldChange: -0.0099805452<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.5370596<br />gene: FBgn0030418","log2FoldChange: -0.2465876232<br />-log10(padj): 2.258934e-01<br />log10(baseMean): 3.0323783<br />gene: FBgn0030420","log2FoldChange: -0.1405562846<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 2.2502820<br />gene: FBgn0030421","log2FoldChange: 0.3049224120<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 0.7968354<br />gene: FBgn0030425","log2FoldChange: -0.1216889005<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.7029990<br />gene: FBgn0030431","log2FoldChange: 0.3211733406<br />-log10(padj): 1.326449e-01<br />log10(baseMean): 1.5230306<br />gene: FBgn0030432","log2FoldChange: 0.3650679363<br />-log10(padj): 6.855618e-01<br />log10(baseMean): 2.6928840<br />gene: FBgn0030433","log2FoldChange: -0.1498593971<br />-log10(padj): 1.588723e-01<br />log10(baseMean): 2.9153370<br />gene: FBgn0030434","log2FoldChange: 0.0417114995<br />-log10(padj): 3.091232e-02<br />log10(baseMean): 2.9003156<br />gene: FBgn0030435","log2FoldChange: 0.1672964647<br />-log10(padj): 1.392150e-01<br />log10(baseMean): 2.7555276<br />gene: FBgn0030447","log2FoldChange: 0.1825918854<br />-log10(padj): 1.490190e-01<br />log10(baseMean): 2.7241408<br />gene: FBgn0030448","log2FoldChange: -0.1911574937<br />-log10(padj): 3.232840e-01<br />log10(baseMean): 3.0084851<br />gene: FBgn0030451","log2FoldChange: -0.7036005578<br />-log10(padj): 6.281127e+00<br />log10(baseMean): 2.9756471<br />gene: FBgn0030452","log2FoldChange: 0.3164914402<br />-log10(padj): 1.024792e+00<br />log10(baseMean): 2.8436698<br />gene: FBgn0030456","log2FoldChange: 0.0546355182<br />-log10(padj): 4.160701e-02<br />log10(baseMean): 2.7122463<br />gene: FBgn0030457","log2FoldChange: 0.1348581009<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 0.9002607<br />gene: FBgn0030459","log2FoldChange: 0.1742266229<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.6312260<br />gene: FBgn0030460","log2FoldChange: -0.1982681103<br />-log10(padj): 2.993339e-01<br />log10(baseMean): 2.8044140<br />gene: FBgn0030465","log2FoldChange: 0.3406058218<br />-log10(padj): 1.208046e+00<br />log10(baseMean): 3.4390992<br />gene: FBgn0030466","log2FoldChange: 0.2851873457<br />-log10(padj): 8.687011e-01<br />log10(baseMean): 2.9142722<br />gene: FBgn0030467","log2FoldChange: -0.0743709105<br />-log10(padj): 6.917231e-02<br />log10(baseMean): 3.2297730<br />gene: FBgn0030468","log2FoldChange: 0.2198491216<br />-log10(padj): 2.158958e-01<br />log10(baseMean): 3.3407463<br />gene: FBgn0030469","log2FoldChange: 0.1661768392<br />-log10(padj): 3.551050e-01<br />log10(baseMean): 2.9843801<br />gene: FBgn0030474","log2FoldChange: -0.1442593193<br />-log10(padj): 2.124755e-01<br />log10(baseMean): 3.1673686<br />gene: FBgn0030478","log2FoldChange: -0.3023683640<br />-log10(padj): 5.092879e-01<br />log10(baseMean): 2.3993824<br />gene: FBgn0030479","log2FoldChange: 0.1858054835<br />-log10(padj): 9.035501e-02<br />log10(baseMean): 2.3305657<br />gene: FBgn0030480","log2FoldChange: 0.1144843160<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 2.6435978<br />gene: FBgn0030481","log2FoldChange: -0.2491393678<br />-log10(padj): 1.366172e+00<br />log10(baseMean): 3.6258872<br />gene: FBgn0030482","log2FoldChange: -0.0091018455<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 3.0759488<br />gene: FBgn0030484","log2FoldChange: 0.4999997810<br />-log10(padj): 1.636011e+00<br />log10(baseMean): 2.5059236<br />gene: FBgn0030485","log2FoldChange: 0.0817688499<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 3.3494415<br />gene: FBgn0030486","log2FoldChange: 0.0682329667<br />-log10(padj): 4.371691e-02<br />log10(baseMean): 2.8886273<br />gene: FBgn0030499","log2FoldChange: 0.1104099162<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 2.7951411<br />gene: FBgn0030500","log2FoldChange: -0.2719391343<br />-log10(padj): 4.918229e-01<br />log10(baseMean): 3.0978595<br />gene: FBgn0030501","log2FoldChange: -0.2408485603<br />-log10(padj): 7.485074e-01<br />log10(baseMean): 3.4212533<br />gene: FBgn0030502","log2FoldChange: 0.1986814567<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.2050056<br />gene: FBgn0030503","log2FoldChange: -0.0789954181<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 3.4654724<br />gene: FBgn0030504","log2FoldChange: -0.2650375122<br />-log10(padj): 5.944280e-01<br />log10(baseMean): 3.2309478<br />gene: FBgn0030505","log2FoldChange: 0.1123611938<br />-log10(padj): 1.197415e-01<br />log10(baseMean): 2.5953797<br />gene: FBgn0030506","log2FoldChange: -0.2516075978<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.5477172<br />gene: FBgn0030507","log2FoldChange: 0.4174569162<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 0.9140605<br />gene: FBgn0030508","log2FoldChange: 0.2044142059<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 2.6461849<br />gene: FBgn0030510","log2FoldChange: 0.1044786327<br />-log10(padj): 7.240700e-02<br />log10(baseMean): 2.2786608<br />gene: FBgn0030511","log2FoldChange: -0.0059698944<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.1132383<br />gene: FBgn0030512","log2FoldChange: -0.1475466415<br />-log10(padj): 2.444792e-01<br />log10(baseMean): 3.2185351<br />gene: FBgn0030514","log2FoldChange: -0.0119909050<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 2.4404977<br />gene: FBgn0030518","log2FoldChange: -0.2035660473<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 3.0210682<br />gene: FBgn0030519","log2FoldChange: 0.1506018117<br />-log10(padj): 1.942147e-01<br />log10(baseMean): 3.7022526<br />gene: FBgn0030520","log2FoldChange: 0.0010294878<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.6964461<br />gene: FBgn0030521","log2FoldChange: -0.0486388485<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.4219723<br />gene: FBgn0030522","log2FoldChange: -0.4140205851<br />-log10(padj): 7.570770e-02<br />log10(baseMean): 0.9418158<br />gene: FBgn0030524","log2FoldChange: -0.2623499635<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.1409187<br />gene: FBgn0030528","log2FoldChange: -0.5634565451<br />-log10(padj): 6.534016e+00<br />log10(baseMean): 3.9484614<br />gene: FBgn0030529","log2FoldChange: 0.0586903318<br />-log10(padj): 3.176448e-02<br />log10(baseMean): 3.2388604<br />gene: FBgn0030530","log2FoldChange: -0.2784422184<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 3.3156463<br />gene: FBgn0030532","log2FoldChange: 0.4235924970<br />-log10(padj): 1.037479e+00<br />log10(baseMean): 2.4937629<br />gene: FBgn0030545","log2FoldChange: 0.0641072054<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.7029120<br />gene: FBgn0030552","log2FoldChange: -0.2759194946<br />-log10(padj): 3.628965e-01<br />log10(baseMean): 2.4565198<br />gene: FBgn0030554","log2FoldChange: 0.0319858249<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 2.3502900<br />gene: FBgn0030555","log2FoldChange: -0.0560372645<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 3.1523605<br />gene: FBgn0030556","log2FoldChange: -0.0566790996<br />-log10(padj): 1.466187e-02<br />log10(baseMean): 1.5890378<br />gene: FBgn0030562","log2FoldChange: 0.1492060228<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 2.3795997<br />gene: FBgn0030571","log2FoldChange: -0.0853066153<br />-log10(padj): 7.847011e-02<br />log10(baseMean): 2.5156095<br />gene: FBgn0030572","log2FoldChange: 0.1593554057<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 1.5514323<br />gene: FBgn0030573","log2FoldChange: 0.7104539103<br />-log10(padj): 3.592539e-01<br />log10(baseMean): 1.1919389<br />gene: FBgn0030574","log2FoldChange: 0.3688886464<br />-log10(padj): 2.510493e-01<br />log10(baseMean): 1.7378599<br />gene: FBgn0030575","log2FoldChange: -0.5424246627<br />-log10(padj): 2.518353e+00<br />log10(baseMean): 2.4463059<br />gene: FBgn0030576","log2FoldChange: -0.0448425695<br />-log10(padj): 2.703040e-02<br />log10(baseMean): 2.7023012<br />gene: FBgn0030581","log2FoldChange: 0.0020057416<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 3.0129101<br />gene: FBgn0030582","log2FoldChange: 0.5238711668<br />-log10(padj): 4.866456e-01<br />log10(baseMean): 1.6290746<br />gene: FBgn0030583","log2FoldChange: -0.0149393272<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.4748678<br />gene: FBgn0030584","log2FoldChange: 0.5635770301<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 0.8738428<br />gene: FBgn0030587","log2FoldChange: 0.5683665652<br />-log10(padj): 1.543439e+00<br />log10(baseMean): 2.2727805<br />gene: FBgn0030588","log2FoldChange: 0.7243813169<br />-log10(padj): 5.453758e-01<br />log10(baseMean): 1.5206388<br />gene: FBgn0030590","log2FoldChange: 0.2373274947<br />-log10(padj): 3.287411e-02<br />log10(baseMean): 1.0990424<br />gene: FBgn0030592","log2FoldChange: -0.5213869896<br />-log10(padj): 1.331873e-01<br />log10(baseMean): 1.0959851<br />gene: FBgn0030593","log2FoldChange: 0.1102233237<br />-log10(padj): 1.426905e-02<br />log10(baseMean): 1.0833210<br />gene: FBgn0030594","log2FoldChange: -0.4587611441<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 1.3251228<br />gene: FBgn0030596","log2FoldChange: 0.0221664433<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.0129895<br />gene: FBgn0030597","log2FoldChange: 2.0874591189<br />-log10(padj): 7.905228e+00<br />log10(baseMean): 1.5792148<br />gene: FBgn0030598","log2FoldChange: 0.0132584722<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.4039605<br />gene: FBgn0030600","log2FoldChange: -0.0261774095<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.8106074<br />gene: FBgn0030603","log2FoldChange: -0.1172594721<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.8187518<br />gene: FBgn0030605","log2FoldChange: 0.2155468222<br />-log10(padj): 3.097949e-01<br />log10(baseMean): 2.7211089<br />gene: FBgn0030606","log2FoldChange: 0.6071742573<br />-log10(padj): 2.170299e-01<br />log10(baseMean): 1.3747787<br />gene: FBgn0030607","log2FoldChange: -0.0071523602<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.4195555<br />gene: FBgn0030610","log2FoldChange: -0.1643027902<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 2.1071616<br />gene: FBgn0030611","log2FoldChange: -0.5024141728<br />-log10(padj): 1.385909e+00<br />log10(baseMean): 2.5956069<br />gene: FBgn0030612","log2FoldChange: 0.0418061248<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.0308041<br />gene: FBgn0030613","log2FoldChange: -0.1351755668<br />-log10(padj): 6.485116e-02<br />log10(baseMean): 1.8844256<br />gene: FBgn0030615","log2FoldChange: -0.1931327628<br />-log10(padj): 2.265001e-01<br />log10(baseMean): 4.3815962<br />gene: FBgn0030616","log2FoldChange: 0.0900857803<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.4339902<br />gene: FBgn0030625","log2FoldChange: -0.0473491888<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.6478273<br />gene: FBgn0030626","log2FoldChange: -0.0043532193<br />-log10(padj): 2.300912e-03<br />log10(baseMean): 2.9680186<br />gene: FBgn0030627","log2FoldChange: 0.0700596289<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.7212737<br />gene: FBgn0030628","log2FoldChange: 0.4711738713<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 2.1396122<br />gene: FBgn0030629","log2FoldChange: 0.1337757453<br />-log10(padj): 9.410274e-02<br />log10(baseMean): 2.7650101<br />gene: FBgn0030630","log2FoldChange: -0.1586782613<br />-log10(padj): 2.272806e-01<br />log10(baseMean): 3.1037213<br />gene: FBgn0030631","log2FoldChange: -3.2132121827<br />-log10(padj): 2.223543e+00<br />log10(baseMean): 0.9050152<br />gene: FBgn0030634","log2FoldChange: -0.6325632142<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 1.2397024<br />gene: FBgn0030636","log2FoldChange: 0.2122392395<br />-log10(padj): 3.048210e-01<br />log10(baseMean): 2.9911945<br />gene: FBgn0030638","log2FoldChange: 0.1223692971<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.3405652<br />gene: FBgn0030642","log2FoldChange: -0.5864328789<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 0.9771726<br />gene: FBgn0030643","log2FoldChange: 0.0105243455<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.9572078<br />gene: FBgn0030645","log2FoldChange: 0.2717702696<br />-log10(padj): 5.275248e-01<br />log10(baseMean): 2.3529619<br />gene: FBgn0030646","log2FoldChange: -0.3239696817<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 1.0753884<br />gene: FBgn0030647","log2FoldChange: -0.0442912971<br />-log10(padj): 3.197684e-02<br />log10(baseMean): 2.5960944<br />gene: FBgn0030648","log2FoldChange: 0.4290350789<br />-log10(padj): 6.092959e-01<br />log10(baseMean): 2.3185410<br />gene: FBgn0030653","log2FoldChange: -0.2978373567<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 2.0411549<br />gene: FBgn0030654","log2FoldChange: -0.1348232618<br />-log10(padj): 1.476243e-01<br />log10(baseMean): 2.4701869<br />gene: FBgn0030655","log2FoldChange: -0.0396156071<br />-log10(padj): 1.388958e-02<br />log10(baseMean): 1.9199362<br />gene: FBgn0030657","log2FoldChange: -0.1206478080<br />-log10(padj): 1.182827e-01<br />log10(baseMean): 3.0520393<br />gene: FBgn0030658","log2FoldChange: -0.0388702483<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.6564142<br />gene: FBgn0030659","log2FoldChange: 0.3600944528<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.2151402<br />gene: FBgn0030660","log2FoldChange: 0.5890562570<br />-log10(padj): 3.278556e-01<br />log10(baseMean): 1.4099838<br />gene: FBgn0030661","log2FoldChange: 0.5852949940<br />-log10(padj): 1.663613e+00<br />log10(baseMean): 2.8528104<br />gene: FBgn0030662","log2FoldChange: 0.3981885726<br />-log10(padj): 8.483890e-02<br />log10(baseMean): 1.1006629<br />gene: FBgn0030664","log2FoldChange: -0.5254480575<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 0.8066432<br />gene: FBgn0030665","log2FoldChange: -1.0451018694<br />-log10(padj): 4.817784e-01<br />log10(baseMean): 1.1358642<br />gene: FBgn0030666","log2FoldChange: 0.3201077258<br />-log10(padj): 2.764490e-01<br />log10(baseMean): 2.1023494<br />gene: FBgn0030667","log2FoldChange: -0.0325509848<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.3741403<br />gene: FBgn0030668","log2FoldChange: -0.0485485769<br />-log10(padj): 2.528335e-02<br />log10(baseMean): 2.3396741<br />gene: FBgn0030669","log2FoldChange: -0.2776892879<br />-log10(padj): 5.205772e-01<br />log10(baseMean): 2.8096193<br />gene: FBgn0030670","log2FoldChange: -0.0655102027<br />-log10(padj): 4.600680e-02<br />log10(baseMean): 2.4756504<br />gene: FBgn0030671","log2FoldChange: -0.3633272471<br />-log10(padj): 1.482539e+00<br />log10(baseMean): 3.7798628<br />gene: FBgn0030672","log2FoldChange: -0.0575625918<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 1.9694511<br />gene: FBgn0030673","log2FoldChange: -0.0150828621<br />-log10(padj): 6.267132e-03<br />log10(baseMean): 3.9769184<br />gene: FBgn0030674","log2FoldChange: 0.1929753065<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 1.8634483<br />gene: FBgn0030675","log2FoldChange: 0.3772028881<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.0174068<br />gene: FBgn0030676","log2FoldChange: -0.0707076026<br />-log10(padj): 3.642035e-02<br />log10(baseMean): 2.3368067<br />gene: FBgn0030678","log2FoldChange: -0.1789575944<br />-log10(padj): 1.866767e-01<br />log10(baseMean): 2.6391232<br />gene: FBgn0030679","log2FoldChange: -0.0691494815<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.7616989<br />gene: FBgn0030680","log2FoldChange: -0.1837700557<br />-log10(padj): 1.361276e-01<br />log10(baseMean): 2.4042403<br />gene: FBgn0030683","log2FoldChange: -0.3226869341<br />-log10(padj): 1.182231e+00<br />log10(baseMean): 3.3263437<br />gene: FBgn0030685","log2FoldChange: -0.1440831984<br />-log10(padj): 1.590182e-01<br />log10(baseMean): 2.6577786<br />gene: FBgn0030686","log2FoldChange: -0.0094407421<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 2.8938531<br />gene: FBgn0030687","log2FoldChange: 0.2609676679<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 1.4059275<br />gene: FBgn0030691","log2FoldChange: -0.0217153093<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.8949398<br />gene: FBgn0030692","log2FoldChange: -0.1064008121<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 2.5683969<br />gene: FBgn0030693","log2FoldChange: -0.0495438523<br />-log10(padj): 2.778407e-02<br />log10(baseMean): 2.3432825<br />gene: FBgn0030694","log2FoldChange: -0.1580459571<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.3080047<br />gene: FBgn0030695","log2FoldChange: 0.0725104438<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 1.3707316<br />gene: FBgn0030696","log2FoldChange: -0.1826189693<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 2.8573094<br />gene: FBgn0030699","log2FoldChange: -0.0256664059<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.6992523<br />gene: FBgn0030700","log2FoldChange: -0.1620013313<br />-log10(padj): 2.842807e-01<br />log10(baseMean): 3.1129189<br />gene: FBgn0030701","log2FoldChange: -0.1035945458<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 3.2524580<br />gene: FBgn0030703","log2FoldChange: -0.0020756103<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 2.2510815<br />gene: FBgn0030704","log2FoldChange: -0.5349556903<br />-log10(padj): 1.606752e-01<br />log10(baseMean): 1.2089038<br />gene: FBgn0030706","log2FoldChange: 0.2153396604<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 2.3301369<br />gene: FBgn0030710","log2FoldChange: -0.3531349526<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 2.3440107<br />gene: FBgn0030711","log2FoldChange: 0.2462333496<br />-log10(padj): 6.059510e-01<br />log10(baseMean): 2.9054602<br />gene: FBgn0030716","log2FoldChange: -0.0582170498<br />-log10(padj): 3.278870e-02<br />log10(baseMean): 2.5071010<br />gene: FBgn0030717","log2FoldChange: 0.1430511989<br />-log10(padj): 1.404418e-01<br />log10(baseMean): 3.0086491<br />gene: FBgn0030718","log2FoldChange: 0.0237393652<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.4188634<br />gene: FBgn0030719","log2FoldChange: -0.1804961368<br />-log10(padj): 3.551050e-01<br />log10(baseMean): 3.1158332<br />gene: FBgn0030720","log2FoldChange: -0.0178492774<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 1.9095773<br />gene: FBgn0030721","log2FoldChange: 0.3591712085<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 1.4042702<br />gene: FBgn0030722","log2FoldChange: 0.6263788996<br />-log10(padj): 6.056909e-01<br />log10(baseMean): 1.5401641<br />gene: FBgn0030723","log2FoldChange: 0.0295626158<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.6195465<br />gene: FBgn0030724","log2FoldChange: -0.4010487370<br />-log10(padj): 7.795537e-02<br />log10(baseMean): 1.0654500<br />gene: FBgn0030725","log2FoldChange: -0.0761244419<br />-log10(padj): 5.281232e-02<br />log10(baseMean): 3.1398499<br />gene: FBgn0030731","log2FoldChange: -0.1287704866<br />-log10(padj): 1.602979e-01<br />log10(baseMean): 3.5180071<br />gene: FBgn0030734","log2FoldChange: -0.1842752716<br />-log10(padj): 4.699313e-01<br />log10(baseMean): 3.0781474<br />gene: FBgn0030735","log2FoldChange: 0.1818182265<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.0696436<br />gene: FBgn0030737","log2FoldChange: -0.2418968890<br />-log10(padj): 2.580519e-01<br />log10(baseMean): 2.7831743<br />gene: FBgn0030738","log2FoldChange: -0.0200191629<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.5319176<br />gene: FBgn0030739","log2FoldChange: -0.0846217718<br />-log10(padj): 5.646630e-02<br />log10(baseMean): 3.5542319<br />gene: FBgn0030740","log2FoldChange: 0.1788058245<br />-log10(padj): 4.169496e-02<br />log10(baseMean): 1.2738355<br />gene: FBgn0030742","log2FoldChange: 0.0193965085<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.2900713<br />gene: FBgn0030743","log2FoldChange: -0.1968798948<br />-log10(padj): 2.755476e-01<br />log10(baseMean): 2.8183973<br />gene: FBgn0030744","log2FoldChange: 0.0574276086<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.8729197<br />gene: FBgn0030745","log2FoldChange: -0.4516588714<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 1.5711022<br />gene: FBgn0030746","log2FoldChange: 0.0882686529<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 3.1355827<br />gene: FBgn0030747","log2FoldChange: -0.1283624652<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 2.9740466<br />gene: FBgn0030748","log2FoldChange: -0.0041469204<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.0165030<br />gene: FBgn0030749","log2FoldChange: -0.4060248236<br />-log10(padj): 1.959781e+00<br />log10(baseMean): 3.5394125<br />gene: FBgn0030752","log2FoldChange: -0.0558317316<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.2643481<br />gene: FBgn0030753","log2FoldChange: 0.0645166215<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 2.1067666<br />gene: FBgn0030755","log2FoldChange: 0.0909920312<br />-log10(padj): 4.600680e-02<br />log10(baseMean): 2.2551368<br />gene: FBgn0030757","log2FoldChange: -0.2475969561<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 3.1649498<br />gene: FBgn0030758","log2FoldChange: 0.0802440032<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 2.6619926<br />gene: FBgn0030759","log2FoldChange: -0.0168603343<br />-log10(padj): 9.053881e-03<br />log10(baseMean): 2.6348556<br />gene: FBgn0030761","log2FoldChange: -0.3787535907<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 1.2086896<br />gene: FBgn0030764","log2FoldChange: 0.1374240910<br />-log10(padj): 1.069440e-01<br />log10(baseMean): 2.9266176<br />gene: FBgn0030765","log2FoldChange: -0.0449403559<br />-log10(padj): 3.275836e-02<br />log10(baseMean): 2.7637943<br />gene: FBgn0030766","log2FoldChange: 0.4216402290<br />-log10(padj): 1.018875e+00<br />log10(baseMean): 2.8294414<br />gene: FBgn0030768","log2FoldChange: -0.1979733230<br />-log10(padj): 2.704878e-01<br />log10(baseMean): 2.8181892<br />gene: FBgn0030786","log2FoldChange: 0.1119699917<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 2.2801368<br />gene: FBgn0030787","log2FoldChange: -0.1078720852<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 2.6613450<br />gene: FBgn0030788","log2FoldChange: 0.0957414337<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 2.3828453<br />gene: FBgn0030789","log2FoldChange: 0.0266660215<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.8237168<br />gene: FBgn0030790","log2FoldChange: -0.1209802256<br />-log10(padj): 1.078691e-01<br />log10(baseMean): 2.8404731<br />gene: FBgn0030791","log2FoldChange: 0.1502723755<br />-log10(padj): 1.420115e-01<br />log10(baseMean): 2.9089670<br />gene: FBgn0030792","log2FoldChange: 0.0358411456<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 2.7025351<br />gene: FBgn0030793","log2FoldChange: 0.1106154539<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 1.8347488<br />gene: FBgn0030794","log2FoldChange: 0.0430952823<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.1413708<br />gene: FBgn0030795","log2FoldChange: 0.2131522713<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 3.9895293<br />gene: FBgn0030796","log2FoldChange: -0.1976729102<br />-log10(padj): 2.901409e-01<br />log10(baseMean): 2.6355333<br />gene: FBgn0030797","log2FoldChange: 0.5069053445<br />-log10(padj): 1.208505e+00<br />log10(baseMean): 2.1403339<br />gene: FBgn0030799","log2FoldChange: -0.1536129618<br />-log10(padj): 1.896155e-01<br />log10(baseMean): 2.5805916<br />gene: FBgn0030800","log2FoldChange: -0.1217004024<br />-log10(padj): 6.117917e-02<br />log10(baseMean): 2.0985129<br />gene: FBgn0030801","log2FoldChange: 0.0737176132<br />-log10(padj): 3.727518e-02<br />log10(baseMean): 2.3875020<br />gene: FBgn0030802","log2FoldChange: -0.0670674778<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 2.5874092<br />gene: FBgn0030803","log2FoldChange: 1.0758289321<br />-log10(padj): 7.570622e+00<br />log10(baseMean): 2.5326476<br />gene: FBgn0030805","log2FoldChange: 0.1659799418<br />-log10(padj): 1.788178e-01<br />log10(baseMean): 2.4356471<br />gene: FBgn0030806","log2FoldChange: 0.4323046426<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 1.0353355<br />gene: FBgn0030807","log2FoldChange: -0.1146848789<br />-log10(padj): 1.320282e-01<br />log10(baseMean): 3.4230932<br />gene: FBgn0030808","log2FoldChange: -0.0621221182<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 3.5380509<br />gene: FBgn0030809","log2FoldChange: -0.1358875634<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 3.3026020<br />gene: FBgn0030812","log2FoldChange: 0.2088204889<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 1.8135472<br />gene: FBgn0030813","log2FoldChange: -0.3255061897<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 2.0159519<br />gene: FBgn0030816","log2FoldChange: 0.4645681453<br />-log10(padj): 7.084185e-02<br />log10(baseMean): 0.8851181<br />gene: FBgn0030817","log2FoldChange: -0.0743503097<br />-log10(padj): 1.400974e-02<br />log10(baseMean): 1.2876515<br />gene: FBgn0030828","log2FoldChange: -0.0261762926<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.2031635<br />gene: FBgn0030833","log2FoldChange: -0.0694837060<br />-log10(padj): 5.763448e-02<br />log10(baseMean): 2.5904784<br />gene: FBgn0030834","log2FoldChange: -0.1266834007<br />-log10(padj): 1.797241e-01<br />log10(baseMean): 2.7402310<br />gene: FBgn0030838","log2FoldChange: -0.5549319647<br />-log10(padj): 1.315522e+00<br />log10(baseMean): 2.2027879<br />gene: FBgn0030839","log2FoldChange: 0.0032014213<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 3.0981805<br />gene: FBgn0030842","log2FoldChange: 0.0940597837<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 1.3759341<br />gene: FBgn0030847","log2FoldChange: 0.0259970722<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.0096795<br />gene: FBgn0030850","log2FoldChange: 0.1419804535<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 2.1560658<br />gene: FBgn0030851","log2FoldChange: 0.2366013833<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 1.4588135<br />gene: FBgn0030852","log2FoldChange: 0.0384077035<br />-log10(padj): 1.897739e-02<br />log10(baseMean): 2.9785474<br />gene: FBgn0030853","log2FoldChange: 0.2445121855<br />-log10(padj): 3.338230e-01<br />log10(baseMean): 2.8500794<br />gene: FBgn0030854","log2FoldChange: -0.0274483531<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.8932089<br />gene: FBgn0030855","log2FoldChange: 0.0963981185<br />-log10(padj): 7.260138e-02<br />log10(baseMean): 3.0603661<br />gene: FBgn0030858","log2FoldChange: 0.6176228526<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.0099911<br />gene: FBgn0030859","log2FoldChange: 0.3503078334<br />-log10(padj): 9.900167e-01<br />log10(baseMean): 3.2112566<br />gene: FBgn0030863","log2FoldChange: 0.2494173895<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 2.7155707<br />gene: FBgn0030864","log2FoldChange: 0.0332147054<br />-log10(padj): 1.609026e-02<br />log10(baseMean): 3.2825013<br />gene: FBgn0030869","log2FoldChange: 0.3611550867<br />-log10(padj): 7.629574e-01<br />log10(baseMean): 2.8307954<br />gene: FBgn0030871","log2FoldChange: 0.1819517991<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 2.6272227<br />gene: FBgn0030872","log2FoldChange: -0.0822558234<br />-log10(padj): 8.409693e-02<br />log10(baseMean): 3.1745907<br />gene: FBgn0030873","log2FoldChange: 0.0557093900<br />-log10(padj): 2.603321e-02<br />log10(baseMean): 2.2750892<br />gene: FBgn0030874","log2FoldChange: -0.0679046386<br />-log10(padj): 2.528335e-02<br />log10(baseMean): 2.0210999<br />gene: FBgn0030876","log2FoldChange: 0.1294446480<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8713703<br />gene: FBgn0030877","log2FoldChange: 0.0123401764<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.0145475<br />gene: FBgn0030878","log2FoldChange: 0.0493108674<br />-log10(padj): 2.579879e-02<br />log10(baseMean): 2.2435657<br />gene: FBgn0030883","log2FoldChange: 0.7619010837<br />-log10(padj): 1.979463e+00<br />log10(baseMean): 1.9082510<br />gene: FBgn0030884","log2FoldChange: 0.0908990975<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 3.0186337<br />gene: FBgn0030890","log2FoldChange: 0.0975219899<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 2.7653963<br />gene: FBgn0030891","log2FoldChange: -0.1590761519<br />-log10(padj): 2.334264e-01<br />log10(baseMean): 2.5657332<br />gene: FBgn0030892","log2FoldChange: 0.1871966944<br />-log10(padj): 3.324575e-01<br />log10(baseMean): 3.2796094<br />gene: FBgn0030893","log2FoldChange: -0.0832312540<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.0010326<br />gene: FBgn0030894","log2FoldChange: -1.4848739799<br />-log10(padj): 1.003309e+00<br />log10(baseMean): 1.0959008<br />gene: FBgn0030904","log2FoldChange: 0.1226331201<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.3374143<br />gene: FBgn0030915","log2FoldChange: 1.0011328864<br />-log10(padj): 5.038769e-01<br />log10(baseMean): 1.0261878<br />gene: FBgn0030918","log2FoldChange: 0.3828936646<br />-log10(padj): 1.863749e-01<br />log10(baseMean): 1.5266563<br />gene: FBgn0030925","log2FoldChange: 0.0739456700<br />-log10(padj): 1.324813e-02<br />log10(baseMean): 1.2552707<br />gene: FBgn0030927","log2FoldChange: 0.7217475270<br />-log10(padj): 7.176060e-01<br />log10(baseMean): 1.4714945<br />gene: FBgn0030928","log2FoldChange: 0.1023830968<br />-log10(padj): 1.224480e-02<br />log10(baseMean): 0.8606453<br />gene: FBgn0030929","log2FoldChange: 0.1158625445<br />-log10(padj): 1.670142e-01<br />log10(baseMean): 3.2536479<br />gene: FBgn0030930","log2FoldChange: 0.0795326958<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 1.3806235<br />gene: FBgn0030931","log2FoldChange: 0.4211016289<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 1.0072268<br />gene: FBgn0030932","log2FoldChange: 0.2518940761<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 0.8168330<br />gene: FBgn0030933","log2FoldChange: 0.9576449796<br />-log10(padj): 1.000222e+00<br />log10(baseMean): 1.3945634<br />gene: FBgn0030938","log2FoldChange: -0.1794659348<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 3.1366905<br />gene: FBgn0030941","log2FoldChange: 0.0718728089<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 2.8412689<br />gene: FBgn0030943","log2FoldChange: 0.0856692227<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.7385182<br />gene: FBgn0030944","log2FoldChange: -0.0798160060<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.5407128<br />gene: FBgn0030945","log2FoldChange: 0.1408511786<br />-log10(padj): 1.230588e-01<br />log10(baseMean): 3.3876775<br />gene: FBgn0030946","log2FoldChange: 1.5853963399<br />-log10(padj): 1.983319e+00<br />log10(baseMean): 1.2306824<br />gene: FBgn0030954","log2FoldChange: 0.0615966228<br />-log10(padj): 2.774356e-02<br />log10(baseMean): 3.6214657<br />gene: FBgn0030955","log2FoldChange: 0.1899206624<br />-log10(padj): 1.409127e-01<br />log10(baseMean): 2.1355304<br />gene: FBgn0030956","log2FoldChange: -0.5327653206<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 1.4143015<br />gene: FBgn0030958","log2FoldChange: 0.3135311219<br />-log10(padj): 1.605136e-01<br />log10(baseMean): 2.4067894<br />gene: FBgn0030959","log2FoldChange: -0.4762121305<br />-log10(padj): 1.620701e+00<br />log10(baseMean): 2.4025033<br />gene: FBgn0030960","log2FoldChange: 0.5381374776<br />-log10(padj): 2.249970e-01<br />log10(baseMean): 1.2565258<br />gene: FBgn0030961","log2FoldChange: -0.0960306102<br />-log10(padj): 3.177149e-02<br />log10(baseMean): 2.0126135<br />gene: FBgn0030963","log2FoldChange: 0.6562919761<br />-log10(padj): 2.466783e+00<br />log10(baseMean): 2.4079856<br />gene: FBgn0030966","log2FoldChange: 0.2641140274<br />-log10(padj): 1.583967e-01<br />log10(baseMean): 2.0237485<br />gene: FBgn0030968","log2FoldChange: 0.0531785024<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.7528638<br />gene: FBgn0030969","log2FoldChange: 0.0630100156<br />-log10(padj): 4.497479e-02<br />log10(baseMean): 2.6469189<br />gene: FBgn0030970","log2FoldChange: 0.0681829787<br />-log10(padj): 4.586647e-02<br />log10(baseMean): 2.9214907<br />gene: FBgn0030973","log2FoldChange: -0.2667376770<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 3.1627553<br />gene: FBgn0030974","log2FoldChange: 0.5583890781<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 1.4688358<br />gene: FBgn0030985","log2FoldChange: -0.0341405951<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 1.3352400<br />gene: FBgn0030989","log2FoldChange: -0.1418704886<br />-log10(padj): 1.994480e-01<br />log10(baseMean): 3.3553863<br />gene: FBgn0030990","log2FoldChange: 0.1650394564<br />-log10(padj): 2.200835e-01<br />log10(baseMean): 3.5603589<br />gene: FBgn0030991","log2FoldChange: -0.1446901573<br />-log10(padj): 7.995419e-02<br />log10(baseMean): 2.2289794<br />gene: FBgn0030992","log2FoldChange: -0.3574839324<br />-log10(padj): 5.470330e-01<br />log10(baseMean): 3.5598404<br />gene: FBgn0030993","log2FoldChange: -0.0026972604<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.8490698<br />gene: FBgn0030994","log2FoldChange: 0.0492513163<br />-log10(padj): 1.767870e-02<br />log10(baseMean): 2.0203561<br />gene: FBgn0030995","log2FoldChange: 0.1332720979<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.9995193<br />gene: FBgn0030996","log2FoldChange: 0.6064628387<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 1.0104771<br />gene: FBgn0030997","log2FoldChange: 0.1780281297<br />-log10(padj): 8.749732e-02<br />log10(baseMean): 2.7325734<br />gene: FBgn0031002","log2FoldChange: 0.3057353962<br />-log10(padj): 8.078685e-01<br />log10(baseMean): 2.4964061<br />gene: FBgn0031003","log2FoldChange: -0.1321540284<br />-log10(padj): 1.230588e-01<br />log10(baseMean): 3.2357647<br />gene: FBgn0031006","log2FoldChange: -0.0380984101<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.8470461<br />gene: FBgn0031008","log2FoldChange: -0.9378575524<br />-log10(padj): 2.455541e-01<br />log10(baseMean): 0.8316502<br />gene: FBgn0031010","log2FoldChange: -0.0973162931<br />-log10(padj): 1.084218e-01<br />log10(baseMean): 3.6441970<br />gene: FBgn0031011","log2FoldChange: -0.4739395103<br />-log10(padj): 3.622028e+00<br />log10(baseMean): 2.9728733<br />gene: FBgn0031012","log2FoldChange: -0.5009532864<br />-log10(padj): 8.901217e-01<br />log10(baseMean): 2.0895266<br />gene: FBgn0031016","log2FoldChange: 0.9496722542<br />-log10(padj): 1.650294e+00<br />log10(baseMean): 1.6142365<br />gene: FBgn0031018","log2FoldChange: -0.0379146936<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 3.3381387<br />gene: FBgn0031020","log2FoldChange: -0.2945056528<br />-log10(padj): 4.677072e-01<br />log10(baseMean): 2.5894981<br />gene: FBgn0031021","log2FoldChange: 0.0867540906<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.3764230<br />gene: FBgn0031022","log2FoldChange: 0.1539243388<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 3.3283295<br />gene: FBgn0031023","log2FoldChange: -0.0685371204<br />-log10(padj): 5.612991e-02<br />log10(baseMean): 3.6489351<br />gene: FBgn0031030","log2FoldChange: -0.1025077299<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.5292351<br />gene: FBgn0031031","log2FoldChange: -0.1382595424<br />-log10(padj): 5.702578e-02<br />log10(baseMean): 1.6868517<br />gene: FBgn0031032","log2FoldChange: 0.2421399176<br />-log10(padj): 7.555807e-02<br />log10(baseMean): 1.4838853<br />gene: FBgn0031033","log2FoldChange: -0.3582760543<br />-log10(padj): 2.869994e-01<br />log10(baseMean): 1.9145390<br />gene: FBgn0031034","log2FoldChange: -0.2403621060<br />-log10(padj): 2.996071e-01<br />log10(baseMean): 4.4507283<br />gene: FBgn0031035","log2FoldChange: -0.1880182532<br />-log10(padj): 1.782026e-01<br />log10(baseMean): 2.5544725<br />gene: FBgn0031036","log2FoldChange: 0.1631181870<br />-log10(padj): 2.085617e-01<br />log10(baseMean): 2.6727210<br />gene: FBgn0031037","log2FoldChange: -0.5468607669<br />-log10(padj): 1.433881e-01<br />log10(baseMean): 1.3414330<br />gene: FBgn0031039","log2FoldChange: -0.2886816941<br />-log10(padj): 1.483338e-01<br />log10(baseMean): 2.1118116<br />gene: FBgn0031040","log2FoldChange: -0.3809946754<br />-log10(padj): 2.648558e-01<br />log10(baseMean): 1.7734266<br />gene: FBgn0031042","log2FoldChange: -0.0942401184<br />-log10(padj): 6.605863e-02<br />log10(baseMean): 2.6980001<br />gene: FBgn0031043","log2FoldChange: -0.1921992108<br />-log10(padj): 3.544783e-01<br />log10(baseMean): 2.6394130<br />gene: FBgn0031044","log2FoldChange: -0.4126881559<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 1.8094518<br />gene: FBgn0031045","log2FoldChange: 0.3721211882<br />-log10(padj): 1.417955e+00<br />log10(baseMean): 3.1861105<br />gene: FBgn0031047","log2FoldChange: 0.4093432000<br />-log10(padj): 1.100773e+00<br />log10(baseMean): 2.9633957<br />gene: FBgn0031048","log2FoldChange: 0.6928665175<br />-log10(padj): 1.324846e+00<br />log10(baseMean): 2.6640420<br />gene: FBgn0031049","log2FoldChange: -0.0133620136<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 2.6769504<br />gene: FBgn0031050","log2FoldChange: 0.0714216636<br />-log10(padj): 4.881401e-02<br />log10(baseMean): 3.2782640<br />gene: FBgn0031051","log2FoldChange: 0.1015568286<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 3.2594696<br />gene: FBgn0031052","log2FoldChange: 0.2396041675<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.5741587<br />gene: FBgn0031053","log2FoldChange: -0.1172842820<br />-log10(padj): 1.467275e-01<br />log10(baseMean): 2.7282689<br />gene: FBgn0031054","log2FoldChange: -0.5502182723<br />-log10(padj): 1.979463e+00<br />log10(baseMean): 3.4647216<br />gene: FBgn0031055","log2FoldChange: -0.0837510387<br />-log10(padj): 9.253663e-02<br />log10(baseMean): 3.5788808<br />gene: FBgn0031057","log2FoldChange: -0.0879381829<br />-log10(padj): 4.186484e-02<br />log10(baseMean): 1.9855545<br />gene: FBgn0031058","log2FoldChange: 0.3053565141<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 2.1354997<br />gene: FBgn0031059","log2FoldChange: 0.0105385823<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 2.5167187<br />gene: FBgn0031060","log2FoldChange: 0.0046146505<br />-log10(padj): 2.222923e-03<br />log10(baseMean): 2.7489240<br />gene: FBgn0031061","log2FoldChange: -0.1747093684<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 2.6680921<br />gene: FBgn0031062","log2FoldChange: 0.0472760688<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 3.4364255<br />gene: FBgn0031066","log2FoldChange: 0.3646814257<br />-log10(padj): 7.592377e-01<br />log10(baseMean): 2.6305791<br />gene: FBgn0031068","log2FoldChange: 0.0460840430<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.4301575<br />gene: FBgn0031069","log2FoldChange: 0.5440456267<br />-log10(padj): 1.636011e+00<br />log10(baseMean): 2.8938146<br />gene: FBgn0031070","log2FoldChange: -0.3158109924<br />-log10(padj): 5.749248e-02<br />log10(baseMean): 1.0983873<br />gene: FBgn0031074","log2FoldChange: 0.0695167608<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 3.1276154<br />gene: FBgn0031077","log2FoldChange: 0.0834759166<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.7344466<br />gene: FBgn0031078","log2FoldChange: 0.3906823571<br />-log10(padj): 1.042149e+00<br />log10(baseMean): 3.5857131<br />gene: FBgn0031079","log2FoldChange: 0.2203844594<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 2.9879342<br />gene: FBgn0031080","log2FoldChange: -0.2572731689<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 1.1175262<br />gene: FBgn0031081","log2FoldChange: 0.1933098385<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 0.7921826<br />gene: FBgn0031088","log2FoldChange: -0.3192767630<br />-log10(padj): 1.798294e-01<br />log10(baseMean): 2.3444697<br />gene: FBgn0031089","log2FoldChange: -0.0734624645<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.2938678<br />gene: FBgn0031090","log2FoldChange: 0.1315501403<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.7344044<br />gene: FBgn0031091","log2FoldChange: -0.0488338088<br />-log10(padj): 3.993822e-02<br />log10(baseMean): 2.7148904<br />gene: FBgn0031092","log2FoldChange: 0.1018489570<br />-log10(padj): 6.900827e-02<br />log10(baseMean): 2.6143254<br />gene: FBgn0031093","log2FoldChange: 0.2567880656<br />-log10(padj): 3.592083e-01<br />log10(baseMean): 2.3067911<br />gene: FBgn0031094","log2FoldChange: -0.6027464084<br />-log10(padj): 1.232503e+00<br />log10(baseMean): 1.9678396<br />gene: FBgn0031097","log2FoldChange: -0.0912917333<br />-log10(padj): 4.996635e-02<br />log10(baseMean): 2.6471394<br />gene: FBgn0031098","log2FoldChange: 0.2135444634<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 1.7146731<br />gene: FBgn0031099","log2FoldChange: -0.0937409202<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.6681090<br />gene: FBgn0031106","log2FoldChange: 0.1748235438<br />-log10(padj): 1.392150e-01<br />log10(baseMean): 3.4560052<br />gene: FBgn0031107","log2FoldChange: 0.1823478508<br />-log10(padj): 2.338570e-01<br />log10(baseMean): 3.0016619<br />gene: FBgn0031114","log2FoldChange: 0.1447980053<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 2.6908845<br />gene: FBgn0031115","log2FoldChange: -0.2216454408<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 1.3394463<br />gene: FBgn0031116","log2FoldChange: 1.0226724887<br />-log10(padj): 6.496165e+00<br />log10(baseMean): 2.5139254<br />gene: FBgn0031117","log2FoldChange: -0.1106865968<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 3.4028892<br />gene: FBgn0031118","log2FoldChange: 0.2495501499<br />-log10(padj): 4.434813e-01<br />log10(baseMean): 2.4067200<br />gene: FBgn0031119","log2FoldChange: -0.4720702467<br />-log10(padj): 2.557526e+00<br />log10(baseMean): 2.5504707<br />gene: FBgn0031126","log2FoldChange: -1.6059594571<br />-log10(padj): 1.053160e+00<br />log10(baseMean): 0.9961535<br />gene: FBgn0031127","log2FoldChange: -0.2027053462<br />-log10(padj): 2.781304e-02<br />log10(baseMean): 0.9330936<br />gene: FBgn0031128","log2FoldChange: 0.3765359858<br />-log10(padj): 1.997566e-01<br />log10(baseMean): 1.6292385<br />gene: FBgn0031142","log2FoldChange: 0.3547119798<br />-log10(padj): 1.021690e+00<br />log10(baseMean): 2.8059505<br />gene: FBgn0031143","log2FoldChange: 0.0740280478<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.8975836<br />gene: FBgn0031144","log2FoldChange: -0.1218387792<br />-log10(padj): 8.129830e-02<br />log10(baseMean): 3.2606198<br />gene: FBgn0031145","log2FoldChange: 0.2915416461<br />-log10(padj): 2.713173e-01<br />log10(baseMean): 1.9554788<br />gene: FBgn0031146","log2FoldChange: 0.5596806379<br />-log10(padj): 1.877712e+00<br />log10(baseMean): 3.4376102<br />gene: FBgn0031148","log2FoldChange: 0.1584341631<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 3.6831364<br />gene: FBgn0031149","log2FoldChange: 1.1813499467<br />-log10(padj): 1.041794e+01<br />log10(baseMean): 2.9005724<br />gene: FBgn0031150","log2FoldChange: 0.0087994764<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 1.0034646<br />gene: FBgn0031155","log2FoldChange: 0.2824091500<br />-log10(padj): 6.406388e-01<br />log10(baseMean): 2.5647041<br />gene: FBgn0031157","log2FoldChange: 0.1966754671<br />-log10(padj): 5.303612e-01<br />log10(baseMean): 2.8075624<br />gene: FBgn0031161","log2FoldChange: -0.5043725898<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.1169048<br />gene: FBgn0031168","log2FoldChange: -0.0082341117<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.3195341<br />gene: FBgn0031169","log2FoldChange: -0.3912892483<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 2.9725515<br />gene: FBgn0031170","log2FoldChange: -0.0772269858<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 3.2908031<br />gene: FBgn0031174","log2FoldChange: 0.1305330603<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 1.1733133<br />gene: FBgn0031182","log2FoldChange: -0.3471467510<br />-log10(padj): 9.978468e-01<br />log10(baseMean): 2.6319716<br />gene: FBgn0031183","log2FoldChange: 0.4476425888<br />-log10(padj): 6.990361e-01<br />log10(baseMean): 1.9242025<br />gene: FBgn0031184","log2FoldChange: -0.1373142397<br />-log10(padj): 2.362813e-01<br />log10(baseMean): 3.1798608<br />gene: FBgn0031186","log2FoldChange: 0.0784278276<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 3.4256641<br />gene: FBgn0031187","log2FoldChange: -0.0526832398<br />-log10(padj): 3.861693e-02<br />log10(baseMean): 2.5915947<br />gene: FBgn0031188","log2FoldChange: 0.0533721519<br />-log10(padj): 2.535083e-02<br />log10(baseMean): 2.1698862<br />gene: FBgn0031189","log2FoldChange: -0.1446507039<br />-log10(padj): 3.630513e-01<br />log10(baseMean): 3.1733530<br />gene: FBgn0031190","log2FoldChange: -0.1092642910<br />-log10(padj): 1.094620e-01<br />log10(baseMean): 2.6134753<br />gene: FBgn0031191","log2FoldChange: -0.0639377044<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.8899444<br />gene: FBgn0031194","log2FoldChange: 0.1468359050<br />-log10(padj): 1.052752e-01<br />log10(baseMean): 2.2449063<br />gene: FBgn0031195","log2FoldChange: 0.2560798422<br />-log10(padj): 7.157400e-02<br />log10(baseMean): 1.3684381<br />gene: FBgn0031196","log2FoldChange: -0.1541546898<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 1.7262903<br />gene: FBgn0031201","log2FoldChange: -0.0143482535<br />-log10(padj): 5.286459e-03<br />log10(baseMean): 2.8826845<br />gene: FBgn0031213","log2FoldChange: 0.2306589735<br />-log10(padj): 3.197684e-02<br />log10(baseMean): 0.9130651<br />gene: FBgn0031214","log2FoldChange: -0.1282135209<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.8424920<br />gene: FBgn0031216","log2FoldChange: 0.0173036373<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.9076141<br />gene: FBgn0031217","log2FoldChange: -0.2879866795<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 1.5087082<br />gene: FBgn0031219","log2FoldChange: 0.2827201305<br />-log10(padj): 5.357615e-01<br />log10(baseMean): 3.4228500<br />gene: FBgn0031220","log2FoldChange: 0.0820187804<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 2.5246489<br />gene: FBgn0031224","log2FoldChange: -0.2069006944<br />-log10(padj): 7.765385e-02<br />log10(baseMean): 2.0225525<br />gene: FBgn0031227","log2FoldChange: 0.0477378560<br />-log10(padj): 3.436728e-02<br />log10(baseMean): 2.6513878<br />gene: FBgn0031228","log2FoldChange: -0.1188321068<br />-log10(padj): 7.750817e-02<br />log10(baseMean): 2.6287583<br />gene: FBgn0031229","log2FoldChange: 0.1582600390<br />-log10(padj): 1.095359e-01<br />log10(baseMean): 2.2910988<br />gene: FBgn0031231","log2FoldChange: -0.1433994534<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.4546955<br />gene: FBgn0031233","log2FoldChange: 0.1136773033<br />-log10(padj): 9.634103e-02<br />log10(baseMean): 2.3881318<br />gene: FBgn0031238","log2FoldChange: -0.0638723694<br />-log10(padj): 7.316617e-03<br />log10(baseMean): 1.0546152<br />gene: FBgn0031239","log2FoldChange: 0.3610875894<br />-log10(padj): 5.526083e-02<br />log10(baseMean): 0.8136358<br />gene: FBgn0031240","log2FoldChange: -0.0093652006<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.2750873<br />gene: FBgn0031244","log2FoldChange: 0.6848201487<br />-log10(padj): 3.124079e+00<br />log10(baseMean): 2.2857317<br />gene: FBgn0031245","log2FoldChange: 0.4311320785<br />-log10(padj): 7.126996e-01<br />log10(baseMean): 2.1439782<br />gene: FBgn0031247","log2FoldChange: -0.1189772206<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.6742309<br />gene: FBgn0031250","log2FoldChange: -0.1476382268<br />-log10(padj): 1.337431e-01<br />log10(baseMean): 2.6056468<br />gene: FBgn0031252","log2FoldChange: 0.0498125020<br />-log10(padj): 1.804487e-02<br />log10(baseMean): 2.2526027<br />gene: FBgn0031253","log2FoldChange: -0.2609047916<br />-log10(padj): 1.504910e-01<br />log10(baseMean): 1.8871262<br />gene: FBgn0031254","log2FoldChange: -0.7058279875<br />-log10(padj): 6.052329e-01<br />log10(baseMean): 1.5127776<br />gene: FBgn0031255","log2FoldChange: 0.0794870360<br />-log10(padj): 6.180702e-02<br />log10(baseMean): 3.1585568<br />gene: FBgn0031256","log2FoldChange: 0.1389407451<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 3.1448982<br />gene: FBgn0031257","log2FoldChange: -1.3215866450<br />-log10(padj): 5.448852e+00<br />log10(baseMean): 2.0316836<br />gene: FBgn0031258","log2FoldChange: 0.0580632053<br />-log10(padj): 4.169496e-02<br />log10(baseMean): 3.3527540<br />gene: FBgn0031260","log2FoldChange: -0.5938833889<br />-log10(padj): 1.608910e+00<br />log10(baseMean): 3.4387558<br />gene: FBgn0031261","log2FoldChange: 0.3575618421<br />-log10(padj): 1.732862e+00<br />log10(baseMean): 3.1743703<br />gene: FBgn0031263","log2FoldChange: -0.7240904361<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 0.8302843<br />gene: FBgn0031264","log2FoldChange: 0.0434270727<br />-log10(padj): 3.243344e-02<br />log10(baseMean): 2.6734516<br />gene: FBgn0031265","log2FoldChange: -0.1022064988<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 3.1426538<br />gene: FBgn0031266","log2FoldChange: -0.2124797161<br />-log10(padj): 4.195221e-01<br />log10(baseMean): 2.6876247<br />gene: FBgn0031267","log2FoldChange: 0.8262197397<br />-log10(padj): 3.184652e+00<br />log10(baseMean): 2.1011217<br />gene: FBgn0031268","log2FoldChange: -0.0390779550<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.5384010<br />gene: FBgn0031270","log2FoldChange: 0.1616365530<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.2716036<br />gene: FBgn0031281","log2FoldChange: -0.1706294550<br />-log10(padj): 1.235517e-01<br />log10(baseMean): 2.1114953<br />gene: FBgn0031282","log2FoldChange: -0.3844897048<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 0.9101971<br />gene: FBgn0031283","log2FoldChange: 0.1332875192<br />-log10(padj): 7.794305e-02<br />log10(baseMean): 2.2343284<br />gene: FBgn0031284","log2FoldChange: -0.4388774860<br />-log10(padj): 3.200783e+00<br />log10(baseMean): 3.5131616<br />gene: FBgn0031285","log2FoldChange: 0.0486624839<br />-log10(padj): 2.571083e-02<br />log10(baseMean): 2.4990021<br />gene: FBgn0031286","log2FoldChange: -0.0650466306<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 2.3694440<br />gene: FBgn0031287","log2FoldChange: 0.1896267923<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 1.1946222<br />gene: FBgn0031292","log2FoldChange: 0.3457972818<br />-log10(padj): 9.877629e-02<br />log10(baseMean): 1.2723017<br />gene: FBgn0031296","log2FoldChange: 0.0901033377<br />-log10(padj): 8.678154e-02<br />log10(baseMean): 2.9777132<br />gene: FBgn0031298","log2FoldChange: -0.0690739053<br />-log10(padj): 2.496210e-02<br />log10(baseMean): 1.9060561<br />gene: FBgn0031299","log2FoldChange: 0.0436192062<br />-log10(padj): 4.371691e-02<br />log10(baseMean): 3.0133572<br />gene: FBgn0031300","log2FoldChange: 0.4197065935<br />-log10(padj): 3.566542e-01<br />log10(baseMean): 1.8316531<br />gene: FBgn0031302","log2FoldChange: -0.2440375268<br />-log10(padj): 4.456353e-01<br />log10(baseMean): 3.2080322<br />gene: FBgn0031304","log2FoldChange: 0.8101963433<br />-log10(padj): 3.683004e+00<br />log10(baseMean): 3.2538006<br />gene: FBgn0031307","log2FoldChange: 0.0089691056<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.3653409<br />gene: FBgn0031308","log2FoldChange: -0.0069669705<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.6275044<br />gene: FBgn0031309","log2FoldChange: 0.1051560600<br />-log10(padj): 1.105617e-01<br />log10(baseMean): 2.9305304<br />gene: FBgn0031310","log2FoldChange: 0.0373476181<br />-log10(padj): 1.797597e-02<br />log10(baseMean): 3.1333568<br />gene: FBgn0031312","log2FoldChange: -0.6714046012<br />-log10(padj): 3.438441e+00<br />log10(baseMean): 3.1654919<br />gene: FBgn0031313","log2FoldChange: -0.0180492898<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.5979380<br />gene: FBgn0031314","log2FoldChange: 0.1115229608<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 2.3188436<br />gene: FBgn0031315","log2FoldChange: 0.1457210414<br />-log10(padj): 1.238255e-01<br />log10(baseMean): 2.8531406<br />gene: FBgn0031317","log2FoldChange: 0.0724290493<br />-log10(padj): 6.238437e-02<br />log10(baseMean): 3.1576514<br />gene: FBgn0031318","log2FoldChange: 0.0966525876<br />-log10(padj): 9.661768e-02<br />log10(baseMean): 2.8174330<br />gene: FBgn0031319","log2FoldChange: 0.0635837629<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 2.2969479<br />gene: FBgn0031320","log2FoldChange: -0.1887071562<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.4308011<br />gene: FBgn0031321","log2FoldChange: -1.4114919640<br />-log10(padj): 1.065607e+01<br />log10(baseMean): 2.5985794<br />gene: FBgn0031322","log2FoldChange: 1.2259240828<br />-log10(padj): 9.595653e+00<br />log10(baseMean): 2.8787446<br />gene: FBgn0031327","log2FoldChange: -0.1834558608<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.2953524<br />gene: FBgn0031344","log2FoldChange: 0.2837271331<br />-log10(padj): 1.711819e-01<br />log10(baseMean): 1.9195870<br />gene: FBgn0031351","log2FoldChange: -0.0434261111<br />-log10(padj): 2.774356e-02<br />log10(baseMean): 3.1074838<br />gene: FBgn0031356","log2FoldChange: 0.0275544226<br />-log10(padj): 1.079808e-02<br />log10(baseMean): 2.4130807<br />gene: FBgn0031357","log2FoldChange: -0.8551502250<br />-log10(padj): 5.336247e+00<br />log10(baseMean): 2.7434699<br />gene: FBgn0031359","log2FoldChange: 0.1832810769<br />-log10(padj): 1.457607e-01<br />log10(baseMean): 2.8165987<br />gene: FBgn0031360","log2FoldChange: -0.0479115385<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.0260911<br />gene: FBgn0031361","log2FoldChange: 0.2719389700<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.4367948<br />gene: FBgn0031362","log2FoldChange: -0.8158151831<br />-log10(padj): 8.197179e+00<br />log10(baseMean): 3.2998710<br />gene: FBgn0031374","log2FoldChange: -0.1295307857<br />-log10(padj): 1.235145e-01<br />log10(baseMean): 3.0751253<br />gene: FBgn0031376","log2FoldChange: -0.0765867023<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.7867421<br />gene: FBgn0031377","log2FoldChange: 0.0541881135<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.0546980<br />gene: FBgn0031378","log2FoldChange: -0.0485532235<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 3.0375275<br />gene: FBgn0031379","log2FoldChange: 0.0966451659<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 3.5747950<br />gene: FBgn0031381","log2FoldChange: 0.3569148049<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 1.9435553<br />gene: FBgn0031384","log2FoldChange: -0.4542771835<br />-log10(padj): 5.963745e-01<br />log10(baseMean): 3.2981795<br />gene: FBgn0031389","log2FoldChange: -0.0522889039<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 3.3885058<br />gene: FBgn0031390","log2FoldChange: 0.2266029830<br />-log10(padj): 2.113080e-01<br />log10(baseMean): 2.3687415<br />gene: FBgn0031391","log2FoldChange: -0.0366061775<br />-log10(padj): 2.814936e-02<br />log10(baseMean): 2.9444405<br />gene: FBgn0031392","log2FoldChange: 0.2006892854<br />-log10(padj): 4.918229e-01<br />log10(baseMean): 2.7600813<br />gene: FBgn0031395","log2FoldChange: -0.8201072378<br />-log10(padj): 6.418970e-01<br />log10(baseMean): 1.3463564<br />gene: FBgn0031397","log2FoldChange: 0.1555357374<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.0000594<br />gene: FBgn0031398","log2FoldChange: -0.2741430708<br />-log10(padj): 9.411121e-01<br />log10(baseMean): 2.8786864<br />gene: FBgn0031399","log2FoldChange: 0.0892075206<br />-log10(padj): 1.059078e-01<br />log10(baseMean): 2.9359480<br />gene: FBgn0031401","log2FoldChange: 0.2050129737<br />-log10(padj): 8.619087e-02<br />log10(baseMean): 2.1037578<br />gene: FBgn0031403","log2FoldChange: -0.5937401228<br />-log10(padj): 1.085542e+00<br />log10(baseMean): 2.2380597<br />gene: FBgn0031405","log2FoldChange: -0.0516205455<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 3.5728090<br />gene: FBgn0031408","log2FoldChange: 0.0669610013<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.0189028<br />gene: FBgn0031413","log2FoldChange: 0.2373522306<br />-log10(padj): 7.848184e-02<br />log10(baseMean): 1.5468891<br />gene: FBgn0031414","log2FoldChange: -0.1498235633<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 3.1353909<br />gene: FBgn0031418","log2FoldChange: -0.3871937434<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 1.4940735<br />gene: FBgn0031419","log2FoldChange: -0.1045743646<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.7795590<br />gene: FBgn0031420","log2FoldChange: 0.2803609076<br />-log10(padj): 5.717553e-01<br />log10(baseMean): 2.4450796<br />gene: FBgn0031421","log2FoldChange: 0.0965339687<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.7656924<br />gene: FBgn0031422","log2FoldChange: 0.2550261154<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 1.9016248<br />gene: FBgn0031428","log2FoldChange: -0.0963248136<br />-log10(padj): 1.077674e-02<br />log10(baseMean): 0.9222304<br />gene: FBgn0031429","log2FoldChange: -0.0093006010<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.5702417<br />gene: FBgn0031434","log2FoldChange: 0.2830691426<br />-log10(padj): 1.156207e-01<br />log10(baseMean): 1.5757625<br />gene: FBgn0031435","log2FoldChange: 0.2367266624<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.8273347<br />gene: FBgn0031436","log2FoldChange: -0.0412864980<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 3.0138506<br />gene: FBgn0031437","log2FoldChange: -0.8530226381<br />-log10(padj): 1.360508e+01<br />log10(baseMean): 2.9640807<br />gene: FBgn0031449","log2FoldChange: -0.0243271659<br />-log10(padj): 1.398029e-02<br />log10(baseMean): 3.2150010<br />gene: FBgn0031450","log2FoldChange: 0.7023586087<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 1.1934326<br />gene: FBgn0031451","log2FoldChange: -0.2255106869<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.6964261<br />gene: FBgn0031452","log2FoldChange: 0.1941451572<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 3.2537978<br />gene: FBgn0031453","log2FoldChange: 0.0387073103<br />-log10(padj): 2.552821e-02<br />log10(baseMean): 2.6908257<br />gene: FBgn0031456","log2FoldChange: 0.1685701290<br />-log10(padj): 2.262589e-01<br />log10(baseMean): 2.7002236<br />gene: FBgn0031457","log2FoldChange: 0.0746594086<br />-log10(padj): 5.286181e-02<br />log10(baseMean): 2.8244480<br />gene: FBgn0031458","log2FoldChange: 0.2899008371<br />-log10(padj): 8.399860e-01<br />log10(baseMean): 3.1154315<br />gene: FBgn0031459","log2FoldChange: 0.5460546730<br />-log10(padj): 5.491635e-01<br />log10(baseMean): 1.7300866<br />gene: FBgn0031460","log2FoldChange: 0.6240795800<br />-log10(padj): 4.180247e+00<br />log10(baseMean): 2.7379607<br />gene: FBgn0031461","log2FoldChange: -1.0734496160<br />-log10(padj): 1.480124e+00<br />log10(baseMean): 1.4648531<br />gene: FBgn0031464","log2FoldChange: -0.2243098497<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.2306787<br />gene: FBgn0031470","log2FoldChange: -0.6201667766<br />-log10(padj): 4.419263e-01<br />log10(baseMean): 1.5419270<br />gene: FBgn0031471","log2FoldChange: -0.0944477070<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 3.4840945<br />gene: FBgn0031474","log2FoldChange: -0.9758979442<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 0.8141330<br />gene: FBgn0031476","log2FoldChange: -0.2236015033<br />-log10(padj): 4.452796e-01<br />log10(baseMean): 2.6906249<br />gene: FBgn0031478","log2FoldChange: 0.0520534900<br />-log10(padj): 3.705232e-02<br />log10(baseMean): 2.4817970<br />gene: FBgn0031479","log2FoldChange: 0.3163525336<br />-log10(padj): 1.103206e+00<br />log10(baseMean): 2.8202405<br />gene: FBgn0031483","log2FoldChange: -0.0819046670<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 2.7426142<br />gene: FBgn0031484","log2FoldChange: 0.0682019624<br />-log10(padj): 3.977426e-02<br />log10(baseMean): 2.3619938<br />gene: FBgn0031485","log2FoldChange: 0.3027512212<br />-log10(padj): 4.452796e-01<br />log10(baseMean): 2.3210477<br />gene: FBgn0031488","log2FoldChange: 0.5104999685<br />-log10(padj): 3.229908e+00<br />log10(baseMean): 3.0092958<br />gene: FBgn0031489","log2FoldChange: 0.3161985176<br />-log10(padj): 6.786429e-01<br />log10(baseMean): 2.9786577<br />gene: FBgn0031490","log2FoldChange: 0.0315161976<br />-log10(padj): 2.096792e-02<br />log10(baseMean): 2.7747241<br />gene: FBgn0031491","log2FoldChange: 0.0353868344<br />-log10(padj): 3.120938e-02<br />log10(baseMean): 3.0017771<br />gene: FBgn0031492","log2FoldChange: -0.0196762914<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 3.1789790<br />gene: FBgn0031493","log2FoldChange: 0.1039440823<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 1.9807852<br />gene: FBgn0031494","log2FoldChange: 0.1341824091<br />-log10(padj): 1.334952e-01<br />log10(baseMean): 3.0356520<br />gene: FBgn0031495","log2FoldChange: 0.1694028153<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 1.2740807<br />gene: FBgn0031496","log2FoldChange: 0.1891226199<br />-log10(padj): 2.334868e-01<br />log10(baseMean): 3.5354698<br />gene: FBgn0031497","log2FoldChange: -0.1771439173<br />-log10(padj): 9.993745e-02<br />log10(baseMean): 2.1318542<br />gene: FBgn0031498","log2FoldChange: 0.1619745710<br />-log10(padj): 9.933931e-02<br />log10(baseMean): 2.0103265<br />gene: FBgn0031500","log2FoldChange: 0.2546505456<br />-log10(padj): 4.197277e-01<br />log10(baseMean): 2.8279321<br />gene: FBgn0031505","log2FoldChange: 0.2864125998<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 0.9993532<br />gene: FBgn0031513","log2FoldChange: -0.3107231185<br />-log10(padj): 5.636755e-02<br />log10(baseMean): 0.9541122<br />gene: FBgn0031515","log2FoldChange: 0.7260748339<br />-log10(padj): 5.816926e+00<br />log10(baseMean): 2.9400475<br />gene: FBgn0031516","log2FoldChange: -0.1263622984<br />-log10(padj): 1.300138e-02<br />log10(baseMean): 0.8490288<br />gene: FBgn0031517","log2FoldChange: 0.3202601028<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 1.2177637<br />gene: FBgn0031518","log2FoldChange: 0.1820574015<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.7584773<br />gene: FBgn0031519","log2FoldChange: 0.1441869827<br />-log10(padj): 1.868422e-01<br />log10(baseMean): 2.6408900<br />gene: FBgn0031529","log2FoldChange: 0.5353402985<br />-log10(padj): 2.007831e-01<br />log10(baseMean): 2.5193141<br />gene: FBgn0031530","log2FoldChange: -0.1097618188<br />-log10(padj): 1.515658e-01<br />log10(baseMean): 3.0566056<br />gene: FBgn0031534","log2FoldChange: -0.0300337744<br />-log10(padj): 1.400974e-02<br />log10(baseMean): 2.5296058<br />gene: FBgn0031535","log2FoldChange: -0.0015253790<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 2.7426348<br />gene: FBgn0031536","log2FoldChange: 0.0792096899<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.7866548<br />gene: FBgn0031537","log2FoldChange: 0.7365711310<br />-log10(padj): 2.467643e+00<br />log10(baseMean): 2.3365374<br />gene: FBgn0031538","log2FoldChange: -0.4880048433<br />-log10(padj): 1.586451e+00<br />log10(baseMean): 2.3371052<br />gene: FBgn0031540","log2FoldChange: -0.1928915243<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 3.1745716<br />gene: FBgn0031544","log2FoldChange: -1.3882934294<br />-log10(padj): 6.531488e+00<br />log10(baseMean): 2.4237161<br />gene: FBgn0031547","log2FoldChange: -1.9311636481<br />-log10(padj): 6.794176e+00<br />log10(baseMean): 1.7296867<br />gene: FBgn0031548","log2FoldChange: -0.0373772030<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.7400695<br />gene: FBgn0031549","log2FoldChange: 0.2579688196<br />-log10(padj): 6.960757e-01<br />log10(baseMean): 2.7023421<br />gene: FBgn0031566","log2FoldChange: -0.6773053908<br />-log10(padj): 6.554094e-01<br />log10(baseMean): 1.5659242<br />gene: FBgn0031568","log2FoldChange: 0.0107009708<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.6061807<br />gene: FBgn0031571","log2FoldChange: 0.0354251477<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.0682480<br />gene: FBgn0031573","log2FoldChange: -0.0501422139<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.4056301<br />gene: FBgn0031574","log2FoldChange: -0.0730459009<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.5379254<br />gene: FBgn0031575","log2FoldChange: 0.1904259334<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 1.0378980<br />gene: FBgn0031580","log2FoldChange: -0.5758779013<br />-log10(padj): 9.252481e-02<br />log10(baseMean): 0.7992004<br />gene: FBgn0031581","log2FoldChange: 0.1170333666<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 3.2049937<br />gene: FBgn0031589","log2FoldChange: -0.0581688680<br />-log10(padj): 5.455412e-02<br />log10(baseMean): 3.3745321<br />gene: FBgn0031590","log2FoldChange: 0.1659724647<br />-log10(padj): 8.199836e-02<br />log10(baseMean): 2.0818993<br />gene: FBgn0031597","log2FoldChange: 0.1648822455<br />-log10(padj): 3.035299e-01<br />log10(baseMean): 2.6522899<br />gene: FBgn0031598","log2FoldChange: 0.0124485527<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.1216586<br />gene: FBgn0031599","log2FoldChange: 0.0550947061<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 2.6392353<br />gene: FBgn0031600","log2FoldChange: 0.0682109686<br />-log10(padj): 2.579879e-02<br />log10(baseMean): 2.2245058<br />gene: FBgn0031601","log2FoldChange: 0.3045501479<br />-log10(padj): 3.247082e-01<br />log10(baseMean): 2.2335439<br />gene: FBgn0031603","log2FoldChange: 0.0120377498<br />-log10(padj): 7.543257e-03<br />log10(baseMean): 3.0474827<br />gene: FBgn0031604","log2FoldChange: 0.1523706613<br />-log10(padj): 1.699820e-01<br />log10(baseMean): 2.9359134<br />gene: FBgn0031606","log2FoldChange: 0.0226534194<br />-log10(padj): 1.069387e-02<br />log10(baseMean): 2.8351706<br />gene: FBgn0031607","log2FoldChange: 0.1507865085<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 2.2187074<br />gene: FBgn0031608","log2FoldChange: 0.2481951811<br />-log10(padj): 3.019219e-01<br />log10(baseMean): 2.3511994<br />gene: FBgn0031609","log2FoldChange: 0.0485431526<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.8971059<br />gene: FBgn0031610","log2FoldChange: 0.1203134194<br />-log10(padj): 7.856885e-02<br />log10(baseMean): 2.4082861<br />gene: FBgn0031611","log2FoldChange: 0.1678841916<br />-log10(padj): 2.404269e-02<br />log10(baseMean): 1.0093619<br />gene: FBgn0031622","log2FoldChange: 0.0676993340<br />-log10(padj): 2.830218e-02<br />log10(baseMean): 1.9502366<br />gene: FBgn0031628","log2FoldChange: 0.7341074788<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 0.9998172<br />gene: FBgn0031630","log2FoldChange: 0.1400497940<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.0398967<br />gene: FBgn0031631","log2FoldChange: -0.0528227083<br />-log10(padj): 2.749859e-02<br />log10(baseMean): 2.9641243<br />gene: FBgn0031633","log2FoldChange: 0.8995252045<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 0.8176475<br />gene: FBgn0031634","log2FoldChange: -0.1017210923<br />-log10(padj): 7.084185e-02<br />log10(baseMean): 2.9114550<br />gene: FBgn0031635","log2FoldChange: -0.3855147399<br />-log10(padj): 2.066376e+00<br />log10(baseMean): 3.0920198<br />gene: FBgn0031637","log2FoldChange: 0.0060282914<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.5630251<br />gene: FBgn0031638","log2FoldChange: 0.0567541509<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.4243397<br />gene: FBgn0031639","log2FoldChange: -0.0802967863<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.6042602<br />gene: FBgn0031640","log2FoldChange: 0.0549094914<br />-log10(padj): 3.921773e-02<br />log10(baseMean): 3.3080092<br />gene: FBgn0031643","log2FoldChange: 0.2065312970<br />-log10(padj): 2.167041e-01<br />log10(baseMean): 2.2834349<br />gene: FBgn0031644","log2FoldChange: 0.1193302083<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.5274231<br />gene: FBgn0031645","log2FoldChange: -0.9300098189<br />-log10(padj): 1.792604e-01<br />log10(baseMean): 0.8043682<br />gene: FBgn0031646","log2FoldChange: -0.0428804206<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.5827905<br />gene: FBgn0031651","log2FoldChange: -0.1974951782<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.9376273<br />gene: FBgn0031652","log2FoldChange: -0.0911750393<br />-log10(padj): 6.229977e-02<br />log10(baseMean): 2.3578510<br />gene: FBgn0031655","log2FoldChange: 0.1075451679<br />-log10(padj): 1.105617e-01<br />log10(baseMean): 2.5518497<br />gene: FBgn0031656","log2FoldChange: -0.0726368477<br />-log10(padj): 4.218202e-02<br />log10(baseMean): 2.5009579<br />gene: FBgn0031657","log2FoldChange: -0.0731245842<br />-log10(padj): 6.453047e-02<br />log10(baseMean): 2.7778880<br />gene: FBgn0031659","log2FoldChange: 0.2630651388<br />-log10(padj): 4.796111e-01<br />log10(baseMean): 3.1254172<br />gene: FBgn0031660","log2FoldChange: -0.0130332101<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.7298874<br />gene: FBgn0031661","log2FoldChange: -0.0997898457<br />-log10(padj): 8.780675e-02<br />log10(baseMean): 2.8936706<br />gene: FBgn0031662","log2FoldChange: -0.1295696064<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.7838315<br />gene: FBgn0031663","log2FoldChange: 0.2971711905<br />-log10(padj): 6.251722e-01<br />log10(baseMean): 2.6464610<br />gene: FBgn0031664","log2FoldChange: -0.1151302962<br />-log10(padj): 1.293714e-01<br />log10(baseMean): 3.1404701<br />gene: FBgn0031670","log2FoldChange: -0.0288524435<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 3.2241223<br />gene: FBgn0031673","log2FoldChange: 0.3777703714<br />-log10(padj): 2.016282e-01<br />log10(baseMean): 1.6786444<br />gene: FBgn0031675","log2FoldChange: 0.0055298817<br />-log10(padj): 3.092711e-03<br />log10(baseMean): 2.4280169<br />gene: FBgn0031676","log2FoldChange: -0.0243822277<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.6187847<br />gene: FBgn0031677","log2FoldChange: 0.0569064286<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 3.2670844<br />gene: FBgn0031678","log2FoldChange: -0.1873172704<br />-log10(padj): 4.729286e-01<br />log10(baseMean): 3.3626479<br />gene: FBgn0031681","log2FoldChange: 0.2259815544<br />-log10(padj): 2.348156e-01<br />log10(baseMean): 2.5357279<br />gene: FBgn0031682","log2FoldChange: -0.0355610315<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.3735948<br />gene: FBgn0031683","log2FoldChange: 0.0209070958<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.6371927<br />gene: FBgn0031684","log2FoldChange: 1.6024294753<br />-log10(padj): 3.346156e+00<br />log10(baseMean): 1.4886048<br />gene: FBgn0031688","log2FoldChange: 0.9572920444<br />-log10(padj): 7.078007e+00<br />log10(baseMean): 2.4380277<br />gene: FBgn0031689","log2FoldChange: 0.1771497757<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 0.9657510<br />gene: FBgn0031690","log2FoldChange: 0.3595294963<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.3528198<br />gene: FBgn0031693","log2FoldChange: 0.2491901712<br />-log10(padj): 5.010955e-02<br />log10(baseMean): 1.1597959<br />gene: FBgn0031694","log2FoldChange: -0.3739537769<br />-log10(padj): 8.264700e-02<br />log10(baseMean): 1.0759923<br />gene: FBgn0031695","log2FoldChange: -0.2007727931<br />-log10(padj): 5.991162e-01<br />log10(baseMean): 3.0059446<br />gene: FBgn0031696","log2FoldChange: -0.1739957332<br />-log10(padj): 1.696066e-01<br />log10(baseMean): 3.5850516<br />gene: FBgn0031698","log2FoldChange: -1.8457115363<br />-log10(padj): 9.032519e+00<br />log10(baseMean): 1.9581786<br />gene: FBgn0031703","log2FoldChange: 0.0934317728<br />-log10(padj): 7.383364e-03<br />log10(baseMean): 0.8302065<br />gene: FBgn0031707","log2FoldChange: 0.1301391730<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.3800176<br />gene: FBgn0031708","log2FoldChange: -0.0801918456<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.5323278<br />gene: FBgn0031710","log2FoldChange: -0.0508603747<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.9066804<br />gene: FBgn0031711","log2FoldChange: 0.2047384654<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.8065256<br />gene: FBgn0031713","log2FoldChange: 0.1443052579<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.7688623<br />gene: FBgn0031717","log2FoldChange: 0.0358178885<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 0.8162715<br />gene: FBgn0031735","log2FoldChange: -0.0394047964<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.9021167<br />gene: FBgn0031736","log2FoldChange: -0.3727912122<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 0.9175015<br />gene: FBgn0031737","log2FoldChange: 0.4718737831<br />-log10(padj): 2.422737e+00<br />log10(baseMean): 3.0019890<br />gene: FBgn0031738","log2FoldChange: -0.1268429547<br />-log10(padj): 8.780675e-02<br />log10(baseMean): 2.3181098<br />gene: FBgn0031739","log2FoldChange: 0.0839339034<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.8321363<br />gene: FBgn0031740","log2FoldChange: -1.0060223446<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 0.8966791<br />gene: FBgn0031747","log2FoldChange: -0.2920271485<br />-log10(padj): 1.345366e+00<br />log10(baseMean): 3.0699511<br />gene: FBgn0031752","log2FoldChange: 0.2541651028<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.2523503<br />gene: FBgn0031756","log2FoldChange: 0.3917739640<br />-log10(padj): 6.187511e-02<br />log10(baseMean): 0.8412496<br />gene: FBgn0031757","log2FoldChange: 0.7140632514<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 1.5209326<br />gene: FBgn0031758","log2FoldChange: -0.0766893488<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.6427275<br />gene: FBgn0031759","log2FoldChange: 0.0453438241<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.6586682<br />gene: FBgn0031760","log2FoldChange: -0.2363986905<br />-log10(padj): 3.019219e-01<br />log10(baseMean): 2.3992693<br />gene: FBgn0031762","log2FoldChange: -0.1012217592<br />-log10(padj): 7.897898e-02<br />log10(baseMean): 2.7140599<br />gene: FBgn0031764","log2FoldChange: 0.1765737077<br />-log10(padj): 1.779246e-01<br />log10(baseMean): 2.5733944<br />gene: FBgn0031768","log2FoldChange: 0.3600287884<br />-log10(padj): 2.502983e+00<br />log10(baseMean): 3.1069643<br />gene: FBgn0031769","log2FoldChange: -0.2375146283<br />-log10(padj): 6.188583e-01<br />log10(baseMean): 3.2533678<br />gene: FBgn0031771","log2FoldChange: 0.0702328896<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.2990558<br />gene: FBgn0031772","log2FoldChange: 0.2217357172<br />-log10(padj): 2.380728e-01<br />log10(baseMean): 2.2231966<br />gene: FBgn0031773","log2FoldChange: 0.2286679953<br />-log10(padj): 1.836853e-01<br />log10(baseMean): 2.2893207<br />gene: FBgn0031774","log2FoldChange: 0.8181274694<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 0.9723521<br />gene: FBgn0031775","log2FoldChange: -0.0035086615<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 2.9088654<br />gene: FBgn0031776","log2FoldChange: 0.1106510288<br />-log10(padj): 5.356785e-02<br />log10(baseMean): 1.9303719<br />gene: FBgn0031777","log2FoldChange: -0.0095637536<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.8376253<br />gene: FBgn0031779","log2FoldChange: -0.1144986249<br />-log10(padj): 6.938076e-02<br />log10(baseMean): 3.1021738<br />gene: FBgn0031781","log2FoldChange: 0.1463831214<br />-log10(padj): 1.208914e-01<br />log10(baseMean): 2.8021007<br />gene: FBgn0031782","log2FoldChange: -0.2520161454<br />-log10(padj): 6.654564e-01<br />log10(baseMean): 2.8571831<br />gene: FBgn0031799","log2FoldChange: -0.8555392972<br />-log10(padj): 5.442078e+00<br />log10(baseMean): 3.4926050<br />gene: FBgn0031805","log2FoldChange: 0.2098206510<br />-log10(padj): 1.290822e-01<br />log10(baseMean): 1.9493324<br />gene: FBgn0031811","log2FoldChange: -0.3096700009<br />-log10(padj): 9.396274e-01<br />log10(baseMean): 2.5184497<br />gene: FBgn0031812","log2FoldChange: -0.1021705498<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 2.6044827<br />gene: FBgn0031813","log2FoldChange: 0.3072608815<br />-log10(padj): 6.813664e-01<br />log10(baseMean): 2.8728229<br />gene: FBgn0031814","log2FoldChange: -0.2568739422<br />-log10(padj): 6.195511e-01<br />log10(baseMean): 2.7960667<br />gene: FBgn0031815","log2FoldChange: 0.6519340257<br />-log10(padj): 1.819872e+00<br />log10(baseMean): 2.1644098<br />gene: FBgn0031816","log2FoldChange: 0.2724337347<br />-log10(padj): 3.244687e-01<br />log10(baseMean): 2.1899956<br />gene: FBgn0031817","log2FoldChange: -0.1279446381<br />-log10(padj): 1.208914e-01<br />log10(baseMean): 2.9566622<br />gene: FBgn0031818","log2FoldChange: 0.1631567362<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 2.3622902<br />gene: FBgn0031822","log2FoldChange: -0.0807281913<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.3839832<br />gene: FBgn0031824","log2FoldChange: -0.2693458982<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 0.8567042<br />gene: FBgn0031829","log2FoldChange: 0.0873430635<br />-log10(padj): 5.079791e-02<br />log10(baseMean): 3.1116474<br />gene: FBgn0031830","log2FoldChange: 0.0932608883<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 1.1355952<br />gene: FBgn0031831","log2FoldChange: 0.0406669218<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 2.4003254<br />gene: FBgn0031832","log2FoldChange: -0.1801035933<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 1.8676342<br />gene: FBgn0031834","log2FoldChange: 0.1406179709<br />-log10(padj): 2.512591e-01<br />log10(baseMean): 3.0198862<br />gene: FBgn0031836","log2FoldChange: -0.1377374948<br />-log10(padj): 3.812981e-01<br />log10(baseMean): 3.3779803<br />gene: FBgn0031842","log2FoldChange: 0.5295303455<br />-log10(padj): 6.004532e-01<br />log10(baseMean): 1.7296637<br />gene: FBgn0031844","log2FoldChange: -0.3102840695<br />-log10(padj): 9.543967e-01<br />log10(baseMean): 2.7847561<br />gene: FBgn0031845","log2FoldChange: -0.1274792327<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 2.1821015<br />gene: FBgn0031848","log2FoldChange: 0.4850068217<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.0153186<br />gene: FBgn0031850","log2FoldChange: -0.1721607411<br />-log10(padj): 2.212443e-01<br />log10(baseMean): 2.7695721<br />gene: FBgn0031851","log2FoldChange: -0.2714333632<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 1.2388494<br />gene: FBgn0031857","log2FoldChange: -0.4033906415<br />-log10(padj): 1.109972e-01<br />log10(baseMean): 1.1978594<br />gene: FBgn0031865","log2FoldChange: -0.0122345510<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.9868810<br />gene: FBgn0031868","log2FoldChange: 0.0570925874<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 0.8220369<br />gene: FBgn0031869","log2FoldChange: 0.2587556891<br />-log10(padj): 7.947596e-01<br />log10(baseMean): 2.7243003<br />gene: FBgn0031871","log2FoldChange: 0.3411971837<br />-log10(padj): 1.670621e+00<br />log10(baseMean): 2.6986232<br />gene: FBgn0031872","log2FoldChange: 0.1326959570<br />-log10(padj): 7.716672e-02<br />log10(baseMean): 2.1912218<br />gene: FBgn0031873","log2FoldChange: -0.1118124971<br />-log10(padj): 4.949434e-02<br />log10(baseMean): 2.0915994<br />gene: FBgn0031874","log2FoldChange: -0.1349720237<br />-log10(padj): 1.241397e-01<br />log10(baseMean): 2.7479611<br />gene: FBgn0031875","log2FoldChange: 0.1685048950<br />-log10(padj): 1.316693e-01<br />log10(baseMean): 2.2977875<br />gene: FBgn0031876","log2FoldChange: -0.1294608002<br />-log10(padj): 5.560148e-02<br />log10(baseMean): 2.0740258<br />gene: FBgn0031877","log2FoldChange: 0.1917706927<br />-log10(padj): 6.133292e-01<br />log10(baseMean): 3.2820302<br />gene: FBgn0031878","log2FoldChange: -0.4914555893<br />-log10(padj): 1.431785e-01<br />log10(baseMean): 1.2478826<br />gene: FBgn0031880","log2FoldChange: 0.0233935233<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.3566451<br />gene: FBgn0031881","log2FoldChange: -0.0604952275<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.4677697<br />gene: FBgn0031882","log2FoldChange: -0.0796746668<br />-log10(padj): 6.507899e-02<br />log10(baseMean): 3.1635942<br />gene: FBgn0031883","log2FoldChange: -0.3945964530<br />-log10(padj): 1.092846e+00<br />log10(baseMean): 2.7615517<br />gene: FBgn0031885","log2FoldChange: 0.1076476233<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.6114102<br />gene: FBgn0031886","log2FoldChange: 0.4927500350<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 1.1482851<br />gene: FBgn0031887","log2FoldChange: 0.4560123329<br />-log10(padj): 4.154926e+00<br />log10(baseMean): 3.4355201<br />gene: FBgn0031888","log2FoldChange: -0.1736219393<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 2.6461371<br />gene: FBgn0031893","log2FoldChange: 0.3092974969<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 2.1760879<br />gene: FBgn0031894","log2FoldChange: 0.4516109237<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 1.8863346<br />gene: FBgn0031895","log2FoldChange: 0.2095331237<br />-log10(padj): 5.061041e-01<br />log10(baseMean): 2.8113451<br />gene: FBgn0031896","log2FoldChange: 0.0370370293<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 3.0271144<br />gene: FBgn0031897","log2FoldChange: 0.0929788971<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 2.6661589<br />gene: FBgn0031904","log2FoldChange: -0.3323263017<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 0.8963522<br />gene: FBgn0031907","log2FoldChange: 0.0782076446<br />-log10(padj): 5.994519e-02<br />log10(baseMean): 2.4530682<br />gene: FBgn0031909","log2FoldChange: -1.0745588720<br />-log10(padj): 1.141165e+01<br />log10(baseMean): 3.1200249<br />gene: FBgn0031912","log2FoldChange: -0.7340811187<br />-log10(padj): 4.461610e+00<br />log10(baseMean): 2.9793974<br />gene: FBgn0031913","log2FoldChange: -0.5390876129<br />-log10(padj): 1.636443e+00<br />log10(baseMean): 2.6184612<br />gene: FBgn0031914","log2FoldChange: -0.9732792427<br />-log10(padj): 5.303612e-01<br />log10(baseMean): 1.1922725<br />gene: FBgn0031920","log2FoldChange: 0.9073779115<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 0.9852768<br />gene: FBgn0031936","log2FoldChange: 0.3409511036<br />-log10(padj): 3.214573e-01<br />log10(baseMean): 2.6548634<br />gene: FBgn0031937","log2FoldChange: -0.0549514596<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 3.0048716<br />gene: FBgn0031947","log2FoldChange: -0.1161249127<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.0424772<br />gene: FBgn0031948","log2FoldChange: -0.1085441744<br />-log10(padj): 1.217867e-01<br />log10(baseMean): 2.8106004<br />gene: FBgn0031950","log2FoldChange: -0.0302990119<br />-log10(padj): 1.609026e-02<br />log10(baseMean): 2.5593964<br />gene: FBgn0031951","log2FoldChange: -0.4112735649<br />-log10(padj): 1.732883e+00<br />log10(baseMean): 2.6143906<br />gene: FBgn0031952","log2FoldChange: -0.4718834943<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 1.1057193<br />gene: FBgn0031955","log2FoldChange: -0.9137738359<br />-log10(padj): 1.133596e+00<br />log10(baseMean): 1.5081124<br />gene: FBgn0031957","log2FoldChange: 0.2168800137<br />-log10(padj): 2.763200e-02<br />log10(baseMean): 0.8334821<br />gene: FBgn0031961","log2FoldChange: 0.1261487486<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 2.7828611<br />gene: FBgn0031962","log2FoldChange: 0.8690017300<br />-log10(padj): 2.630439e+00<br />log10(baseMean): 1.8688046<br />gene: FBgn0031968","log2FoldChange: -0.1246581713<br />-log10(padj): 1.669714e-01<br />log10(baseMean): 3.6401040<br />gene: FBgn0031969","log2FoldChange: -0.0628507837<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 1.2056826<br />gene: FBgn0031970","log2FoldChange: 0.1067206783<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.4221265<br />gene: FBgn0031971","log2FoldChange: -0.3138099790<br />-log10(padj): 6.140041e-01<br />log10(baseMean): 2.5378572<br />gene: FBgn0031972","log2FoldChange: -0.2446970049<br />-log10(padj): 3.234208e-01<br />log10(baseMean): 3.0931912<br />gene: FBgn0031973","log2FoldChange: 0.0598483207<br />-log10(padj): 1.882499e-02<br />log10(baseMean): 1.8374925<br />gene: FBgn0031974","log2FoldChange: -0.3404621128<br />-log10(padj): 1.111215e-01<br />log10(baseMean): 1.3920892<br />gene: FBgn0031975","log2FoldChange: -0.6369389467<br />-log10(padj): 9.682541e-02<br />log10(baseMean): 1.8063757<br />gene: FBgn0031976","log2FoldChange: 0.1835357485<br />-log10(padj): 1.933346e-01<br />log10(baseMean): 3.1859968<br />gene: FBgn0031977","log2FoldChange: 0.4261087975<br />-log10(padj): 2.244500e-01<br />log10(baseMean): 1.7741700<br />gene: FBgn0031979","log2FoldChange: 0.0691114096<br />-log10(padj): 3.987579e-02<br />log10(baseMean): 4.2255081<br />gene: FBgn0031980","log2FoldChange: 0.1611381644<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.9590937<br />gene: FBgn0031981","log2FoldChange: 0.0585710261<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 3.2746039<br />gene: FBgn0031985","log2FoldChange: -0.3678762026<br />-log10(padj): 8.224991e-01<br />log10(baseMean): 2.4476352<br />gene: FBgn0031987","log2FoldChange: -0.3635967862<br />-log10(padj): 2.185871e+00<br />log10(baseMean): 3.2718869<br />gene: FBgn0031988","log2FoldChange: -0.3222195901<br />-log10(padj): 8.676516e-01<br />log10(baseMean): 3.6164013<br />gene: FBgn0031990","log2FoldChange: 0.6179876840<br />-log10(padj): 8.212087e-01<br />log10(baseMean): 2.2797758<br />gene: FBgn0031992","log2FoldChange: -0.2580927743<br />-log10(padj): 7.434783e-01<br />log10(baseMean): 4.1059631<br />gene: FBgn0031993","log2FoldChange: 0.1014043928<br />-log10(padj): 1.408800e-01<br />log10(baseMean): 3.0913374<br />gene: FBgn0031995","log2FoldChange: 0.3228305692<br />-log10(padj): 1.171945e+00<br />log10(baseMean): 2.7163850<br />gene: FBgn0031996","log2FoldChange: 0.0843245291<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 2.4696554<br />gene: FBgn0031997","log2FoldChange: -0.1115908936<br />-log10(padj): 7.348271e-02<br />log10(baseMean): 2.8237266<br />gene: FBgn0031998","log2FoldChange: 0.0311678615<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.8768127<br />gene: FBgn0031999","log2FoldChange: -0.0407481564<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 2.6218207<br />gene: FBgn0032000","log2FoldChange: 0.0767277671<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.5068698<br />gene: FBgn0032001","log2FoldChange: -0.1190766625<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 1.7194973<br />gene: FBgn0032002","log2FoldChange: -0.5480082256<br />-log10(padj): 1.666483e-01<br />log10(baseMean): 1.2201294<br />gene: FBgn0032003","log2FoldChange: -1.1855552886<br />-log10(padj): 1.077421e+01<br />log10(baseMean): 2.9829835<br />gene: FBgn0032004","log2FoldChange: -0.3997191586<br />-log10(padj): 3.125300e+00<br />log10(baseMean): 3.6076529<br />gene: FBgn0032005","log2FoldChange: 0.1758521403<br />-log10(padj): 4.196281e-01<br />log10(baseMean): 4.0916121<br />gene: FBgn0032006","log2FoldChange: 0.4404289352<br />-log10(padj): 2.778236e-01<br />log10(baseMean): 1.5204449<br />gene: FBgn0032010","log2FoldChange: 0.4669422242<br />-log10(padj): 9.152652e-01<br />log10(baseMean): 2.0008861<br />gene: FBgn0032013","log2FoldChange: -0.0505409405<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.7362075<br />gene: FBgn0032014","log2FoldChange: 0.0368076394<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 3.3379121<br />gene: FBgn0032015","log2FoldChange: -0.0295516527<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.6409923<br />gene: FBgn0032016","log2FoldChange: 0.2061302403<br />-log10(padj): 1.111215e-01<br />log10(baseMean): 1.9875948<br />gene: FBgn0032017","log2FoldChange: -0.3126401766<br />-log10(padj): 1.007882e+00<br />log10(baseMean): 3.2390648<br />gene: FBgn0032018","log2FoldChange: 0.4747630942<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 1.9988769<br />gene: FBgn0032019","log2FoldChange: 0.1493655998<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.7051692<br />gene: FBgn0032020","log2FoldChange: -0.2506535925<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 3.0292787<br />gene: FBgn0032024","log2FoldChange: -0.0546686186<br />-log10(padj): 1.913344e-02<br />log10(baseMean): 2.3907829<br />gene: FBgn0032025","log2FoldChange: 0.3298439476<br />-log10(padj): 1.678801e+00<br />log10(baseMean): 3.4114956<br />gene: FBgn0032026","log2FoldChange: -0.9358428261<br />-log10(padj): 7.679285e+00<br />log10(baseMean): 2.5314574<br />gene: FBgn0032029","log2FoldChange: -0.0036942086<br />-log10(padj): 2.753780e-03<br />log10(baseMean): 2.8229643<br />gene: FBgn0032030","log2FoldChange: 0.0370916533<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.4137576<br />gene: FBgn0032031","log2FoldChange: 0.2908855639<br />-log10(padj): 3.928304e-01<br />log10(baseMean): 2.4124926<br />gene: FBgn0032032","log2FoldChange: -0.1072419693<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.0113082<br />gene: FBgn0032033","log2FoldChange: -0.1892435998<br />-log10(padj): 1.097356e-01<br />log10(baseMean): 2.2503862<br />gene: FBgn0032034","log2FoldChange: 0.0647855196<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 3.0249058<br />gene: FBgn0032035","log2FoldChange: -0.5603357538<br />-log10(padj): 5.827646e+00<br />log10(baseMean): 3.8784575<br />gene: FBgn0032036","log2FoldChange: 0.3047509256<br />-log10(padj): 8.409143e-01<br />log10(baseMean): 3.0144215<br />gene: FBgn0032040","log2FoldChange: -0.2025009829<br />-log10(padj): 1.727186e-01<br />log10(baseMean): 2.6866070<br />gene: FBgn0032042","log2FoldChange: 0.4207776760<br />-log10(padj): 4.056397e-01<br />log10(baseMean): 1.7843501<br />gene: FBgn0032047","log2FoldChange: -0.1257363476<br />-log10(padj): 1.665815e-01<br />log10(baseMean): 3.3348169<br />gene: FBgn0032050","log2FoldChange: -0.1592617687<br />-log10(padj): 2.704925e-01<br />log10(baseMean): 3.0571060<br />gene: FBgn0032051","log2FoldChange: -0.0027706102<br />-log10(padj): 1.958023e-03<br />log10(baseMean): 2.9997868<br />gene: FBgn0032052","log2FoldChange: 0.1452581049<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 2.4408088<br />gene: FBgn0032053","log2FoldChange: -0.0303054656<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.7962175<br />gene: FBgn0032054","log2FoldChange: 0.0386925888<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.4984355<br />gene: FBgn0032059","log2FoldChange: 0.1923256061<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 0.8063080<br />gene: FBgn0032061","log2FoldChange: 0.3956242529<br />-log10(padj): 2.864518e+00<br />log10(baseMean): 3.6752433<br />gene: FBgn0032078","log2FoldChange: -0.3503530278<br />-log10(padj): 4.965331e-01<br />log10(baseMean): 2.7263081<br />gene: FBgn0032079","log2FoldChange: -0.2396375215<br />-log10(padj): 3.889025e-02<br />log10(baseMean): 0.9963595<br />gene: FBgn0032080","log2FoldChange: 0.4175428803<br />-log10(padj): 9.152652e-01<br />log10(baseMean): 2.5679925<br />gene: FBgn0032082","log2FoldChange: 0.6998330903<br />-log10(padj): 4.547191e-01<br />log10(baseMean): 1.4278486<br />gene: FBgn0032083","log2FoldChange: 0.3935621960<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 1.4864998<br />gene: FBgn0032084","log2FoldChange: 0.4983143748<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 1.0346245<br />gene: FBgn0032085","log2FoldChange: 0.0499034964<br />-log10(padj): 2.533598e-02<br />log10(baseMean): 2.8034911<br />gene: FBgn0032101","log2FoldChange: 0.5228783464<br />-log10(padj): 3.124079e+00<br />log10(baseMean): 2.7576488<br />gene: FBgn0032105","log2FoldChange: -0.0284426222<br />-log10(padj): 9.302360e-03<br />log10(baseMean): 2.0911373<br />gene: FBgn0032117","log2FoldChange: 0.0830822290<br />-log10(padj): 7.716672e-02<br />log10(baseMean): 3.3142889<br />gene: FBgn0032120","log2FoldChange: 0.1721915186<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.6026248<br />gene: FBgn0032123","log2FoldChange: -1.0605695874<br />-log10(padj): 3.188201e-01<br />log10(baseMean): 0.8886890<br />gene: FBgn0032129","log2FoldChange: -0.3134615940<br />-log10(padj): 8.797138e-01<br />log10(baseMean): 2.6151917<br />gene: FBgn0032130","log2FoldChange: 0.2281942387<br />-log10(padj): 1.869441e-01<br />log10(baseMean): 2.2149049<br />gene: FBgn0032135","log2FoldChange: 0.4463951056<br />-log10(padj): 5.127314e-01<br />log10(baseMean): 2.4890460<br />gene: FBgn0032136","log2FoldChange: -0.1578064714<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 3.6653567<br />gene: FBgn0032138","log2FoldChange: -0.4118945581<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 2.0122305<br />gene: FBgn0032139","log2FoldChange: 0.3824981251<br />-log10(padj): 1.408219e+00<br />log10(baseMean): 3.3322941<br />gene: FBgn0032140","log2FoldChange: -0.2015963410<br />-log10(padj): 3.162310e-02<br />log10(baseMean): 1.2569034<br />gene: FBgn0032142","log2FoldChange: -0.1536874840<br />-log10(padj): 1.490190e-01<br />log10(baseMean): 2.4981135<br />gene: FBgn0032147","log2FoldChange: 0.1385624643<br />-log10(padj): 1.420115e-01<br />log10(baseMean): 2.5094237<br />gene: FBgn0032149","log2FoldChange: -0.2589180248<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 1.6942529<br />gene: FBgn0032150","log2FoldChange: 0.0937708650<br />-log10(padj): 1.885426e-02<br />log10(baseMean): 1.3937504<br />gene: FBgn0032153","log2FoldChange: 0.0512619166<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.3449534<br />gene: FBgn0032154","log2FoldChange: -0.4297061058<br />-log10(padj): 3.225730e+00<br />log10(baseMean): 3.1581181<br />gene: FBgn0032156","log2FoldChange: 0.1002409971<br />-log10(padj): 1.168489e-01<br />log10(baseMean): 2.9913507<br />gene: FBgn0032157","log2FoldChange: -0.0151994052<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 2.7631189<br />gene: FBgn0032160","log2FoldChange: 0.1298348664<br />-log10(padj): 4.745188e-02<br />log10(baseMean): 1.6547573<br />gene: FBgn0032161","log2FoldChange: 0.0768017646<br />-log10(padj): 3.091232e-02<br />log10(baseMean): 1.9565083<br />gene: FBgn0032162","log2FoldChange: -0.7348268727<br />-log10(padj): 2.396890e-01<br />log10(baseMean): 1.1331208<br />gene: FBgn0032163","log2FoldChange: -0.1075892506<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.4646195<br />gene: FBgn0032166","log2FoldChange: -0.3481830266<br />-log10(padj): 8.563559e-01<br />log10(baseMean): 2.8642621<br />gene: FBgn0032167","log2FoldChange: -0.0218711713<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.3123914<br />gene: FBgn0032168","log2FoldChange: 0.0990023725<br />-log10(padj): 6.974744e-02<br />log10(baseMean): 2.3153575<br />gene: FBgn0032169","log2FoldChange: -0.2492768638<br />-log10(padj): 4.334437e-01<br />log10(baseMean): 2.8953921<br />gene: FBgn0032170","log2FoldChange: -0.0722310533<br />-log10(padj): 3.434146e-02<br />log10(baseMean): 2.1193044<br />gene: FBgn0032171","log2FoldChange: -0.1338179842<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.2373784<br />gene: FBgn0032172","log2FoldChange: -0.0239440406<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 1.6750551<br />gene: FBgn0032178","log2FoldChange: -0.1192260932<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.2127129<br />gene: FBgn0032189","log2FoldChange: -0.0928503196<br />-log10(padj): 7.196422e-02<br />log10(baseMean): 3.0818518<br />gene: FBgn0032191","log2FoldChange: -0.0820221875<br />-log10(padj): 2.055674e-02<br />log10(baseMean): 1.6450189<br />gene: FBgn0032192","log2FoldChange: 0.0260631394<br />-log10(padj): 5.431473e-03<br />log10(baseMean): 1.9850353<br />gene: FBgn0032193","log2FoldChange: 0.0136476819<br />-log10(padj): 6.455890e-03<br />log10(baseMean): 2.7255602<br />gene: FBgn0032194","log2FoldChange: -0.0084626360<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.2241767<br />gene: FBgn0032195","log2FoldChange: -0.0660420764<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.5317826<br />gene: FBgn0032196","log2FoldChange: 0.0171782197<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.6232807<br />gene: FBgn0032197","log2FoldChange: 0.1305004569<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 3.3138606<br />gene: FBgn0032198","log2FoldChange: 0.2079580127<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 2.6329687<br />gene: FBgn0032200","log2FoldChange: -0.4173611209<br />-log10(padj): 6.261658e-01<br />log10(baseMean): 2.6773364<br />gene: FBgn0032202","log2FoldChange: 0.1308310447<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.3099778<br />gene: FBgn0032204","log2FoldChange: -0.2491962082<br />-log10(padj): 1.177909e-01<br />log10(baseMean): 1.9994957<br />gene: FBgn0032205","log2FoldChange: -0.1080909352<br />-log10(padj): 1.638394e-01<br />log10(baseMean): 3.8920044<br />gene: FBgn0032208","log2FoldChange: -0.3238001487<br />-log10(padj): 2.713173e-01<br />log10(baseMean): 2.5585089<br />gene: FBgn0032210","log2FoldChange: -0.1223200435<br />-log10(padj): 2.823212e-01<br />log10(baseMean): 3.6478440<br />gene: FBgn0032213","log2FoldChange: 0.0884777326<br />-log10(padj): 4.675296e-02<br />log10(baseMean): 2.6093540<br />gene: FBgn0032214","log2FoldChange: 0.1907149959<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.4121898<br />gene: FBgn0032215","log2FoldChange: 0.0555627886<br />-log10(padj): 5.286807e-02<br />log10(baseMean): 3.2243081<br />gene: FBgn0032216","log2FoldChange: -0.0245632994<br />-log10(padj): 8.278629e-03<br />log10(baseMean): 3.1141638<br />gene: FBgn0032217","log2FoldChange: -0.3514649109<br />-log10(padj): 3.473641e-01<br />log10(baseMean): 2.5958756<br />gene: FBgn0032218","log2FoldChange: 0.5592789591<br />-log10(padj): 1.441906e-01<br />log10(baseMean): 1.0121743<br />gene: FBgn0032219","log2FoldChange: 0.0571746117<br />-log10(padj): 1.466187e-02<br />log10(baseMean): 1.6441002<br />gene: FBgn0032221","log2FoldChange: -0.2083870474<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.7419816<br />gene: FBgn0032222","log2FoldChange: -0.0805469619<br />-log10(padj): 3.436600e-02<br />log10(baseMean): 2.7390755<br />gene: FBgn0032223","log2FoldChange: 0.1734736330<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.4640222<br />gene: FBgn0032224","log2FoldChange: 0.0409773175<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.6275555<br />gene: FBgn0032228","log2FoldChange: -0.0288236636<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.8687747<br />gene: FBgn0032229","log2FoldChange: -0.2534631771<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 1.0513041<br />gene: FBgn0032230","log2FoldChange: -0.0590091605<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.4509507<br />gene: FBgn0032231","log2FoldChange: 0.3735953073<br />-log10(padj): 1.177783e-01<br />log10(baseMean): 1.2788116<br />gene: FBgn0032233","log2FoldChange: -0.0003395776<br />-log10(padj): 3.308367e-04<br />log10(baseMean): 2.6256913<br />gene: FBgn0032234","log2FoldChange: 0.0874404928<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 1.3920793<br />gene: FBgn0032235","log2FoldChange: -0.0409715269<br />-log10(padj): 1.882090e-02<br />log10(baseMean): 2.7651748<br />gene: FBgn0032236","log2FoldChange: 0.3711452610<br />-log10(padj): 8.853984e-01<br />log10(baseMean): 3.4330990<br />gene: FBgn0032237","log2FoldChange: 0.0203522440<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 3.2359285<br />gene: FBgn0032242","log2FoldChange: -0.0502761663<br />-log10(padj): 2.373932e-02<br />log10(baseMean): 2.5780412<br />gene: FBgn0032243","log2FoldChange: -0.0622268801<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.4223864<br />gene: FBgn0032244","log2FoldChange: 0.0661556078<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.4183529<br />gene: FBgn0032246","log2FoldChange: -0.0133629235<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 1.9465749<br />gene: FBgn0032247","log2FoldChange: 0.3865478511<br />-log10(padj): 9.774826e-01<br />log10(baseMean): 2.4594615<br />gene: FBgn0032248","log2FoldChange: 0.0569643377<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 1.9994930<br />gene: FBgn0032249","log2FoldChange: 0.1437851784<br />-log10(padj): 9.544956e-02<br />log10(baseMean): 2.3136882<br />gene: FBgn0032250","log2FoldChange: -0.2819619465<br />-log10(padj): 2.287577e-01<br />log10(baseMean): 2.0349591<br />gene: FBgn0032251","log2FoldChange: 0.4080974335<br />-log10(padj): 7.005836e-01<br />log10(baseMean): 2.0418371<br />gene: FBgn0032252","log2FoldChange: 0.3050724621<br />-log10(padj): 4.579421e-01<br />log10(baseMean): 2.5353189<br />gene: FBgn0032253","log2FoldChange: -0.2223540404<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.3085626<br />gene: FBgn0032256","log2FoldChange: 0.1299753079<br />-log10(padj): 1.874626e-01<br />log10(baseMean): 3.0423015<br />gene: FBgn0032258","log2FoldChange: 0.1359496503<br />-log10(padj): 7.971213e-02<br />log10(baseMean): 2.0868995<br />gene: FBgn0032259","log2FoldChange: 0.0323335095<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.1410610<br />gene: FBgn0032261","log2FoldChange: 0.1473781854<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 1.2523852<br />gene: FBgn0032262","log2FoldChange: 0.1226438734<br />-log10(padj): 1.238949e-01<br />log10(baseMean): 3.1126583<br />gene: FBgn0032264","log2FoldChange: 0.1061360935<br />-log10(padj): 8.492934e-02<br />log10(baseMean): 2.4294778<br />gene: FBgn0032290","log2FoldChange: -0.0984027697<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.8810120<br />gene: FBgn0032292","log2FoldChange: -0.0455749710<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 2.3575669<br />gene: FBgn0032293","log2FoldChange: 0.0503418412<br />-log10(padj): 3.115612e-02<br />log10(baseMean): 3.0212551<br />gene: FBgn0032295","log2FoldChange: 0.2259192045<br />-log10(padj): 3.214573e-01<br />log10(baseMean): 2.7489135<br />gene: FBgn0032296","log2FoldChange: -0.0161074077<br />-log10(padj): 5.064052e-03<br />log10(baseMean): 2.2541202<br />gene: FBgn0032297","log2FoldChange: -0.1671291314<br />-log10(padj): 1.751247e-01<br />log10(baseMean): 2.9077516<br />gene: FBgn0032298","log2FoldChange: -0.1705402601<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 3.2369629<br />gene: FBgn0032305","log2FoldChange: -0.4104314338<br />-log10(padj): 8.962574e-02<br />log10(baseMean): 1.0297142<br />gene: FBgn0032310","log2FoldChange: -0.1660912741<br />-log10(padj): 2.060188e-01<br />log10(baseMean): 2.7493127<br />gene: FBgn0032321","log2FoldChange: 0.3796468599<br />-log10(padj): 7.514536e-01<br />log10(baseMean): 2.3834564<br />gene: FBgn0032322","log2FoldChange: -0.1121110396<br />-log10(padj): 7.947646e-02<br />log10(baseMean): 2.3804363<br />gene: FBgn0032329","log2FoldChange: -0.6574430602<br />-log10(padj): 2.049371e+00<br />log10(baseMean): 2.5357235<br />gene: FBgn0032330","log2FoldChange: -0.0979860691<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.6534707<br />gene: FBgn0032339","log2FoldChange: 0.1160183599<br />-log10(padj): 2.018527e-01<br />log10(baseMean): 3.4672730<br />gene: FBgn0032340","log2FoldChange: 0.0425627992<br />-log10(padj): 3.366415e-02<br />log10(baseMean): 3.1025721<br />gene: FBgn0032341","log2FoldChange: -0.1175296730<br />-log10(padj): 4.897514e-02<br />log10(baseMean): 1.7716238<br />gene: FBgn0032343","log2FoldChange: -0.3072197571<br />-log10(padj): 4.497479e-02<br />log10(baseMean): 0.9428133<br />gene: FBgn0032345","log2FoldChange: -0.0924902995<br />-log10(padj): 4.284657e-02<br />log10(baseMean): 2.2645523<br />gene: FBgn0032346","log2FoldChange: 0.1064289024<br />-log10(padj): 1.249646e-01<br />log10(baseMean): 3.0460761<br />gene: FBgn0032348","log2FoldChange: 0.7605145253<br />-log10(padj): 3.765035e-01<br />log10(baseMean): 1.1449647<br />gene: FBgn0032350","log2FoldChange: 0.0962382734<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.2211357<br />gene: FBgn0032354","log2FoldChange: -0.0922417448<br />-log10(padj): 3.792700e-02<br />log10(baseMean): 1.9807251<br />gene: FBgn0032358","log2FoldChange: -0.1557909722<br />-log10(padj): 1.445256e-01<br />log10(baseMean): 2.4441429<br />gene: FBgn0032361","log2FoldChange: -0.1040848092<br />-log10(padj): 1.331873e-01<br />log10(baseMean): 3.4808815<br />gene: FBgn0032363","log2FoldChange: -0.1018984793<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.8244973<br />gene: FBgn0032373","log2FoldChange: 0.4305979958<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 1.5527506<br />gene: FBgn0032377","log2FoldChange: -0.1052238085<br />-log10(padj): 1.434008e-01<br />log10(baseMean): 3.3051395<br />gene: FBgn0032378","log2FoldChange: -0.2591395848<br />-log10(padj): 4.413021e-01<br />log10(baseMean): 2.3149532<br />gene: FBgn0032382","log2FoldChange: -0.0231212761<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.6404423<br />gene: FBgn0032387","log2FoldChange: -0.0479053846<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 3.3376592<br />gene: FBgn0032388","log2FoldChange: 0.2517512285<br />-log10(padj): 5.060940e-01<br />log10(baseMean): 2.6070388<br />gene: FBgn0032390","log2FoldChange: 0.1756401060<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 2.1569282<br />gene: FBgn0032391","log2FoldChange: 0.1131824984<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 2.9519637<br />gene: FBgn0032393","log2FoldChange: 0.1111811429<br />-log10(padj): 6.917231e-02<br />log10(baseMean): 2.8336434<br />gene: FBgn0032394","log2FoldChange: -0.2621802270<br />-log10(padj): 7.506008e-01<br />log10(baseMean): 2.7308688<br />gene: FBgn0032395","log2FoldChange: 0.0214180083<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 2.8938724<br />gene: FBgn0032397","log2FoldChange: -0.0785546763<br />-log10(padj): 8.273621e-02<br />log10(baseMean): 3.0864692<br />gene: FBgn0032398","log2FoldChange: -0.0020220714<br />-log10(padj): 1.175523e-03<br />log10(baseMean): 1.9033840<br />gene: FBgn0032399","log2FoldChange: -0.8201954224<br />-log10(padj): 1.955312e+00<br />log10(baseMean): 2.6397779<br />gene: FBgn0032400","log2FoldChange: 0.1081871619<br />-log10(padj): 3.759752e-02<br />log10(baseMean): 1.7815132<br />gene: FBgn0032401","log2FoldChange: 0.3247779140<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 1.6328592<br />gene: FBgn0032402","log2FoldChange: 2.6727386583<br />-log10(padj): 2.547902e+01<br />log10(baseMean): 1.9010003<br />gene: FBgn0032405","log2FoldChange: 0.1702705893<br />-log10(padj): 3.048210e-01<br />log10(baseMean): 2.9761686<br />gene: FBgn0032407","log2FoldChange: -0.2543554813<br />-log10(padj): 4.579421e-01<br />log10(baseMean): 2.6964995<br />gene: FBgn0032408","log2FoldChange: 0.3205304339<br />-log10(padj): 1.506432e+00<br />log10(baseMean): 2.9532466<br />gene: FBgn0032409","log2FoldChange: 0.7882775973<br />-log10(padj): 3.423274e+00<br />log10(baseMean): 2.1285425<br />gene: FBgn0032420","log2FoldChange: 0.7158417156<br />-log10(padj): 2.242441e+00<br />log10(baseMean): 2.2565012<br />gene: FBgn0032421","log2FoldChange: 0.9580534596<br />-log10(padj): 1.366942e+00<br />log10(baseMean): 1.6290931<br />gene: FBgn0032422","log2FoldChange: -0.0794501999<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 1.2732229<br />gene: FBgn0032428","log2FoldChange: 0.1967316232<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 2.8765828<br />gene: FBgn0032429","log2FoldChange: -0.0429842569<br />-log10(padj): 3.243344e-02<br />log10(baseMean): 2.8194331<br />gene: FBgn0032430","log2FoldChange: 0.1790718682<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 1.2582117<br />gene: FBgn0032433","log2FoldChange: 2.3502629901<br />-log10(padj): 3.807875e+00<br />log10(baseMean): 1.2375343<br />gene: FBgn0032436","log2FoldChange: 0.2368244934<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.7539303<br />gene: FBgn0032444","log2FoldChange: 0.0359954602<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.3180139<br />gene: FBgn0032445","log2FoldChange: 0.2261960084<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 1.7178909<br />gene: FBgn0032446","log2FoldChange: 0.3743879670<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 1.6644511<br />gene: FBgn0032447","log2FoldChange: 0.0620436095<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 1.6970334<br />gene: FBgn0032449","log2FoldChange: -0.0602050148<br />-log10(padj): 3.520160e-02<br />log10(baseMean): 2.5709895<br />gene: FBgn0032450","log2FoldChange: 0.7596939726<br />-log10(padj): 4.180247e+00<br />log10(baseMean): 2.7669268<br />gene: FBgn0032451","log2FoldChange: 0.2303399482<br />-log10(padj): 9.443517e-02<br />log10(baseMean): 1.5675123<br />gene: FBgn0032452","log2FoldChange: 0.2438932976<br />-log10(padj): 8.367808e-01<br />log10(baseMean): 3.1057596<br />gene: FBgn0032453","log2FoldChange: 0.1528046794<br />-log10(padj): 3.996402e-01<br />log10(baseMean): 3.3777822<br />gene: FBgn0032454","log2FoldChange: -0.0584265618<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.9180111<br />gene: FBgn0032455","log2FoldChange: -0.1300846135<br />-log10(padj): 2.666229e-01<br />log10(baseMean): 3.8814932<br />gene: FBgn0032456","log2FoldChange: -0.0634460102<br />-log10(padj): 4.306243e-02<br />log10(baseMean): 2.9818864<br />gene: FBgn0032465","log2FoldChange: 0.0165494284<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 3.2404066<br />gene: FBgn0032467","log2FoldChange: -0.2667172515<br />-log10(padj): 3.753221e-01<br />log10(baseMean): 3.4612804<br />gene: FBgn0032469","log2FoldChange: -0.1069294767<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.3283152<br />gene: FBgn0032470","log2FoldChange: 0.1217261447<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 0.9970085<br />gene: FBgn0032473","log2FoldChange: 0.1872696593<br />-log10(padj): 2.388550e-01<br />log10(baseMean): 2.5017935<br />gene: FBgn0032474","log2FoldChange: -0.2014617977<br />-log10(padj): 7.918610e-01<br />log10(baseMean): 3.1954323<br />gene: FBgn0032475","log2FoldChange: -0.0166215025<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.8531718<br />gene: FBgn0032476","log2FoldChange: -0.1477320068<br />-log10(padj): 1.841669e-01<br />log10(baseMean): 2.6977477<br />gene: FBgn0032477","log2FoldChange: -0.3978484434<br />-log10(padj): 3.168145e-01<br />log10(baseMean): 1.7292990<br />gene: FBgn0032478","log2FoldChange: 0.0167465166<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.6457127<br />gene: FBgn0032479","log2FoldChange: -0.0743642813<br />-log10(padj): 7.795537e-02<br />log10(baseMean): 3.1202996<br />gene: FBgn0032480","log2FoldChange: -0.1182139118<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8017897<br />gene: FBgn0032481","log2FoldChange: -0.3421746736<br />-log10(padj): 2.284579e+00<br />log10(baseMean): 3.5602579<br />gene: FBgn0032482","log2FoldChange: -0.1826065486<br />-log10(padj): 2.945997e-02<br />log10(baseMean): 1.0604573<br />gene: FBgn0032483","log2FoldChange: 0.1577234392<br />-log10(padj): 2.111034e-01<br />log10(baseMean): 2.8786644<br />gene: FBgn0032485","log2FoldChange: 0.0674509341<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 2.2209550<br />gene: FBgn0032486","log2FoldChange: -0.0974857496<br />-log10(padj): 5.153919e-02<br />log10(baseMean): 2.1144654<br />gene: FBgn0032487","log2FoldChange: 0.0350705983<br />-log10(padj): 2.116949e-02<br />log10(baseMean): 2.7338879<br />gene: FBgn0032488","log2FoldChange: 0.2013804161<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 2.3242971<br />gene: FBgn0032497","log2FoldChange: 1.0537927200<br />-log10(padj): 2.418834e+00<br />log10(baseMean): 1.7585832<br />gene: FBgn0032498","log2FoldChange: -0.0166796262<br />-log10(padj): 8.451393e-03<br />log10(baseMean): 2.7328943<br />gene: FBgn0032499","log2FoldChange: -0.2715297193<br />-log10(padj): 2.733646e-02<br />log10(baseMean): 1.0274633<br />gene: FBgn0032504","log2FoldChange: 0.1633794135<br />-log10(padj): 3.116702e-01<br />log10(baseMean): 2.8787341<br />gene: FBgn0032509","log2FoldChange: 0.0721731200<br />-log10(padj): 7.638315e-02<br />log10(baseMean): 2.9214753<br />gene: FBgn0032511","log2FoldChange: 0.0655432174<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.9605345<br />gene: FBgn0032512","log2FoldChange: -0.1320649727<br />-log10(padj): 1.854143e-01<br />log10(baseMean): 2.8216395<br />gene: FBgn0032513","log2FoldChange: 0.1135539337<br />-log10(padj): 1.757891e-01<br />log10(baseMean): 3.3922039<br />gene: FBgn0032514","log2FoldChange: 0.0279296703<br />-log10(padj): 1.396258e-02<br />log10(baseMean): 2.3021319<br />gene: FBgn0032515","log2FoldChange: 0.0070657292<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.4775725<br />gene: FBgn0032516","log2FoldChange: -0.0243245665<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.7463743<br />gene: FBgn0032517","log2FoldChange: -0.1645649696<br />-log10(padj): 1.060516e-01<br />log10(baseMean): 4.3222031<br />gene: FBgn0032518","log2FoldChange: -0.5701350542<br />-log10(padj): 8.111198e-01<br />log10(baseMean): 1.8161274<br />gene: FBgn0032520","log2FoldChange: 0.0864158632<br />-log10(padj): 1.005749e-01<br />log10(baseMean): 2.9847427<br />gene: FBgn0032521","log2FoldChange: 0.2701951204<br />-log10(padj): 4.731339e-01<br />log10(baseMean): 2.4800943<br />gene: FBgn0032524","log2FoldChange: 1.0797076642<br />-log10(padj): 3.396391e-01<br />log10(baseMean): 0.9359588<br />gene: FBgn0032530","log2FoldChange: 0.7245525370<br />-log10(padj): 1.512124e+00<br />log10(baseMean): 1.8209180<br />gene: FBgn0032533","log2FoldChange: -0.7204050090<br />-log10(padj): 2.016282e-01<br />log10(baseMean): 0.9381757<br />gene: FBgn0032535","log2FoldChange: 0.0089988053<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.5642440<br />gene: FBgn0032586","log2FoldChange: -0.2719511400<br />-log10(padj): 5.840400e-01<br />log10(baseMean): 3.3886558<br />gene: FBgn0032587","log2FoldChange: 0.0280671788<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 1.8793503<br />gene: FBgn0032590","log2FoldChange: 0.1232483956<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.1798320<br />gene: FBgn0032595","log2FoldChange: 0.2045364725<br />-log10(padj): 1.944603e-01<br />log10(baseMean): 3.0772362<br />gene: FBgn0032596","log2FoldChange: 0.1891493361<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 2.8540766<br />gene: FBgn0032597","log2FoldChange: 1.0871657166<br />-log10(padj): 1.162498e+01<br />log10(baseMean): 2.4987576<br />gene: FBgn0032598","log2FoldChange: -0.1631509770<br />-log10(padj): 1.653558e-01<br />log10(baseMean): 3.0905656<br />gene: FBgn0032600","log2FoldChange: -0.8951978939<br />-log10(padj): 2.341847e+00<br />log10(baseMean): 2.4031703<br />gene: FBgn0032601","log2FoldChange: 0.1946728459<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 1.0449111<br />gene: FBgn0032602","log2FoldChange: -0.4195877161<br />-log10(padj): 1.128857e+00<br />log10(baseMean): 2.3922030<br />gene: FBgn0032603","log2FoldChange: -0.1726805781<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.7495324<br />gene: FBgn0032613","log2FoldChange: 0.0847231952<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.4550890<br />gene: FBgn0032620","log2FoldChange: -0.2526183947<br />-log10(padj): 6.261658e-01<br />log10(baseMean): 3.1298186<br />gene: FBgn0032621","log2FoldChange: -0.1825350635<br />-log10(padj): 4.426189e-01<br />log10(baseMean): 3.3652007<br />gene: FBgn0032622","log2FoldChange: -0.0746658400<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 3.4058954<br />gene: FBgn0032633","log2FoldChange: 0.0859982299<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.5807737<br />gene: FBgn0032634","log2FoldChange: 0.6009165424<br />-log10(padj): 5.678910e+00<br />log10(baseMean): 2.7858776<br />gene: FBgn0032635","log2FoldChange: 0.1318534996<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 1.6294792<br />gene: FBgn0032637","log2FoldChange: -1.4357692272<br />-log10(padj): 7.020595e+00<br />log10(baseMean): 2.1203593<br />gene: FBgn0032638","log2FoldChange: 0.0575633230<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 3.3320925<br />gene: FBgn0032640","log2FoldChange: -0.1712751074<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 2.4619503<br />gene: FBgn0032642","log2FoldChange: 0.1723610441<br />-log10(padj): 3.145853e-01<br />log10(baseMean): 3.7854598<br />gene: FBgn0032643","log2FoldChange: -0.1531782981<br />-log10(padj): 1.052021e-01<br />log10(baseMean): 2.3473134<br />gene: FBgn0032644","log2FoldChange: -0.0222545304<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.8955900<br />gene: FBgn0032646","log2FoldChange: 0.3765091305<br />-log10(padj): 1.229309e-01<br />log10(baseMean): 1.4354236<br />gene: FBgn0032647","log2FoldChange: -0.0722553836<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 2.6756705<br />gene: FBgn0032656","log2FoldChange: 0.5315783846<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 1.0228097<br />gene: FBgn0032666","log2FoldChange: 0.1968328739<br />-log10(padj): 7.845574e-02<br />log10(baseMean): 1.8155298<br />gene: FBgn0032670","log2FoldChange: 0.1255293877<br />-log10(padj): 7.066721e-02<br />log10(baseMean): 2.3115065<br />gene: FBgn0032671","log2FoldChange: -0.0224629929<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 3.4983989<br />gene: FBgn0032679","log2FoldChange: -0.6423270298<br />-log10(padj): 7.716674e-02<br />log10(baseMean): 1.8308913<br />gene: FBgn0032680","log2FoldChange: 0.7689885303<br />-log10(padj): 1.204759e+00<br />log10(baseMean): 1.7876185<br />gene: FBgn0032681","log2FoldChange: -0.1198532358<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 3.0010265<br />gene: FBgn0032683","log2FoldChange: -0.2943019999<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 0.8158080<br />gene: FBgn0032684","log2FoldChange: 0.2746356813<br />-log10(padj): 5.688062e-02<br />log10(baseMean): 1.1538908<br />gene: FBgn0032685","log2FoldChange: -0.1249124631<br />-log10(padj): 1.420115e-01<br />log10(baseMean): 2.7878664<br />gene: FBgn0032688","log2FoldChange: -0.0812541111<br />-log10(padj): 7.421765e-02<br />log10(baseMean): 3.1080135<br />gene: FBgn0032689","log2FoldChange: 0.0600473543<br />-log10(padj): 5.936353e-02<br />log10(baseMean): 3.0083998<br />gene: FBgn0032690","log2FoldChange: 0.2978468311<br />-log10(padj): 7.484801e-01<br />log10(baseMean): 2.6226967<br />gene: FBgn0032691","log2FoldChange: -0.6792354662<br />-log10(padj): 5.817648e-01<br />log10(baseMean): 1.8255133<br />gene: FBgn0032693","log2FoldChange: -0.0593258676<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 3.2777309<br />gene: FBgn0032694","log2FoldChange: -0.4836753757<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 2.6069420<br />gene: FBgn0032698","log2FoldChange: 0.3343160109<br />-log10(padj): 1.398865e+00<br />log10(baseMean): 3.2192831<br />gene: FBgn0032699","log2FoldChange: -0.0685141208<br />-log10(padj): 4.221267e-02<br />log10(baseMean): 2.5122935<br />gene: FBgn0032700","log2FoldChange: -0.2050738318<br />-log10(padj): 4.564987e-01<br />log10(baseMean): 2.7405260<br />gene: FBgn0032701","log2FoldChange: -0.1284163514<br />-log10(padj): 1.353758e-01<br />log10(baseMean): 3.2160588<br />gene: FBgn0032702","log2FoldChange: -0.0076855030<br />-log10(padj): 3.581171e-03<br />log10(baseMean): 2.3466276<br />gene: FBgn0032703","log2FoldChange: -0.1573215920<br />-log10(padj): 1.444780e-01<br />log10(baseMean): 3.3637807<br />gene: FBgn0032704","log2FoldChange: -0.1618255420<br />-log10(padj): 1.940626e-01<br />log10(baseMean): 2.6076964<br />gene: FBgn0032705","log2FoldChange: -0.5959192905<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 0.8742988<br />gene: FBgn0032713","log2FoldChange: 0.0470990856<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.2000622<br />gene: FBgn0032715","log2FoldChange: -0.2508403069<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 3.2642104<br />gene: FBgn0032717","log2FoldChange: 0.3302614646<br />-log10(padj): 1.075092e-01<br />log10(baseMean): 1.3376448<br />gene: FBgn0032719","log2FoldChange: 0.0709759286<br />-log10(padj): 3.566099e-02<br />log10(baseMean): 2.0654666<br />gene: FBgn0032720","log2FoldChange: 0.1251603904<br />-log10(padj): 1.588723e-01<br />log10(baseMean): 3.0091049<br />gene: FBgn0032721","log2FoldChange: 0.1327139053<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.6934428<br />gene: FBgn0032723","log2FoldChange: -0.0617248544<br />-log10(padj): 2.851854e-02<br />log10(baseMean): 2.4475797<br />gene: FBgn0032724","log2FoldChange: 0.2438196883<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 2.4530436<br />gene: FBgn0032725","log2FoldChange: -0.4833075607<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 1.5400077<br />gene: FBgn0032726","log2FoldChange: -0.2472155195<br />-log10(padj): 4.421245e-01<br />log10(baseMean): 2.3432569<br />gene: FBgn0032727","log2FoldChange: -0.1527530263<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.3290163<br />gene: FBgn0032728","log2FoldChange: -0.3831956400<br />-log10(padj): 1.221486e+00<br />log10(baseMean): 2.4935201<br />gene: FBgn0032729","log2FoldChange: 0.8610420626<br />-log10(padj): 1.826383e-01<br />log10(baseMean): 1.8093907<br />gene: FBgn0032730","log2FoldChange: 0.2194849062<br />-log10(padj): 3.536785e-01<br />log10(baseMean): 3.6115155<br />gene: FBgn0032731","log2FoldChange: -0.2652285164<br />-log10(padj): 2.333681e-01<br />log10(baseMean): 2.1879344<br />gene: FBgn0032732","log2FoldChange: 0.4286619068<br />-log10(padj): 4.510611e-01<br />log10(baseMean): 1.9555534<br />gene: FBgn0032744","log2FoldChange: 0.0463768764<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.4854193<br />gene: FBgn0032746","log2FoldChange: -0.2139245649<br />-log10(padj): 6.406388e-01<br />log10(baseMean): 3.2489059<br />gene: FBgn0032748","log2FoldChange: 0.2204473205<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 2.0981296<br />gene: FBgn0032750","log2FoldChange: -0.1703294178<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 1.7807121<br />gene: FBgn0032751","log2FoldChange: 0.5163991749<br />-log10(padj): 8.335775e-01<br />log10(baseMean): 1.9559658<br />gene: FBgn0032752","log2FoldChange: 0.1255538488<br />-log10(padj): 7.166896e-02<br />log10(baseMean): 2.0168660<br />gene: FBgn0032763","log2FoldChange: 0.4687318938<br />-log10(padj): 1.011182e+00<br />log10(baseMean): 2.0367453<br />gene: FBgn0032772","log2FoldChange: 0.0751687308<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 3.4006678<br />gene: FBgn0032773","log2FoldChange: -0.1747823193<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 2.4657699<br />gene: FBgn0032774","log2FoldChange: -0.7617288677<br />-log10(padj): 4.978754e+00<br />log10(baseMean): 3.4957516<br />gene: FBgn0032775","log2FoldChange: -0.2071522444<br />-log10(padj): 3.101635e-01<br />log10(baseMean): 3.3570136<br />gene: FBgn0032779","log2FoldChange: -0.7943328464<br />-log10(padj): 6.288426e-01<br />log10(baseMean): 1.9301511<br />gene: FBgn0032780","log2FoldChange: 0.1542244706<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 3.1383186<br />gene: FBgn0032781","log2FoldChange: -0.3419239073<br />-log10(padj): 7.699618e-01<br />log10(baseMean): 2.4893859<br />gene: FBgn0032782","log2FoldChange: -0.3215799416<br />-log10(padj): 2.303000e-01<br />log10(baseMean): 1.9464899<br />gene: FBgn0032783","log2FoldChange: -0.5579381077<br />-log10(padj): 2.024058e+00<br />log10(baseMean): 2.3266032<br />gene: FBgn0032785","log2FoldChange: 0.1316112595<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 2.3328724<br />gene: FBgn0032787","log2FoldChange: 0.0439864391<br />-log10(padj): 2.122202e-02<br />log10(baseMean): 2.1913886<br />gene: FBgn0032790","log2FoldChange: 0.2418593525<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 1.8136460<br />gene: FBgn0032791","log2FoldChange: 0.3039721081<br />-log10(padj): 5.956716e-01<br />log10(baseMean): 2.5647146<br />gene: FBgn0032793","log2FoldChange: -0.2135410108<br />-log10(padj): 4.726491e-01<br />log10(baseMean): 2.8175689<br />gene: FBgn0032796","log2FoldChange: 0.0837241606<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.5419169<br />gene: FBgn0032797","log2FoldChange: 0.4817060711<br />-log10(padj): 6.002928e-01<br />log10(baseMean): 1.9120438<br />gene: FBgn0032798","log2FoldChange: 0.0064500053<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 2.7982971<br />gene: FBgn0032799","log2FoldChange: 0.0898626795<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.4092493<br />gene: FBgn0032800","log2FoldChange: 0.0288014557<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 2.3306815<br />gene: FBgn0032801","log2FoldChange: 0.4407916079<br />-log10(padj): 3.867314e-01<br />log10(baseMean): 2.2911925<br />gene: FBgn0032805","log2FoldChange: -1.2147508090<br />-log10(padj): 1.564340e+00<br />log10(baseMean): 1.4524730<br />gene: FBgn0032808","log2FoldChange: 0.0273543848<br />-log10(padj): 3.433775e-03<br />log10(baseMean): 0.8405004<br />gene: FBgn0032809","log2FoldChange: 0.8179585812<br />-log10(padj): 1.344186e+00<br />log10(baseMean): 1.8678708<br />gene: FBgn0032810","log2FoldChange: 0.0221487055<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.3662181<br />gene: FBgn0032811","log2FoldChange: 0.0332469383<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.7655822<br />gene: FBgn0032812","log2FoldChange: -0.2086244088<br />-log10(padj): 6.780135e-02<br />log10(baseMean): 1.4979199<br />gene: FBgn0032813","log2FoldChange: 0.0455156047<br />-log10(padj): 2.965646e-02<br />log10(baseMean): 2.6362534<br />gene: FBgn0032814","log2FoldChange: -0.1500111917<br />-log10(padj): 1.331873e-01<br />log10(baseMean): 3.1780809<br />gene: FBgn0032815","log2FoldChange: 0.2102711954<br />-log10(padj): 1.512365e-01<br />log10(baseMean): 2.3934810<br />gene: FBgn0032816","log2FoldChange: -0.2166771261<br />-log10(padj): 5.490181e-01<br />log10(baseMean): 3.4315536<br />gene: FBgn0032817","log2FoldChange: 0.2201711526<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 2.5486459<br />gene: FBgn0032818","log2FoldChange: -0.1079829917<br />-log10(padj): 8.709772e-02<br />log10(baseMean): 2.4971022<br />gene: FBgn0032819","log2FoldChange: 0.6169758699<br />-log10(padj): 4.714871e+00<br />log10(baseMean): 2.5217161<br />gene: FBgn0032820","log2FoldChange: 0.0893841765<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 3.3664382<br />gene: FBgn0032821","log2FoldChange: -0.0845004814<br />-log10(padj): 2.451126e-02<br />log10(baseMean): 1.8650868<br />gene: FBgn0032822","log2FoldChange: 0.0793686003<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.5528185<br />gene: FBgn0032833","log2FoldChange: -0.0620992194<br />-log10(padj): 1.353168e-02<br />log10(baseMean): 1.5149923<br />gene: FBgn0032843","log2FoldChange: -0.2784986001<br />-log10(padj): 2.364454e-01<br />log10(baseMean): 2.0303083<br />gene: FBgn0032845","log2FoldChange: 0.0072811266<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 2.3263905<br />gene: FBgn0032846","log2FoldChange: 0.0476618158<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.7201578<br />gene: FBgn0032847","log2FoldChange: 0.0581617963<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.9740171<br />gene: FBgn0032848","log2FoldChange: 0.0856695295<br />-log10(padj): 6.868318e-02<br />log10(baseMean): 2.4833014<br />gene: FBgn0032849","log2FoldChange: 0.0217447360<br />-log10(padj): 1.296089e-02<br />log10(baseMean): 2.4231016<br />gene: FBgn0032858","log2FoldChange: -0.4896428451<br />-log10(padj): 2.938265e+00<br />log10(baseMean): 3.5351848<br />gene: FBgn0032859","log2FoldChange: 0.3011646312<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 2.6228035<br />gene: FBgn0032863","log2FoldChange: -0.1088105425<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 3.9890160<br />gene: FBgn0032864","log2FoldChange: -0.1256859437<br />-log10(padj): 5.105255e-02<br />log10(baseMean): 1.7924008<br />gene: FBgn0032869","log2FoldChange: -0.0298610201<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 2.7117943<br />gene: FBgn0032870","log2FoldChange: 0.1995157695<br />-log10(padj): 1.855676e-01<br />log10(baseMean): 2.1980696<br />gene: FBgn0032871","log2FoldChange: -0.0782241996<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 3.2957352<br />gene: FBgn0032872","log2FoldChange: -0.0194636875<br />-log10(padj): 9.014029e-03<br />log10(baseMean): 2.7821955<br />gene: FBgn0032873","log2FoldChange: 0.1074263432<br />-log10(padj): 1.022245e-01<br />log10(baseMean): 2.7160501<br />gene: FBgn0032876","log2FoldChange: 0.2526040931<br />-log10(padj): 2.475425e-01<br />log10(baseMean): 2.1723308<br />gene: FBgn0032877","log2FoldChange: 0.9425375214<br />-log10(padj): 7.368835e-01<br />log10(baseMean): 1.3697370<br />gene: FBgn0032879","log2FoldChange: 0.0212416337<br />-log10(padj): 1.349861e-02<br />log10(baseMean): 3.5708340<br />gene: FBgn0032880","log2FoldChange: 0.6493831876<br />-log10(padj): 3.048863e+00<br />log10(baseMean): 2.3473754<br />gene: FBgn0032881","log2FoldChange: 0.1933148379<br />-log10(padj): 3.294375e-01<br />log10(baseMean): 2.8711753<br />gene: FBgn0032882","log2FoldChange: -0.1746062972<br />-log10(padj): 3.664125e-01<br />log10(baseMean): 2.8566089<br />gene: FBgn0032883","log2FoldChange: 0.1636223796<br />-log10(padj): 2.996071e-01<br />log10(baseMean): 3.0311150<br />gene: FBgn0032884","log2FoldChange: -0.3089944132<br />-log10(padj): 8.666742e-01<br />log10(baseMean): 3.2567967<br />gene: FBgn0032886","log2FoldChange: 0.1064800030<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.5048827<br />gene: FBgn0032889","log2FoldChange: 0.2251741285<br />-log10(padj): 1.079031e-01<br />log10(baseMean): 1.7289395<br />gene: FBgn0032891","log2FoldChange: 0.6853750353<br />-log10(padj): 1.687731e-01<br />log10(baseMean): 0.9779267<br />gene: FBgn0032895","log2FoldChange: -0.2474817421<br />-log10(padj): 1.245608e-01<br />log10(baseMean): 1.7765597<br />gene: FBgn0032897","log2FoldChange: -0.5261812342<br />-log10(padj): 1.840628e+00<br />log10(baseMean): 3.0409507<br />gene: FBgn0032899","log2FoldChange: -0.8715649127<br />-log10(padj): 1.003309e+00<br />log10(baseMean): 1.4704655<br />gene: FBgn0032900","log2FoldChange: 0.1516938598<br />-log10(padj): 1.009422e-01<br />log10(baseMean): 3.4188672<br />gene: FBgn0032901","log2FoldChange: 0.2119093488<br />-log10(padj): 4.842655e-01<br />log10(baseMean): 2.7721677<br />gene: FBgn0032904","log2FoldChange: -0.0356184637<br />-log10(padj): 2.498915e-02<br />log10(baseMean): 3.1420858<br />gene: FBgn0032906","log2FoldChange: 0.1632086279<br />-log10(padj): 1.238255e-01<br />log10(baseMean): 2.3542088<br />gene: FBgn0032907","log2FoldChange: 0.7704131197<br />-log10(padj): 8.085934e-01<br />log10(baseMean): 1.5790885<br />gene: FBgn0032908","log2FoldChange: 0.2250871535<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 1.0040510<br />gene: FBgn0032910","log2FoldChange: -0.2424778880<br />-log10(padj): 8.012310e-01<br />log10(baseMean): 3.1946721<br />gene: FBgn0032915","log2FoldChange: -0.0411442558<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 2.5243741<br />gene: FBgn0032916","log2FoldChange: -0.1377901670<br />-log10(padj): 1.975194e-01<br />log10(baseMean): 3.1395128<br />gene: FBgn0032919","log2FoldChange: -0.0155340784<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 2.0329037<br />gene: FBgn0032921","log2FoldChange: 0.2576988405<br />-log10(padj): 2.074624e-01<br />log10(baseMean): 2.0675244<br />gene: FBgn0032922","log2FoldChange: 0.0997271534<br />-log10(padj): 1.434008e-01<br />log10(baseMean): 3.2570631<br />gene: FBgn0032923","log2FoldChange: -0.0539950052<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 2.6989833<br />gene: FBgn0032924","log2FoldChange: -0.2099387855<br />-log10(padj): 3.423478e-01<br />log10(baseMean): 3.0802243<br />gene: FBgn0032925","log2FoldChange: 0.1936630638<br />-log10(padj): 4.125902e-01<br />log10(baseMean): 2.7879067<br />gene: FBgn0032926","log2FoldChange: -0.2542779740<br />-log10(padj): 3.473641e-01<br />log10(baseMean): 2.7246543<br />gene: FBgn0032929","log2FoldChange: 0.2635525294<br />-log10(padj): 4.240436e-01<br />log10(baseMean): 2.2511367<br />gene: FBgn0032934","log2FoldChange: -0.3148236204<br />-log10(padj): 1.198945e+00<br />log10(baseMean): 2.7658443<br />gene: FBgn0032935","log2FoldChange: 0.2778314468<br />-log10(padj): 6.731530e-01<br />log10(baseMean): 3.1842595<br />gene: FBgn0032938","log2FoldChange: -0.1029052916<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 3.4896121<br />gene: FBgn0032940","log2FoldChange: 0.1625654883<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.7533317<br />gene: FBgn0032943","log2FoldChange: 0.3611540959<br />-log10(padj): 1.648969e-01<br />log10(baseMean): 1.9925186<br />gene: FBgn0032946","log2FoldChange: -0.1265820101<br />-log10(padj): 6.717052e-02<br />log10(baseMean): 3.9183976<br />gene: FBgn0032949","log2FoldChange: 0.0545678190<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.7796258<br />gene: FBgn0032955","log2FoldChange: 0.5822648118<br />-log10(padj): 1.906722e+00<br />log10(baseMean): 3.4468718<br />gene: FBgn0032956","log2FoldChange: -0.2082650839<br />-log10(padj): 5.098456e-01<br />log10(baseMean): 3.0564174<br />gene: FBgn0032957","log2FoldChange: -0.0812664895<br />-log10(padj): 6.504494e-02<br />log10(baseMean): 3.4870378<br />gene: FBgn0032961","log2FoldChange: 0.7127773190<br />-log10(padj): 1.207488e-01<br />log10(baseMean): 0.8148786<br />gene: FBgn0032964","log2FoldChange: -0.1622571417<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 1.9396196<br />gene: FBgn0032965","log2FoldChange: -0.4466509658<br />-log10(padj): 4.373670e-01<br />log10(baseMean): 1.8257476<br />gene: FBgn0032967","log2FoldChange: -0.1251198452<br />-log10(padj): 4.631374e-02<br />log10(baseMean): 2.0242718<br />gene: FBgn0032968","log2FoldChange: 0.2932642510<br />-log10(padj): 6.528536e-02<br />log10(baseMean): 1.2687185<br />gene: FBgn0032972","log2FoldChange: -0.0469937954<br />-log10(padj): 8.164022e-03<br />log10(baseMean): 1.3344140<br />gene: FBgn0032973","log2FoldChange: 0.0275982130<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 3.3607025<br />gene: FBgn0032974","log2FoldChange: -0.0853504720<br />-log10(padj): 1.111215e-01<br />log10(baseMean): 3.5170594<br />gene: FBgn0032976","log2FoldChange: -0.0376520075<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.4069928<br />gene: FBgn0032978","log2FoldChange: -0.2447025799<br />-log10(padj): 7.932671e-01<br />log10(baseMean): 3.4337008<br />gene: FBgn0032979","log2FoldChange: 0.0882691772<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 2.9109772<br />gene: FBgn0032981","log2FoldChange: 0.0128604100<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.9971343<br />gene: FBgn0032986","log2FoldChange: -0.1236742438<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 4.5695388<br />gene: FBgn0032987","log2FoldChange: 0.0453367793<br />-log10(padj): 1.415695e-02<br />log10(baseMean): 3.1452094<br />gene: FBgn0032988","log2FoldChange: -0.3447792396<br />-log10(padj): 2.016282e-01<br />log10(baseMean): 2.6259264<br />gene: FBgn0032997","log2FoldChange: 0.4473321072<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 2.2641070<br />gene: FBgn0033000","log2FoldChange: -0.0798646034<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 3.0506397<br />gene: FBgn0033005","log2FoldChange: 0.0452341033<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.6274456<br />gene: FBgn0033010","log2FoldChange: 0.0064714992<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.8828841<br />gene: FBgn0033015","log2FoldChange: 0.1079507859<br />-log10(padj): 2.055674e-02<br />log10(baseMean): 2.7775277<br />gene: FBgn0033017","log2FoldChange: 0.1698495723<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.0858654<br />gene: FBgn0033019","log2FoldChange: 0.1401729981<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.1364727<br />gene: FBgn0033021","log2FoldChange: 0.4532242975<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 1.3252536<br />gene: FBgn0033027","log2FoldChange: -0.1149925370<br />-log10(padj): 9.682541e-02<br />log10(baseMean): 3.2036825<br />gene: FBgn0033028","log2FoldChange: -0.1196023207<br />-log10(padj): 1.502119e-01<br />log10(baseMean): 3.4196948<br />gene: FBgn0033029","log2FoldChange: 0.3821986431<br />-log10(padj): 5.027731e-01<br />log10(baseMean): 2.4783856<br />gene: FBgn0033032","log2FoldChange: -0.3744663800<br />-log10(padj): 2.467589e+00<br />log10(baseMean): 3.2520185<br />gene: FBgn0033033","log2FoldChange: 0.2858487291<br />-log10(padj): 1.040319e+00<br />log10(baseMean): 2.8222111<br />gene: FBgn0033038","log2FoldChange: -0.1907573406<br />-log10(padj): 1.858855e-01<br />log10(baseMean): 3.5706789<br />gene: FBgn0033039","log2FoldChange: -0.1616522441<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.4314279<br />gene: FBgn0033049","log2FoldChange: -0.2230965403<br />-log10(padj): 7.779423e-01<br />log10(baseMean): 3.1150905<br />gene: FBgn0033050","log2FoldChange: -0.1114987026<br />-log10(padj): 1.511863e-01<br />log10(baseMean): 2.8784876<br />gene: FBgn0033051","log2FoldChange: -0.1716687903<br />-log10(padj): 3.278556e-01<br />log10(baseMean): 3.5787216<br />gene: FBgn0033052","log2FoldChange: -0.0901963695<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 2.9497646<br />gene: FBgn0033055","log2FoldChange: -0.1597714973<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 2.6148406<br />gene: FBgn0033059","log2FoldChange: -0.2093762465<br />-log10(padj): 1.693284e-01<br />log10(baseMean): 2.0723511<br />gene: FBgn0033060","log2FoldChange: 0.2417588274<br />-log10(padj): 5.079791e-02<br />log10(baseMean): 1.1294914<br />gene: FBgn0033061","log2FoldChange: -0.0531771997<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 3.3384874<br />gene: FBgn0033062","log2FoldChange: 3.0221515543<br />-log10(padj): 1.030655e+01<br />log10(baseMean): 1.4763720<br />gene: FBgn0033065","log2FoldChange: 0.1276060153<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.0133082<br />gene: FBgn0033068","log2FoldChange: 0.0015214096<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 3.4601300<br />gene: FBgn0033073","log2FoldChange: -0.3630191408<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.0385232<br />gene: FBgn0033074","log2FoldChange: -0.1638400758<br />-log10(padj): 3.739047e-01<br />log10(baseMean): 2.8665927<br />gene: FBgn0033075","log2FoldChange: -0.6553312426<br />-log10(padj): 4.496560e+00<br />log10(baseMean): 2.5786427<br />gene: FBgn0033079","log2FoldChange: -0.1174611769<br />-log10(padj): 1.766420e-01<br />log10(baseMean): 3.0283040<br />gene: FBgn0033081","log2FoldChange: 0.0215808701<br />-log10(padj): 1.224480e-02<br />log10(baseMean): 2.9469716<br />gene: FBgn0033083","log2FoldChange: -0.0850008731<br />-log10(padj): 7.196422e-02<br />log10(baseMean): 2.4791139<br />gene: FBgn0033087","log2FoldChange: 0.1679691562<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 2.5181745<br />gene: FBgn0033088","log2FoldChange: 0.3223170613<br />-log10(padj): 5.593255e-01<br />log10(baseMean): 2.2542448<br />gene: FBgn0033089","log2FoldChange: -0.5157448588<br />-log10(padj): 1.138111e-01<br />log10(baseMean): 0.9512081<br />gene: FBgn0033090","log2FoldChange: -0.2844433554<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 2.0733921<br />gene: FBgn0033092","log2FoldChange: -0.0224684555<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.5734775<br />gene: FBgn0033093","log2FoldChange: 0.7645724180<br />-log10(padj): 1.344128e+01<br />log10(baseMean): 3.1729285<br />gene: FBgn0033095","log2FoldChange: -0.0214044248<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.2264914<br />gene: FBgn0033100","log2FoldChange: 0.2125801629<br />-log10(padj): 2.820483e-01<br />log10(baseMean): 2.8933226<br />gene: FBgn0033101","log2FoldChange: -0.0306465058<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.2085745<br />gene: FBgn0033104","log2FoldChange: 0.0791283226<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 2.7984453<br />gene: FBgn0033107","log2FoldChange: 0.7624461938<br />-log10(padj): 3.247082e-01<br />log10(baseMean): 1.0941643<br />gene: FBgn0033108","log2FoldChange: 0.1192030572<br />-log10(padj): 2.397553e-01<br />log10(baseMean): 3.3245627<br />gene: FBgn0033109","log2FoldChange: 0.5561693022<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 1.4065334<br />gene: FBgn0033110","log2FoldChange: 0.9812551873<br />-log10(padj): 3.727861e+00<br />log10(baseMean): 2.5727448<br />gene: FBgn0033113","log2FoldChange: 0.5414088795<br />-log10(padj): 1.685196e+00<br />log10(baseMean): 2.2276219<br />gene: FBgn0033115","log2FoldChange: 0.0482868671<br />-log10(padj): 2.778407e-02<br />log10(baseMean): 2.3594041<br />gene: FBgn0033117","log2FoldChange: 0.0191226952<br />-log10(padj): 1.244639e-02<br />log10(baseMean): 2.6313128<br />gene: FBgn0033121","log2FoldChange: 0.0157847664<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.7675423<br />gene: FBgn0033122","log2FoldChange: -0.3052419395<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 0.9008840<br />gene: FBgn0033124","log2FoldChange: 0.1688046145<br />-log10(padj): 2.255230e-01<br />log10(baseMean): 2.9952713<br />gene: FBgn0033127","log2FoldChange: -0.5784786033<br />-log10(padj): 2.647498e+00<br />log10(baseMean): 2.6542263<br />gene: FBgn0033128","log2FoldChange: -0.7275244330<br />-log10(padj): 1.280015e-01<br />log10(baseMean): 0.8308903<br />gene: FBgn0033130","log2FoldChange: -0.2193982303<br />-log10(padj): 8.264700e-02<br />log10(baseMean): 1.6389152<br />gene: FBgn0033132","log2FoldChange: -0.5667508108<br />-log10(padj): 1.468821e-01<br />log10(baseMean): 1.0567098<br />gene: FBgn0033133","log2FoldChange: -0.5294058288<br />-log10(padj): 1.041504e+00<br />log10(baseMean): 2.1327258<br />gene: FBgn0033134","log2FoldChange: 0.7684342863<br />-log10(padj): 2.925016e-01<br />log10(baseMean): 1.0903476<br />gene: FBgn0033136","log2FoldChange: -0.1178328053<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.1049447<br />gene: FBgn0033137","log2FoldChange: 0.6525461193<br />-log10(padj): 2.565293e+00<br />log10(baseMean): 2.4104137<br />gene: FBgn0033153","log2FoldChange: -0.1583076975<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 3.1653238<br />gene: FBgn0033155","log2FoldChange: 0.0498185869<br />-log10(padj): 3.075722e-02<br />log10(baseMean): 3.5623606<br />gene: FBgn0033159","log2FoldChange: -0.1748885538<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 3.2110016<br />gene: FBgn0033160","log2FoldChange: 0.2699248746<br />-log10(padj): 4.957541e-01<br />log10(baseMean): 3.0152922<br />gene: FBgn0033162","log2FoldChange: -0.0743664636<br />-log10(padj): 6.633266e-02<br />log10(baseMean): 3.0470880<br />gene: FBgn0033166","log2FoldChange: -0.0785022796<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.9617760<br />gene: FBgn0033169","log2FoldChange: 0.7167216698<br />-log10(padj): 7.729714e-01<br />log10(baseMean): 1.5944597<br />gene: FBgn0033170","log2FoldChange: -0.2608849763<br />-log10(padj): 4.181491e-01<br />log10(baseMean): 2.6195464<br />gene: FBgn0033174","log2FoldChange: -0.0608416056<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 3.1070140<br />gene: FBgn0033177","log2FoldChange: 0.1092444001<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.0458507<br />gene: FBgn0033178","log2FoldChange: 0.0162460262<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.3744151<br />gene: FBgn0033179","log2FoldChange: -0.0641851196<br />-log10(padj): 3.635924e-02<br />log10(baseMean): 2.4576411<br />gene: FBgn0033182","log2FoldChange: 0.2646828715<br />-log10(padj): 6.514033e-01<br />log10(baseMean): 3.0400527<br />gene: FBgn0033183","log2FoldChange: 0.0385472231<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.5417506<br />gene: FBgn0033184","log2FoldChange: 0.0629437166<br />-log10(padj): 4.881401e-02<br />log10(baseMean): 2.4801675<br />gene: FBgn0033185","log2FoldChange: 0.2294686058<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 1.9970994<br />gene: FBgn0033186","log2FoldChange: -0.0499354409<br />-log10(padj): 3.113986e-02<br />log10(baseMean): 2.8331724<br />gene: FBgn0033187","log2FoldChange: -0.5360328190<br />-log10(padj): 4.450481e+00<br />log10(baseMean): 3.1341752<br />gene: FBgn0033188","log2FoldChange: -0.1308258721<br />-log10(padj): 1.753139e-01<br />log10(baseMean): 3.2041671<br />gene: FBgn0033189","log2FoldChange: 0.2388138806<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 1.6697118<br />gene: FBgn0033190","log2FoldChange: 0.1960509938<br />-log10(padj): 2.640659e-01<br />log10(baseMean): 2.8221610<br />gene: FBgn0033191","log2FoldChange: -0.3912482979<br />-log10(padj): 7.624052e-01<br />log10(baseMean): 2.6107524<br />gene: FBgn0033192","log2FoldChange: 0.0719393658<br />-log10(padj): 7.211131e-02<br />log10(baseMean): 3.5210579<br />gene: FBgn0033194","log2FoldChange: -0.2979751527<br />-log10(padj): 2.674797e-01<br />log10(baseMean): 1.9093792<br />gene: FBgn0033195","log2FoldChange: 0.2037990635<br />-log10(padj): 3.810279e-01<br />log10(baseMean): 3.2598678<br />gene: FBgn0033196","log2FoldChange: -0.0868148413<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.3464943<br />gene: FBgn0033199","log2FoldChange: -0.4298133209<br />-log10(padj): 1.900692e+00<br />log10(baseMean): 2.5253541<br />gene: FBgn0033204","log2FoldChange: 0.3494191178<br />-log10(padj): 1.969127e+00<br />log10(baseMean): 3.1884802<br />gene: FBgn0033205","log2FoldChange: -0.1168441564<br />-log10(padj): 1.165047e-01<br />log10(baseMean): 2.6715755<br />gene: FBgn0033206","log2FoldChange: 0.2534646888<br />-log10(padj): 3.621771e-01<br />log10(baseMean): 2.3641407<br />gene: FBgn0033208","log2FoldChange: 0.0143341014<br />-log10(padj): 7.204181e-03<br />log10(baseMean): 2.5490078<br />gene: FBgn0033209","log2FoldChange: -0.0923664830<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 2.6504882<br />gene: FBgn0033210","log2FoldChange: 0.2420061178<br />-log10(padj): 9.130654e-01<br />log10(baseMean): 3.3780670<br />gene: FBgn0033212","log2FoldChange: 0.1505095420<br />-log10(padj): 2.005583e-02<br />log10(baseMean): 0.9579075<br />gene: FBgn0033214","log2FoldChange: 0.4074545867<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 1.1783041<br />gene: FBgn0033221","log2FoldChange: -0.3218978706<br />-log10(padj): 1.123533e+00<br />log10(baseMean): 3.1711081<br />gene: FBgn0033224","log2FoldChange: 0.1394212429<br />-log10(padj): 1.445256e-01<br />log10(baseMean): 2.8523642<br />gene: FBgn0033225","log2FoldChange: 0.1261134816<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 3.0952382<br />gene: FBgn0033226","log2FoldChange: 0.1351883467<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 2.6423490<br />gene: FBgn0033232","log2FoldChange: 0.0618394203<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.1995267<br />gene: FBgn0033233","log2FoldChange: -0.3799289037<br />-log10(padj): 1.173238e-01<br />log10(baseMean): 1.2516931<br />gene: FBgn0033234","log2FoldChange: -0.0114790264<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 3.1612020<br />gene: FBgn0033235","log2FoldChange: -0.1877852227<br />-log10(padj): 2.803235e-01<br />log10(baseMean): 2.8979158<br />gene: FBgn0033236","log2FoldChange: 0.2141820830<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 2.0790131<br />gene: FBgn0033240","log2FoldChange: -0.0171238345<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.3397746<br />gene: FBgn0033241","log2FoldChange: -0.6815310997<br />-log10(padj): 2.404598e-01<br />log10(baseMean): 1.0705889<br />gene: FBgn0033243","log2FoldChange: -0.0435783685<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.8508118<br />gene: FBgn0033244","log2FoldChange: -0.2019958741<br />-log10(padj): 4.275921e-01<br />log10(baseMean): 3.8652535<br />gene: FBgn0033246","log2FoldChange: -0.1694563228<br />-log10(padj): 2.683111e-01<br />log10(baseMean): 2.7924259<br />gene: FBgn0033247","log2FoldChange: -0.0082793640<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 0.8884827<br />gene: FBgn0033252","log2FoldChange: -0.5139779848<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.1444943<br />gene: FBgn0033257","log2FoldChange: -0.5193946840<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 1.8207551<br />gene: FBgn0033258","log2FoldChange: -0.2158159532<br />-log10(padj): 5.093919e-01<br />log10(baseMean): 2.9954988<br />gene: FBgn0033259","log2FoldChange: -0.0337474425<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 3.1110540<br />gene: FBgn0033260","log2FoldChange: -0.3318793928<br />-log10(padj): 2.703222e-01<br />log10(baseMean): 2.3512456<br />gene: FBgn0033261","log2FoldChange: -0.0594237673<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 2.9540890<br />gene: FBgn0033264","log2FoldChange: -0.0296792597<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 2.4344822<br />gene: FBgn0033265","log2FoldChange: -0.2786765811<br />-log10(padj): 1.103097e+00<br />log10(baseMean): 2.7818614<br />gene: FBgn0033266","log2FoldChange: 1.0770778766<br />-log10(padj): 3.146817e+00<br />log10(baseMean): 1.9160326<br />gene: FBgn0033268","log2FoldChange: -0.3113507411<br />-log10(padj): 9.518441e-01<br />log10(baseMean): 3.3123797<br />gene: FBgn0033269","log2FoldChange: -0.0374607672<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.8494946<br />gene: FBgn0033272","log2FoldChange: 0.1987447763<br />-log10(padj): 1.303420e-01<br />log10(baseMean): 2.0073284<br />gene: FBgn0033273","log2FoldChange: 0.1807679480<br />-log10(padj): 7.406088e-02<br />log10(baseMean): 1.7371638<br />gene: FBgn0033274","log2FoldChange: -1.3255541540<br />-log10(padj): 6.582255e-01<br />log10(baseMean): 1.1735180<br />gene: FBgn0033277","log2FoldChange: 1.0031474448<br />-log10(padj): 2.272806e-01<br />log10(baseMean): 0.8128401<br />gene: FBgn0033282","log2FoldChange: 0.0216252672<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 0.8918503<br />gene: FBgn0033286","log2FoldChange: 1.4180526127<br />-log10(padj): 1.443637e+00<br />log10(baseMean): 1.1790024<br />gene: FBgn0033288","log2FoldChange: -0.9139450121<br />-log10(padj): 2.312075e+00<br />log10(baseMean): 1.9168295<br />gene: FBgn0033292","log2FoldChange: -0.3235743763<br />-log10(padj): 1.829102e+00<br />log10(baseMean): 2.9122143<br />gene: FBgn0033302","log2FoldChange: 0.1223231016<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.7642460<br />gene: FBgn0033304","log2FoldChange: 0.3403451090<br />-log10(padj): 1.977972e-01<br />log10(baseMean): 1.6709550<br />gene: FBgn0033308","log2FoldChange: 0.0374946206<br />-log10(padj): 2.108847e-02<br />log10(baseMean): 2.7943491<br />gene: FBgn0033309","log2FoldChange: 0.2702596640<br />-log10(padj): 4.591891e-01<br />log10(baseMean): 3.0868308<br />gene: FBgn0033310","log2FoldChange: 0.5008615672<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 1.3610722<br />gene: FBgn0033312","log2FoldChange: 0.2504042333<br />-log10(padj): 5.163237e-01<br />log10(baseMean): 3.5041131<br />gene: FBgn0033313","log2FoldChange: -0.2083648890<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 2.2298213<br />gene: FBgn0033315","log2FoldChange: -0.0460125531<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.6511337<br />gene: FBgn0033316","log2FoldChange: 0.0443001141<br />-log10(padj): 3.739424e-02<br />log10(baseMean): 3.0135418<br />gene: FBgn0033317","log2FoldChange: 0.4589239088<br />-log10(padj): 1.074640e-01<br />log10(baseMean): 1.0248830<br />gene: FBgn0033320","log2FoldChange: 0.0383131146<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 1.0557840<br />gene: FBgn0033321","log2FoldChange: -0.0694345339<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 2.2079272<br />gene: FBgn0033337","log2FoldChange: 0.0385935405<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 3.6659453<br />gene: FBgn0033339","log2FoldChange: -0.0004828119<br />-log10(padj): 3.453894e-04<br />log10(baseMean): 2.7280238<br />gene: FBgn0033340","log2FoldChange: -0.0557694599<br />-log10(padj): 3.275836e-02<br />log10(baseMean): 2.5737859<br />gene: FBgn0033341","log2FoldChange: 0.3155484966<br />-log10(padj): 8.676516e-01<br />log10(baseMean): 3.8016234<br />gene: FBgn0033342","log2FoldChange: -0.6813635204<br />-log10(padj): 1.807875e-01<br />log10(baseMean): 0.9575294<br />gene: FBgn0033344","log2FoldChange: -0.2603727289<br />-log10(padj): 4.202573e-02<br />log10(baseMean): 0.9792849<br />gene: FBgn0033347","log2FoldChange: 0.1005903972<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.2536024<br />gene: FBgn0033348","log2FoldChange: -0.0567448350<br />-log10(padj): 5.444957e-02<br />log10(baseMean): 3.0959817<br />gene: FBgn0033349","log2FoldChange: 0.1279272781<br />-log10(padj): 1.410758e-01<br />log10(baseMean): 2.6745658<br />gene: FBgn0033350","log2FoldChange: 0.1088508288<br />-log10(padj): 9.998393e-02<br />log10(baseMean): 2.7411141<br />gene: FBgn0033351","log2FoldChange: -0.0984401692<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 3.3031199<br />gene: FBgn0033352","log2FoldChange: -0.7261912351<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 1.5374021<br />gene: FBgn0033353","log2FoldChange: -0.0558769076<br />-log10(padj): 3.122636e-02<br />log10(baseMean): 2.8068935<br />gene: FBgn0033354","log2FoldChange: -0.4723695850<br />-log10(padj): 1.166502e+00<br />log10(baseMean): 2.9375467<br />gene: FBgn0033356","log2FoldChange: 0.3291161609<br />-log10(padj): 6.871396e-01<br />log10(baseMean): 2.6588100<br />gene: FBgn0033357","log2FoldChange: -0.7706307095<br />-log10(padj): 3.358359e+00<br />log10(baseMean): 2.5663651<br />gene: FBgn0033359","log2FoldChange: -0.8697211090<br />-log10(padj): 2.966478e+00<br />log10(baseMean): 2.1105667<br />gene: FBgn0033362","log2FoldChange: -0.9420230279<br />-log10(padj): 1.875051e+00<br />log10(baseMean): 1.8465192<br />gene: FBgn0033363","log2FoldChange: -0.7716746364<br />-log10(padj): 2.083457e+00<br />log10(baseMean): 1.9404130<br />gene: FBgn0033364","log2FoldChange: 0.2364648531<br />-log10(padj): 1.001337e-01<br />log10(baseMean): 1.6941981<br />gene: FBgn0033366","log2FoldChange: 1.4549323923<br />-log10(padj): 9.072719e+00<br />log10(baseMean): 4.1853997<br />gene: FBgn0033367","log2FoldChange: 1.3259425145<br />-log10(padj): 4.242743e+00<br />log10(baseMean): 1.8049299<br />gene: FBgn0033368","log2FoldChange: -0.0202202707<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.4796266<br />gene: FBgn0033372","log2FoldChange: -0.2402573562<br />-log10(padj): 3.492311e-01<br />log10(baseMean): 2.6416216<br />gene: FBgn0033373","log2FoldChange: 0.0070398403<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 2.1887501<br />gene: FBgn0033375","log2FoldChange: -0.0804340960<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.5960937<br />gene: FBgn0033376","log2FoldChange: 0.2121761619<br />-log10(padj): 7.629574e-01<br />log10(baseMean): 3.2779961<br />gene: FBgn0033377","log2FoldChange: 0.0671561563<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.9138866<br />gene: FBgn0033378","log2FoldChange: -0.1523283116<br />-log10(padj): 3.056808e-01<br />log10(baseMean): 2.9488100<br />gene: FBgn0033379","log2FoldChange: -0.0272626557<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.5943694<br />gene: FBgn0033380","log2FoldChange: 0.2834144296<br />-log10(padj): 3.566542e-01<br />log10(baseMean): 2.3038686<br />gene: FBgn0033381","log2FoldChange: -0.0052160685<br />-log10(padj): 2.725201e-03<br />log10(baseMean): 2.6321624<br />gene: FBgn0033382","log2FoldChange: -0.2239330046<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 2.0398065<br />gene: FBgn0033387","log2FoldChange: -0.2100890046<br />-log10(padj): 2.036646e-01<br />log10(baseMean): 2.1847977<br />gene: FBgn0033388","log2FoldChange: 0.2543634878<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.2239556<br />gene: FBgn0033389","log2FoldChange: -0.0093596527<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.1531459<br />gene: FBgn0033391","log2FoldChange: -0.0316637969<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.1799391<br />gene: FBgn0033392","log2FoldChange: -0.0994868826<br />-log10(padj): 1.105617e-01<br />log10(baseMean): 3.5046174<br />gene: FBgn0033393","log2FoldChange: -0.4061415037<br />-log10(padj): 2.168900e-01<br />log10(baseMean): 1.7531163<br />gene: FBgn0033397","log2FoldChange: 0.0340379977<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.6498620<br />gene: FBgn0033400","log2FoldChange: 0.1568314486<br />-log10(padj): 1.770380e-01<br />log10(baseMean): 2.6261921<br />gene: FBgn0033401","log2FoldChange: -0.4117922512<br />-log10(padj): 2.912713e+00<br />log10(baseMean): 3.0440123<br />gene: FBgn0033402","log2FoldChange: -0.8196975806<br />-log10(padj): 1.911162e-01<br />log10(baseMean): 0.8404870<br />gene: FBgn0033403","log2FoldChange: -0.0962284156<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.0584947<br />gene: FBgn0033405","log2FoldChange: -0.0238748985<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.8928690<br />gene: FBgn0033413","log2FoldChange: 0.3304856523<br />-log10(padj): 8.029754e-01<br />log10(baseMean): 2.7703919<br />gene: FBgn0033421","log2FoldChange: -0.3256205548<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 1.5112564<br />gene: FBgn0033422","log2FoldChange: -0.6660470711<br />-log10(padj): 4.661415e-01<br />log10(baseMean): 1.4019519<br />gene: FBgn0033423","log2FoldChange: -0.9842393021<br />-log10(padj): 9.194029e+00<br />log10(baseMean): 2.5477085<br />gene: FBgn0033426","log2FoldChange: 0.2552062846<br />-log10(padj): 2.942306e-01<br />log10(baseMean): 2.2459531<br />gene: FBgn0033427","log2FoldChange: 0.2099313263<br />-log10(padj): 3.543435e-01<br />log10(baseMean): 2.9534965<br />gene: FBgn0033428","log2FoldChange: 0.5392740150<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 1.8666580<br />gene: FBgn0033429","log2FoldChange: 0.0876014365<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.5534615<br />gene: FBgn0033431","log2FoldChange: -0.0713433925<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 2.9926818<br />gene: FBgn0033434","log2FoldChange: 0.1165745216<br />-log10(padj): 4.497479e-02<br />log10(baseMean): 1.6970067<br />gene: FBgn0033437","log2FoldChange: 0.2132416706<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 3.1576766<br />gene: FBgn0033438","log2FoldChange: -1.1211013732<br />-log10(padj): 4.027263e+00<br />log10(baseMean): 1.9113951<br />gene: FBgn0033446","log2FoldChange: -0.5583163491<br />-log10(padj): 2.932910e+00<br />log10(baseMean): 2.9171609<br />gene: FBgn0033448","log2FoldChange: -0.2777731833<br />-log10(padj): 2.397917e-01<br />log10(baseMean): 1.9759343<br />gene: FBgn0033449","log2FoldChange: 0.1267598335<br />-log10(padj): 3.665809e-02<br />log10(baseMean): 1.7048560<br />gene: FBgn0033450","log2FoldChange: 0.2513670636<br />-log10(padj): 3.541955e-01<br />log10(baseMean): 2.5575308<br />gene: FBgn0033451","log2FoldChange: 0.1478086284<br />-log10(padj): 2.327447e-01<br />log10(baseMean): 2.9061347<br />gene: FBgn0033452","log2FoldChange: 0.1956464386<br />-log10(padj): 3.115478e-01<br />log10(baseMean): 2.8990307<br />gene: FBgn0033453","log2FoldChange: 0.0187709202<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.9287707<br />gene: FBgn0033454","log2FoldChange: 0.1426385740<br />-log10(padj): 1.133007e-01<br />log10(baseMean): 2.3605857<br />gene: FBgn0033457","log2FoldChange: 1.0976476451<br />-log10(padj): 1.886983e+00<br />log10(baseMean): 1.6139524<br />gene: FBgn0033458","log2FoldChange: 0.4389440003<br />-log10(padj): 4.357382e-01<br />log10(baseMean): 1.9029498<br />gene: FBgn0033459","log2FoldChange: -0.0996720670<br />-log10(padj): 1.680682e-01<br />log10(baseMean): 3.4078403<br />gene: FBgn0033460","log2FoldChange: -0.0743997899<br />-log10(padj): 7.964138e-02<br />log10(baseMean): 3.2302569<br />gene: FBgn0033463","log2FoldChange: -0.1651725335<br />-log10(padj): 3.251341e-01<br />log10(baseMean): 2.7556531<br />gene: FBgn0033465","log2FoldChange: -0.2477764666<br />-log10(padj): 6.225587e-01<br />log10(baseMean): 2.8341125<br />gene: FBgn0033466","log2FoldChange: 0.1434003341<br />-log10(padj): 1.685188e-02<br />log10(baseMean): 1.2534011<br />gene: FBgn0033467","log2FoldChange: -0.0271354523<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.7369827<br />gene: FBgn0033468","log2FoldChange: -0.4311459999<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.1591300<br />gene: FBgn0033469","log2FoldChange: -0.0317065559<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.6270139<br />gene: FBgn0033471","log2FoldChange: -0.1726013811<br />-log10(padj): 2.591363e-01<br />log10(baseMean): 2.5650508<br />gene: FBgn0033473","log2FoldChange: -0.0743367565<br />-log10(padj): 5.810400e-02<br />log10(baseMean): 2.9944059<br />gene: FBgn0033474","log2FoldChange: 0.2597460395<br />-log10(padj): 3.001021e-01<br />log10(baseMean): 2.4297483<br />gene: FBgn0033475","log2FoldChange: 0.0453074949<br />-log10(padj): 3.889025e-02<br />log10(baseMean): 3.6467285<br />gene: FBgn0033476","log2FoldChange: -0.0235631654<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.0474358<br />gene: FBgn0033477","log2FoldChange: 0.5799246663<br />-log10(padj): 8.006545e-01<br />log10(baseMean): 1.7470843<br />gene: FBgn0033478","log2FoldChange: 0.0845980736<br />-log10(padj): 4.985877e-02<br />log10(baseMean): 2.4375679<br />gene: FBgn0033479","log2FoldChange: 0.1326025523<br />-log10(padj): 7.355363e-02<br />log10(baseMean): 2.3258513<br />gene: FBgn0033480","log2FoldChange: 1.0867352626<br />-log10(padj): 4.505560e-01<br />log10(baseMean): 0.9357414<br />gene: FBgn0033481","log2FoldChange: -0.1300533281<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 3.4447018<br />gene: FBgn0033482","log2FoldChange: -0.2936688871<br />-log10(padj): 2.432064e+00<br />log10(baseMean): 3.6881535<br />gene: FBgn0033483","log2FoldChange: -0.2472504999<br />-log10(padj): 4.731339e-01<br />log10(baseMean): 2.8219369<br />gene: FBgn0033485","log2FoldChange: -0.1295177982<br />-log10(padj): 1.109972e-01<br />log10(baseMean): 2.9176213<br />gene: FBgn0033486","log2FoldChange: -0.0526727517<br />-log10(padj): 2.202788e-02<br />log10(baseMean): 2.3784433<br />gene: FBgn0033491","log2FoldChange: -0.0616804885<br />-log10(padj): 2.538057e-02<br />log10(baseMean): 2.0546546<br />gene: FBgn0033494","log2FoldChange: -2.0243418262<br />-log10(padj): 5.127384e+00<br />log10(baseMean): 1.8223492<br />gene: FBgn0033495","log2FoldChange: 0.1361529735<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 0.7943816<br />gene: FBgn0033501","log2FoldChange: -0.9015726141<br />-log10(padj): 2.074563e+00<br />log10(baseMean): 1.8682400<br />gene: FBgn0033502","log2FoldChange: -0.6805896218<br />-log10(padj): 3.805585e+00<br />log10(baseMean): 3.3170385<br />gene: FBgn0033504","log2FoldChange: -0.1696954448<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 2.5720832<br />gene: FBgn0033507","log2FoldChange: -1.4762657139<br />-log10(padj): 8.159360e-01<br />log10(baseMean): 1.0182601<br />gene: FBgn0033518","log2FoldChange: 0.3712353659<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 0.9343387<br />gene: FBgn0033519","log2FoldChange: 0.1583382156<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 2.0546235<br />gene: FBgn0033523","log2FoldChange: 0.9151509669<br />-log10(padj): 8.721040e-01<br />log10(baseMean): 1.4759593<br />gene: FBgn0033524","log2FoldChange: -0.2419290139<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 2.7062692<br />gene: FBgn0033526","log2FoldChange: 0.1830085954<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 1.9660708<br />gene: FBgn0033527","log2FoldChange: 0.2843196601<br />-log10(padj): 3.774457e-01<br />log10(baseMean): 2.6452425<br />gene: FBgn0033528","log2FoldChange: 0.1276151291<br />-log10(padj): 1.310012e-01<br />log10(baseMean): 3.1133369<br />gene: FBgn0033529","log2FoldChange: -0.6290136995<br />-log10(padj): 1.135727e+00<br />log10(baseMean): 2.7531234<br />gene: FBgn0033538","log2FoldChange: 0.2862493248<br />-log10(padj): 1.135727e+00<br />log10(baseMean): 2.8786202<br />gene: FBgn0033539","log2FoldChange: 0.2086983358<br />-log10(padj): 2.823653e-01<br />log10(baseMean): 2.5822250<br />gene: FBgn0033540","log2FoldChange: 1.0994213173<br />-log10(padj): 1.460923e+00<br />log10(baseMean): 1.3569446<br />gene: FBgn0033541","log2FoldChange: 0.1938923317<br />-log10(padj): 1.112677e-01<br />log10(baseMean): 1.9326715<br />gene: FBgn0033543","log2FoldChange: -0.0092148257<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.7506186<br />gene: FBgn0033544","log2FoldChange: -0.1425873398<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.0686926<br />gene: FBgn0033547","log2FoldChange: -0.3319395219<br />-log10(padj): 4.765557e-02<br />log10(baseMean): 2.1482196<br />gene: FBgn0033548","log2FoldChange: -0.1548432731<br />-log10(padj): 6.605863e-02<br />log10(baseMean): 1.7234127<br />gene: FBgn0033549","log2FoldChange: -0.1604204489<br />-log10(padj): 1.235517e-01<br />log10(baseMean): 2.1668280<br />gene: FBgn0033550","log2FoldChange: -0.1657222930<br />-log10(padj): 1.696066e-01<br />log10(baseMean): 2.8828353<br />gene: FBgn0033551","log2FoldChange: -0.2015711705<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 1.5061510<br />gene: FBgn0033554","log2FoldChange: 0.0550147646<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 3.7385737<br />gene: FBgn0033555","log2FoldChange: -0.1819040027<br />-log10(padj): 1.646410e-01<br />log10(baseMean): 2.3107308<br />gene: FBgn0033556","log2FoldChange: -0.3671737458<br />-log10(padj): 8.538948e-01<br />log10(baseMean): 2.6491085<br />gene: FBgn0033557","log2FoldChange: -0.2688594192<br />-log10(padj): 8.150812e-01<br />log10(baseMean): 3.0840805<br />gene: FBgn0033562","log2FoldChange: -0.1755017218<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.4801539<br />gene: FBgn0033566","log2FoldChange: 0.0304510351<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.3308129<br />gene: FBgn0033569","log2FoldChange: -0.0209591857<br />-log10(padj): 1.224480e-02<br />log10(baseMean): 2.7524062<br />gene: FBgn0033570","log2FoldChange: -0.0462839674<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 2.8240812<br />gene: FBgn0033571","log2FoldChange: 0.3557513979<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 0.7999332<br />gene: FBgn0033578","log2FoldChange: 0.8950073366<br />-log10(padj): 2.461220e-01<br />log10(baseMean): 0.9173873<br />gene: FBgn0033579","log2FoldChange: -0.2406176028<br />-log10(padj): 8.800593e-01<br />log10(baseMean): 3.0085818<br />gene: FBgn0033581","log2FoldChange: -0.2045390549<br />-log10(padj): 3.904687e-01<br />log10(baseMean): 2.5724331<br />gene: FBgn0033584","log2FoldChange: 0.0027851381<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.8703808<br />gene: FBgn0033605","log2FoldChange: 0.0122703632<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.7686001<br />gene: FBgn0033607","log2FoldChange: 0.0218459601<br />-log10(padj): 1.032380e-02<br />log10(baseMean): 2.6789141<br />gene: FBgn0033608","log2FoldChange: -0.1941448429<br />-log10(padj): 4.386328e-01<br />log10(baseMean): 2.8070297<br />gene: FBgn0033609","log2FoldChange: 0.0522871989<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 1.1091891<br />gene: FBgn0033610","log2FoldChange: 0.0406648703<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.4512380<br />gene: FBgn0033615","log2FoldChange: -0.2096669455<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.3006271<br />gene: FBgn0033616","log2FoldChange: -0.3669720731<br />-log10(padj): 1.207045e+00<br />log10(baseMean): 2.7467813<br />gene: FBgn0033624","log2FoldChange: 0.0948623562<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 1.9338856<br />gene: FBgn0033627","log2FoldChange: 0.3576658121<br />-log10(padj): 9.497493e-02<br />log10(baseMean): 1.1572133<br />gene: FBgn0033631","log2FoldChange: 1.2054589361<br />-log10(padj): 5.515706e+00<br />log10(baseMean): 3.3962204<br />gene: FBgn0033635","log2FoldChange: -0.0910167751<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 3.8351044<br />gene: FBgn0033636","log2FoldChange: -0.0157469613<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.9696649<br />gene: FBgn0033638","log2FoldChange: -0.1443366126<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 2.7854253<br />gene: FBgn0033639","log2FoldChange: 0.1689264031<br />-log10(padj): 1.076328e-02<br />log10(baseMean): 1.4112319<br />gene: FBgn0033644","log2FoldChange: 0.2642894145<br />-log10(padj): 1.522534e-01<br />log10(baseMean): 1.7436880<br />gene: FBgn0033645","log2FoldChange: -0.7382469786<br />-log10(padj): 2.760723e+00<br />log10(baseMean): 2.8338704<br />gene: FBgn0033649","log2FoldChange: -0.1178935096<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.5105655<br />gene: FBgn0033653","log2FoldChange: 1.0050652628<br />-log10(padj): 3.188201e-01<br />log10(baseMean): 0.8591801<br />gene: FBgn0033654","log2FoldChange: 0.0460256937<br />-log10(padj): 2.981727e-02<br />log10(baseMean): 2.8543782<br />gene: FBgn0033656","log2FoldChange: -0.3046925703<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 3.4059402<br />gene: FBgn0033661","log2FoldChange: 0.2939918811<br />-log10(padj): 9.774826e-01<br />log10(baseMean): 4.2843936<br />gene: FBgn0033663","log2FoldChange: -0.1789595336<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 2.3349841<br />gene: FBgn0033665","log2FoldChange: 1.0593295717<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 0.8548862<br />gene: FBgn0033668","log2FoldChange: 0.1074801480<br />-log10(padj): 1.433881e-01<br />log10(baseMean): 3.0172212<br />gene: FBgn0033669","log2FoldChange: 0.2978187905<br />-log10(padj): 8.641833e-01<br />log10(baseMean): 2.6253854<br />gene: FBgn0033672","log2FoldChange: -0.0651307704<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.3642702<br />gene: FBgn0033673","log2FoldChange: 0.3407741431<br />-log10(padj): 5.891446e-01<br />log10(baseMean): 2.2150979<br />gene: FBgn0033677","log2FoldChange: -0.2334161900<br />-log10(padj): 3.628149e-01<br />log10(baseMean): 2.6452004<br />gene: FBgn0033683","log2FoldChange: 0.2090986319<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 1.4048685<br />gene: FBgn0033685","log2FoldChange: -0.1698195521<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 1.9335275<br />gene: FBgn0033686","log2FoldChange: -0.0530015413<br />-log10(padj): 5.290275e-02<br />log10(baseMean): 3.7711640<br />gene: FBgn0033688","log2FoldChange: 0.0021568529<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 2.3668622<br />gene: FBgn0033690","log2FoldChange: 0.1159380329<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.7185530<br />gene: FBgn0033691","log2FoldChange: -0.0462955368<br />-log10(padj): 2.691674e-02<br />log10(baseMean): 2.5723830<br />gene: FBgn0033692","log2FoldChange: 0.4604783461<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 1.3213736<br />gene: FBgn0033696","log2FoldChange: 0.4100851344<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 1.2549962<br />gene: FBgn0033697","log2FoldChange: 0.1080455436<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 3.0736313<br />gene: FBgn0033698","log2FoldChange: -0.1784957919<br />-log10(padj): 2.704925e-01<br />log10(baseMean): 4.4392015<br />gene: FBgn0033699","log2FoldChange: 1.7561300692<br />-log10(padj): 9.847395e-01<br />log10(baseMean): 0.8742616<br />gene: FBgn0033702","log2FoldChange: 0.1035975777<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 1.3490954<br />gene: FBgn0033706","log2FoldChange: -0.2714797248<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 1.9819289<br />gene: FBgn0033712","log2FoldChange: -0.1346569395<br />-log10(padj): 1.475558e-01<br />log10(baseMean): 2.6287735<br />gene: FBgn0033713","log2FoldChange: -0.1262333670<br />-log10(padj): 2.319413e-01<br />log10(baseMean): 3.3330902<br />gene: FBgn0033714","log2FoldChange: 0.2965172000<br />-log10(padj): 5.329073e-01<br />log10(baseMean): 2.4038183<br />gene: FBgn0033715","log2FoldChange: -0.3475232451<br />-log10(padj): 8.533031e-01<br />log10(baseMean): 2.3792757<br />gene: FBgn0033716","log2FoldChange: -0.2451652539<br />-log10(padj): 6.077817e-01<br />log10(baseMean): 3.7587187<br />gene: FBgn0033717","log2FoldChange: 0.0133543520<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 2.8812944<br />gene: FBgn0033718","log2FoldChange: 1.9098023220<br />-log10(padj): 1.169686e+00<br />log10(baseMean): 0.9122884<br />gene: FBgn0033720","log2FoldChange: 1.4243726355<br />-log10(padj): 8.655466e+00<br />log10(baseMean): 2.1842226<br />gene: FBgn0033724","log2FoldChange: 0.9585696805<br />-log10(padj): 8.399860e-01<br />log10(baseMean): 1.3879733<br />gene: FBgn0033725","log2FoldChange: -0.0829494523<br />-log10(padj): 6.267069e-02<br />log10(baseMean): 2.5601374<br />gene: FBgn0033734","log2FoldChange: 0.0196264625<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.4490898<br />gene: FBgn0033735","log2FoldChange: -0.1471432240<br />-log10(padj): 2.325270e-01<br />log10(baseMean): 2.8054386<br />gene: FBgn0033737","log2FoldChange: 0.2473347068<br />-log10(padj): 9.318228e-01<br />log10(baseMean): 3.1266973<br />gene: FBgn0033738","log2FoldChange: 0.0478720426<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.7949154<br />gene: FBgn0033739","log2FoldChange: 0.0813371881<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.6553412<br />gene: FBgn0033740","log2FoldChange: -0.0605725627<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 3.3477927<br />gene: FBgn0033741","log2FoldChange: 0.4995309841<br />-log10(padj): 5.735677e-02<br />log10(baseMean): 1.7396090<br />gene: FBgn0033744","log2FoldChange: 0.4037049559<br />-log10(padj): 1.074074e-01<br />log10(baseMean): 1.2670051<br />gene: FBgn0033748","log2FoldChange: -0.1885303739<br />-log10(padj): 1.185850e-01<br />log10(baseMean): 2.1314264<br />gene: FBgn0033749","log2FoldChange: 0.0664364109<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.3139414<br />gene: FBgn0033750","log2FoldChange: 0.1613885519<br />-log10(padj): 2.782103e-01<br />log10(baseMean): 3.0624039<br />gene: FBgn0033751","log2FoldChange: 0.1406698941<br />-log10(padj): 1.296387e-01<br />log10(baseMean): 2.3577771<br />gene: FBgn0033752","log2FoldChange: -0.5322059262<br />-log10(padj): 1.807875e-01<br />log10(baseMean): 1.5858714<br />gene: FBgn0033754","log2FoldChange: 0.0668718596<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 3.2427158<br />gene: FBgn0033755","log2FoldChange: -0.2527841830<br />-log10(padj): 2.249970e-01<br />log10(baseMean): 2.8990645<br />gene: FBgn0033756","log2FoldChange: 0.1132925775<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 2.3298945<br />gene: FBgn0033757","log2FoldChange: 0.2575811221<br />-log10(padj): 4.731339e-01<br />log10(baseMean): 2.6764681<br />gene: FBgn0033761","log2FoldChange: -0.1213730896<br />-log10(padj): 1.269622e-01<br />log10(baseMean): 2.7018847<br />gene: FBgn0033762","log2FoldChange: -0.5287802819<br />-log10(padj): 6.826567e-01<br />log10(baseMean): 1.8893199<br />gene: FBgn0033763","log2FoldChange: 3.4116384473<br />-log10(padj): 2.765275e+01<br />log10(baseMean): 1.8142871<br />gene: FBgn0033764","log2FoldChange: 0.1167430236<br />-log10(padj): 1.115870e-01<br />log10(baseMean): 3.1648680<br />gene: FBgn0033765","log2FoldChange: -0.0298402365<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.0353647<br />gene: FBgn0033766","log2FoldChange: 0.2191076985<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 1.1829418<br />gene: FBgn0033767","log2FoldChange: 0.4042369672<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 2.0366551<br />gene: FBgn0033769","log2FoldChange: 1.9347744257<br />-log10(padj): 8.863836e+00<br />log10(baseMean): 1.6709966<br />gene: FBgn0033775","log2FoldChange: 0.5125839834<br />-log10(padj): 3.431456e+00<br />log10(baseMean): 2.9673286<br />gene: FBgn0033777","log2FoldChange: -0.0823508788<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.7881347<br />gene: FBgn0033778","log2FoldChange: -0.2443645661<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.2534429<br />gene: FBgn0033781","log2FoldChange: -1.3714157236<br />-log10(padj): 6.327981e+00<br />log10(baseMean): 2.2101207<br />gene: FBgn0033782","log2FoldChange: -0.0274790616<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.0606832<br />gene: FBgn0033783","log2FoldChange: -0.1669403590<br />-log10(padj): 2.213599e-01<br />log10(baseMean): 2.5773373<br />gene: FBgn0033784","log2FoldChange: 0.3267510903<br />-log10(padj): 5.014606e-01<br />log10(baseMean): 2.1514325<br />gene: FBgn0033785","log2FoldChange: 0.2575311174<br />-log10(padj): 4.125182e-01<br />log10(baseMean): 3.4116688<br />gene: FBgn0033786","log2FoldChange: 0.0265336925<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 1.6764426<br />gene: FBgn0033787","log2FoldChange: -0.2874136758<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.2576196<br />gene: FBgn0033798","log2FoldChange: 0.0745332938<br />-log10(padj): 3.792700e-02<br />log10(baseMean): 2.1962428<br />gene: FBgn0033799","log2FoldChange: -0.2058317874<br />-log10(padj): 4.107652e-01<br />log10(baseMean): 2.8719381<br />gene: FBgn0033802","log2FoldChange: -0.0127818054<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 2.4947056<br />gene: FBgn0033806","log2FoldChange: 0.0581811218<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 1.9447997<br />gene: FBgn0033808","log2FoldChange: 0.3430551976<br />-log10(padj): 6.687984e-01<br />log10(baseMean): 2.4988946<br />gene: FBgn0033809","log2FoldChange: 0.0530691992<br />-log10(padj): 2.830218e-02<br />log10(baseMean): 2.4048949<br />gene: FBgn0033810","log2FoldChange: 0.0793904732<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 2.6395108<br />gene: FBgn0033812","log2FoldChange: 0.1109137644<br />-log10(padj): 6.119574e-02<br />log10(baseMean): 2.0162142<br />gene: FBgn0033813","log2FoldChange: 0.1936192951<br />-log10(padj): 2.674797e-01<br />log10(baseMean): 2.8571513<br />gene: FBgn0033814","log2FoldChange: -0.0685288932<br />-log10(padj): 5.455075e-02<br />log10(baseMean): 2.6848969<br />gene: FBgn0033816","log2FoldChange: -0.8366315989<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 0.9182818<br />gene: FBgn0033821","log2FoldChange: 0.1508113456<br />-log10(padj): 6.237762e-02<br />log10(baseMean): 2.3765168<br />gene: FBgn0033836","log2FoldChange: -0.0707246857<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 2.5464850<br />gene: FBgn0033842","log2FoldChange: -0.2595828587<br />-log10(padj): 1.521343e+00<br />log10(baseMean): 3.6918290<br />gene: FBgn0033844","log2FoldChange: 0.0478179755<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 3.1172947<br />gene: FBgn0033845","log2FoldChange: -0.1245594751<br />-log10(padj): 1.475558e-01<br />log10(baseMean): 2.7670065<br />gene: FBgn0033846","log2FoldChange: 0.1258160794<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 2.6900390<br />gene: FBgn0033853","log2FoldChange: 1.6251678896<br />-log10(padj): 1.422262e+00<br />log10(baseMean): 1.2440322<br />gene: FBgn0033857","log2FoldChange: 0.1362395526<br />-log10(padj): 2.025429e-01<br />log10(baseMean): 2.9268811<br />gene: FBgn0033859","log2FoldChange: 0.1840880517<br />-log10(padj): 3.724778e-02<br />log10(baseMean): 1.2006038<br />gene: FBgn0033869","log2FoldChange: -0.0855748065<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.8388137<br />gene: FBgn0033871","log2FoldChange: 0.5883956068<br />-log10(padj): 1.463085e-01<br />log10(baseMean): 0.9650736<br />gene: FBgn0033872","log2FoldChange: 0.2764158375<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 3.2954970<br />gene: FBgn0033875","log2FoldChange: 0.1976199562<br />-log10(padj): 1.162872e-01<br />log10(baseMean): 1.9130134<br />gene: FBgn0033876","log2FoldChange: -0.1311444405<br />-log10(padj): 1.331016e-01<br />log10(baseMean): 3.2663764<br />gene: FBgn0033879","log2FoldChange: -0.3309570166<br />-log10(padj): 1.707397e-01<br />log10(baseMean): 1.6346894<br />gene: FBgn0033880","log2FoldChange: 0.0983891445<br />-log10(padj): 1.007719e-01<br />log10(baseMean): 2.6784456<br />gene: FBgn0033882","log2FoldChange: 0.0741180260<br />-log10(padj): 4.923107e-02<br />log10(baseMean): 2.4373988<br />gene: FBgn0033883","log2FoldChange: -0.1445580362<br />-log10(padj): 1.141528e-01<br />log10(baseMean): 2.2137806<br />gene: FBgn0033884","log2FoldChange: 0.0550215531<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 3.1766761<br />gene: FBgn0033886","log2FoldChange: -0.4881099153<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 1.0373569<br />gene: FBgn0033887","log2FoldChange: -0.7146333845<br />-log10(padj): 1.988070e-01<br />log10(baseMean): 0.9954867<br />gene: FBgn0033888","log2FoldChange: -0.0726349589<br />-log10(padj): 1.019554e-01<br />log10(baseMean): 3.7722086<br />gene: FBgn0033889","log2FoldChange: -0.3116225066<br />-log10(padj): 7.647449e-01<br />log10(baseMean): 3.0724513<br />gene: FBgn0033890","log2FoldChange: 0.0774313074<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.2219176<br />gene: FBgn0033891","log2FoldChange: -0.1226778571<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 1.0993251<br />gene: FBgn0033893","log2FoldChange: 0.0373525394<br />-log10(padj): 3.599372e-02<br />log10(baseMean): 3.1328750<br />gene: FBgn0033897","log2FoldChange: 0.0400481357<br />-log10(padj): 2.327121e-02<br />log10(baseMean): 2.7031354<br />gene: FBgn0033899","log2FoldChange: -0.0279662230<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.1183150<br />gene: FBgn0033900","log2FoldChange: 0.2074762724<br />-log10(padj): 3.904687e-01<br />log10(baseMean): 2.9851640<br />gene: FBgn0033901","log2FoldChange: -0.1054051044<br />-log10(padj): 1.100856e-01<br />log10(baseMean): 3.8206033<br />gene: FBgn0033902","log2FoldChange: 0.0316200966<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.3270339<br />gene: FBgn0033903","log2FoldChange: 0.1453499084<br />-log10(padj): 1.333676e-01<br />log10(baseMean): 2.3005598<br />gene: FBgn0033905","log2FoldChange: 0.0804815253<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 3.0301159<br />gene: FBgn0033906","log2FoldChange: 0.1056714993<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.3612492<br />gene: FBgn0033907","log2FoldChange: 0.0474132024<br />-log10(padj): 2.728113e-02<br />log10(baseMean): 4.4883677<br />gene: FBgn0033912","log2FoldChange: -1.1613116495<br />-log10(padj): 2.479322e+01<br />log10(baseMean): 3.7170845<br />gene: FBgn0033913","log2FoldChange: 0.0988055123<br />-log10(padj): 1.334952e-01<br />log10(baseMean): 3.0573080<br />gene: FBgn0033915","log2FoldChange: -0.1097758363<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.8190130<br />gene: FBgn0033916","log2FoldChange: 0.4045627627<br />-log10(padj): 1.977676e-01<br />log10(baseMean): 1.4685228<br />gene: FBgn0033917","log2FoldChange: 0.1607593458<br />-log10(padj): 5.365798e-01<br />log10(baseMean): 3.2527774<br />gene: FBgn0033918","log2FoldChange: 0.1655915561<br />-log10(padj): 1.410758e-01<br />log10(baseMean): 2.5596344<br />gene: FBgn0033919","log2FoldChange: -0.1720738900<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 1.5182859<br />gene: FBgn0033921","log2FoldChange: 0.4837035983<br />-log10(padj): 1.834009e+00<br />log10(baseMean): 2.5934453<br />gene: FBgn0033924","log2FoldChange: 0.0520733417<br />-log10(padj): 3.287411e-02<br />log10(baseMean): 2.3511231<br />gene: FBgn0033925","log2FoldChange: 0.5455998630<br />-log10(padj): 2.315235e+00<br />log10(baseMean): 3.8002538<br />gene: FBgn0033926","log2FoldChange: -0.1569853120<br />-log10(padj): 2.186085e-01<br />log10(baseMean): 2.8493233<br />gene: FBgn0033928","log2FoldChange: -0.0102843891<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.3824199<br />gene: FBgn0033929","log2FoldChange: 0.0128957394<br />-log10(padj): 7.909689e-03<br />log10(baseMean): 3.0400843<br />gene: FBgn0033934","log2FoldChange: -0.0612417450<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 2.4910406<br />gene: FBgn0033935","log2FoldChange: -0.2940496237<br />-log10(padj): 5.705703e-02<br />log10(baseMean): 1.1242809<br />gene: FBgn0033936","log2FoldChange: 1.3310902199<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 0.8003083<br />gene: FBgn0033943","log2FoldChange: 0.2069650820<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 1.8523496<br />gene: FBgn0033945","log2FoldChange: 0.0185957269<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.2353503<br />gene: FBgn0033948","log2FoldChange: 0.2299932852<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 1.2912453<br />gene: FBgn0033949","log2FoldChange: -0.0307524368<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.5183951<br />gene: FBgn0033951","log2FoldChange: -0.2499172658<br />-log10(padj): 6.470652e-02<br />log10(baseMean): 1.2736832<br />gene: FBgn0033960","log2FoldChange: 0.0421933086<br />-log10(padj): 2.526360e-02<br />log10(baseMean): 2.4348934<br />gene: FBgn0033961","log2FoldChange: 0.0918780471<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.1804772<br />gene: FBgn0033962","log2FoldChange: 0.6771273761<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 0.7917682<br />gene: FBgn0033963","log2FoldChange: -0.1098858809<br />-log10(padj): 1.094967e-01<br />log10(baseMean): 2.5754779<br />gene: FBgn0033971","log2FoldChange: -0.1933858912<br />-log10(padj): 2.942091e-01<br />log10(baseMean): 2.6765605<br />gene: FBgn0033972","log2FoldChange: -0.0969393719<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 2.0631927<br />gene: FBgn0033973","log2FoldChange: -0.1454995776<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.3226199<br />gene: FBgn0033978","log2FoldChange: 0.0191467258<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.1121846<br />gene: FBgn0033979","log2FoldChange: 0.1627317568<br />-log10(padj): 1.165047e-01<br />log10(baseMean): 3.0146396<br />gene: FBgn0033980","log2FoldChange: -0.7071229400<br />-log10(padj): 1.753597e-01<br />log10(baseMean): 0.9429289<br />gene: FBgn0033982","log2FoldChange: 0.2264164282<br />-log10(padj): 3.771626e-01<br />log10(baseMean): 2.8193966<br />gene: FBgn0033984","log2FoldChange: 0.4851605451<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 1.5360285<br />gene: FBgn0033987","log2FoldChange: 0.1146491108<br />-log10(padj): 8.446817e-02<br />log10(baseMean): 2.8810361<br />gene: FBgn0033988","log2FoldChange: 0.0803683223<br />-log10(padj): 7.502956e-02<br />log10(baseMean): 2.6270673<br />gene: FBgn0033989","log2FoldChange: 0.0033410553<br />-log10(padj): 1.611385e-03<br />log10(baseMean): 2.2050998<br />gene: FBgn0033990","log2FoldChange: -0.1813626301<br />-log10(padj): 5.688062e-02<br />log10(baseMean): 1.7958607<br />gene: FBgn0033993","log2FoldChange: -0.2465245095<br />-log10(padj): 2.069623e-01<br />log10(baseMean): 2.1298025<br />gene: FBgn0033994","log2FoldChange: -0.0222700084<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 2.3429087<br />gene: FBgn0033995","log2FoldChange: 0.0537732018<br />-log10(padj): 2.538057e-02<br />log10(baseMean): 2.5866241<br />gene: FBgn0033996","log2FoldChange: -0.0047701381<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.2700911<br />gene: FBgn0033998","log2FoldChange: -0.0654666177<br />-log10(padj): 1.975768e-02<br />log10(baseMean): 2.1035803<br />gene: FBgn0034000","log2FoldChange: 0.0289824954<br />-log10(padj): 1.076328e-02<br />log10(baseMean): 2.0793764<br />gene: FBgn0034001","log2FoldChange: 0.0932716781<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 2.3944937<br />gene: FBgn0034002","log2FoldChange: 0.2725848934<br />-log10(padj): 2.927652e-01<br />log10(baseMean): 2.3764349<br />gene: FBgn0034005","log2FoldChange: 0.0613704497<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 2.0800370<br />gene: FBgn0034008","log2FoldChange: -0.1549290583<br />-log10(padj): 2.978285e-01<br />log10(baseMean): 3.0230288<br />gene: FBgn0034009","log2FoldChange: -1.4517101927<br />-log10(padj): 2.190851e+01<br />log10(baseMean): 2.7381780<br />gene: FBgn0034010","log2FoldChange: -0.6009192268<br />-log10(padj): 6.749954e-01<br />log10(baseMean): 1.6457537<br />gene: FBgn0034011","log2FoldChange: 0.5524689429<br />-log10(padj): 2.309930e-01<br />log10(baseMean): 1.2775954<br />gene: FBgn0034012","log2FoldChange: 0.4083169548<br />-log10(padj): 2.589209e+00<br />log10(baseMean): 3.1914295<br />gene: FBgn0034013","log2FoldChange: 1.2851444199<br />-log10(padj): 7.713787e-01<br />log10(baseMean): 1.0725243<br />gene: FBgn0034021","log2FoldChange: -0.0263402905<br />-log10(padj): 1.970734e-02<br />log10(baseMean): 2.9217018<br />gene: FBgn0034025","log2FoldChange: 0.2037439450<br />-log10(padj): 9.789936e-02<br />log10(baseMean): 2.0333697<br />gene: FBgn0034027","log2FoldChange: 0.4115000995<br />-log10(padj): 1.133596e+00<br />log10(baseMean): 2.6752042<br />gene: FBgn0034029","log2FoldChange: 0.7486226115<br />-log10(padj): 3.202194e-01<br />log10(baseMean): 1.0952070<br />gene: FBgn0034030","log2FoldChange: -0.1920845527<br />-log10(padj): 1.368061e-01<br />log10(baseMean): 2.3710619<br />gene: FBgn0034032","log2FoldChange: 0.0606852600<br />-log10(padj): 2.373932e-02<br />log10(baseMean): 2.0622573<br />gene: FBgn0034033","log2FoldChange: 0.0333195077<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.6473069<br />gene: FBgn0034035","log2FoldChange: 0.6309532705<br />-log10(padj): 7.916322e-01<br />log10(baseMean): 1.7102851<br />gene: FBgn0034037","log2FoldChange: -0.0751733518<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 2.9860910<br />gene: FBgn0034046","log2FoldChange: -0.1642317444<br />-log10(padj): 2.556949e-01<br />log10(baseMean): 3.5174740<br />gene: FBgn0034049","log2FoldChange: 0.1199570470<br />-log10(padj): 1.259071e-01<br />log10(baseMean): 2.8761549<br />gene: FBgn0034050","log2FoldChange: 0.0505797035<br />-log10(padj): 5.688062e-02<br />log10(baseMean): 3.3913456<br />gene: FBgn0034051","log2FoldChange: 0.3817297298<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 0.9340682<br />gene: FBgn0034053","log2FoldChange: 0.3850942695<br />-log10(padj): 5.969497e-01<br />log10(baseMean): 2.0704955<br />gene: FBgn0034054","log2FoldChange: -0.0501832393<br />-log10(padj): 2.157692e-02<br />log10(baseMean): 3.0177200<br />gene: FBgn0034057","log2FoldChange: 0.0149568590<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.4266552<br />gene: FBgn0034058","log2FoldChange: -0.1062430048<br />-log10(padj): 1.653947e-01<br />log10(baseMean): 3.2072511<br />gene: FBgn0034060","log2FoldChange: 0.1060515394<br />-log10(padj): 7.768911e-02<br />log10(baseMean): 2.3561597<br />gene: FBgn0034061","log2FoldChange: 0.1191109724<br />-log10(padj): 6.143645e-02<br />log10(baseMean): 2.0143507<br />gene: FBgn0034062","log2FoldChange: 0.3219013369<br />-log10(padj): 7.427974e-01<br />log10(baseMean): 2.4934276<br />gene: FBgn0034063","log2FoldChange: -0.0712768791<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.1503286<br />gene: FBgn0034065","log2FoldChange: -0.0631502216<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.2341368<br />gene: FBgn0034066","log2FoldChange: 1.0226976370<br />-log10(padj): 5.252143e+00<br />log10(baseMean): 2.1514112<br />gene: FBgn0034067","log2FoldChange: -0.0869167053<br />-log10(padj): 1.300740e-01<br />log10(baseMean): 3.1300527<br />gene: FBgn0034068","log2FoldChange: -0.0739669888<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.2539386<br />gene: FBgn0034071","log2FoldChange: 0.1009961703<br />-log10(padj): 1.690896e-01<br />log10(baseMean): 3.7009897<br />gene: FBgn0034072","log2FoldChange: -0.2273468985<br />-log10(padj): 2.039022e-01<br />log10(baseMean): 2.7017857<br />gene: FBgn0034073","log2FoldChange: -0.1004014266<br />-log10(padj): 6.008733e-02<br />log10(baseMean): 2.1006050<br />gene: FBgn0034075","log2FoldChange: -0.5141459634<br />-log10(padj): 7.502956e-02<br />log10(baseMean): 0.8313511<br />gene: FBgn0034076","log2FoldChange: 0.0534285832<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.1664814<br />gene: FBgn0034082","log2FoldChange: -0.2670015917<br />-log10(padj): 7.160896e-01<br />log10(baseMean): 3.1927107<br />gene: FBgn0034083","log2FoldChange: 0.0952482290<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.4000673<br />gene: FBgn0034084","log2FoldChange: -0.3318332892<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 0.8935687<br />gene: FBgn0034085","log2FoldChange: 0.0370652807<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.3430127<br />gene: FBgn0034086","log2FoldChange: 0.1414633010<br />-log10(padj): 2.262589e-01<br />log10(baseMean): 3.5265726<br />gene: FBgn0034087","log2FoldChange: -0.0008931871<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 2.4250122<br />gene: FBgn0034088","log2FoldChange: -0.1076410514<br />-log10(padj): 1.327424e-01<br />log10(baseMean): 3.2161195<br />gene: FBgn0034089","log2FoldChange: -0.0254382916<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.9236420<br />gene: FBgn0034091","log2FoldChange: 0.2087658287<br />-log10(padj): 1.300740e-01<br />log10(baseMean): 2.2250122<br />gene: FBgn0034093","log2FoldChange: -0.5559638842<br />-log10(padj): 8.721919e-01<br />log10(baseMean): 2.3159332<br />gene: FBgn0034094","log2FoldChange: -0.0728809282<br />-log10(padj): 1.393747e-02<br />log10(baseMean): 1.3196473<br />gene: FBgn0034095","log2FoldChange: -0.0242500819<br />-log10(padj): 1.124980e-02<br />log10(baseMean): 2.8338850<br />gene: FBgn0034098","log2FoldChange: 0.0481421014<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.3370894<br />gene: FBgn0034109","log2FoldChange: -0.1889224485<br />-log10(padj): 2.432168e-01<br />log10(baseMean): 3.1327178<br />gene: FBgn0034110","log2FoldChange: 0.1148824881<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.5977565<br />gene: FBgn0034113","log2FoldChange: -0.0073830254<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.5683799<br />gene: FBgn0034114","log2FoldChange: 0.1348685432<br />-log10(padj): 2.167041e-01<br />log10(baseMean): 2.9460480<br />gene: FBgn0034117","log2FoldChange: 0.0607534356<br />-log10(padj): 4.923107e-02<br />log10(baseMean): 2.8357859<br />gene: FBgn0034118","log2FoldChange: 0.0101907447<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.6225981<br />gene: FBgn0034126","log2FoldChange: -1.1658598201<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 1.2665761<br />gene: FBgn0034127","log2FoldChange: -0.3540826512<br />-log10(padj): 7.349617e-02<br />log10(baseMean): 1.0216581<br />gene: FBgn0034136","log2FoldChange: -0.7222173142<br />-log10(padj): 4.245076e-01<br />log10(baseMean): 1.3038387<br />gene: FBgn0034137","log2FoldChange: -0.0961307748<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 4.2266099<br />gene: FBgn0034138","log2FoldChange: 1.2268953615<br />-log10(padj): 2.312321e+00<br />log10(baseMean): 1.4608988<br />gene: FBgn0034139","log2FoldChange: 0.6756713922<br />-log10(padj): 2.932760e-01<br />log10(baseMean): 1.1627470<br />gene: FBgn0034140","log2FoldChange: 0.0689037866<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.8074796<br />gene: FBgn0034141","log2FoldChange: 0.2973013822<br />-log10(padj): 1.331442e+00<br />log10(baseMean): 2.8993996<br />gene: FBgn0034142","log2FoldChange: 0.3614545198<br />-log10(padj): 5.645306e-02<br />log10(baseMean): 0.8186123<br />gene: FBgn0034154","log2FoldChange: -0.2089276352<br />-log10(padj): 4.031715e-01<br />log10(baseMean): 2.9528195<br />gene: FBgn0034155","log2FoldChange: -0.0758159213<br />-log10(padj): 6.926412e-02<br />log10(baseMean): 3.6531790<br />gene: FBgn0034158","log2FoldChange: -1.1411816707<br />-log10(padj): 8.534688e-01<br />log10(baseMean): 1.1707952<br />gene: FBgn0034159","log2FoldChange: -1.0243350212<br />-log10(padj): 1.238927e+00<br />log10(baseMean): 1.6236356<br />gene: FBgn0034160","log2FoldChange: 0.3883174030<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 1.1160251<br />gene: FBgn0034162","log2FoldChange: -0.0585632476<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.6646667<br />gene: FBgn0034166","log2FoldChange: 0.0551723597<br />-log10(padj): 3.635924e-02<br />log10(baseMean): 2.3916515<br />gene: FBgn0034172","log2FoldChange: -0.5512873854<br />-log10(padj): 4.753765e-01<br />log10(baseMean): 1.7211398<br />gene: FBgn0034175","log2FoldChange: -0.1951767806<br />-log10(padj): 4.452796e-01<br />log10(baseMean): 3.2151070<br />gene: FBgn0034176","log2FoldChange: 0.0998982892<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 1.9974344<br />gene: FBgn0034177","log2FoldChange: 0.0614726015<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.9312244<br />gene: FBgn0034179","log2FoldChange: 0.0484112754<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 3.3142953<br />gene: FBgn0034180","log2FoldChange: 0.0839696813<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.1165699<br />gene: FBgn0034181","log2FoldChange: -0.0243296936<br />-log10(padj): 7.598195e-03<br />log10(baseMean): 1.9251253<br />gene: FBgn0034182","log2FoldChange: 0.1219981783<br />-log10(padj): 2.106022e-01<br />log10(baseMean): 2.9914147<br />gene: FBgn0034184","log2FoldChange: -0.0264553261<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.5300043<br />gene: FBgn0034186","log2FoldChange: 0.2671225790<br />-log10(padj): 1.512365e-01<br />log10(baseMean): 1.7736905<br />gene: FBgn0034187","log2FoldChange: -0.1570583515<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.1183892<br />gene: FBgn0034191","log2FoldChange: 0.0510415663<br />-log10(padj): 3.287411e-02<br />log10(baseMean): 2.6397003<br />gene: FBgn0034194","log2FoldChange: 0.2551000635<br />-log10(padj): 3.135429e-01<br />log10(baseMean): 3.2611241<br />gene: FBgn0034198","log2FoldChange: 0.3803262025<br />-log10(padj): 3.628149e-01<br />log10(baseMean): 2.0906449<br />gene: FBgn0034199","log2FoldChange: 0.5600773857<br />-log10(padj): 6.125478e-01<br />log10(baseMean): 2.0035364<br />gene: FBgn0034200","log2FoldChange: -0.0141757255<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.3923919<br />gene: FBgn0034210","log2FoldChange: 0.1040581485<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.2140463<br />gene: FBgn0034214","log2FoldChange: -0.2280207445<br />-log10(padj): 3.642698e-01<br />log10(baseMean): 2.6072235<br />gene: FBgn0034215","log2FoldChange: -0.2825919646<br />-log10(padj): 1.715808e-01<br />log10(baseMean): 1.9444882<br />gene: FBgn0034217","log2FoldChange: -0.5832628452<br />-log10(padj): 1.600311e+00<br />log10(baseMean): 2.8943884<br />gene: FBgn0034219","log2FoldChange: -0.9909409122<br />-log10(padj): 2.069716e+00<br />log10(baseMean): 2.1488109<br />gene: FBgn0034221","log2FoldChange: 0.0175126953<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.8992458<br />gene: FBgn0034223","log2FoldChange: -1.3014452801<br />-log10(padj): 3.237881e+00<br />log10(baseMean): 1.6910885<br />gene: FBgn0034225","log2FoldChange: 0.1167935478<br />-log10(padj): 1.700659e-01<br />log10(baseMean): 3.1174924<br />gene: FBgn0034230","log2FoldChange: 0.2400810530<br />-log10(padj): 1.602229e-01<br />log10(baseMean): 1.9146592<br />gene: FBgn0034231","log2FoldChange: -0.0157686689<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 2.1997407<br />gene: FBgn0034232","log2FoldChange: -0.1454471039<br />-log10(padj): 4.976196e-01<br />log10(baseMean): 3.9336298<br />gene: FBgn0034237","log2FoldChange: -0.3593953207<br />-log10(padj): 2.368080e-01<br />log10(baseMean): 1.7598178<br />gene: FBgn0034238","log2FoldChange: -0.2958678147<br />-log10(padj): 1.917584e-01<br />log10(baseMean): 1.7797784<br />gene: FBgn0034239","log2FoldChange: -0.2035700291<br />-log10(padj): 2.869994e-01<br />log10(baseMean): 3.3211012<br />gene: FBgn0034240","log2FoldChange: -0.0155894372<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.2298563<br />gene: FBgn0034242","log2FoldChange: -0.0299370240<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.0534909<br />gene: FBgn0034243","log2FoldChange: 0.1692582974<br />-log10(padj): 3.337508e-02<br />log10(baseMean): 2.6163567<br />gene: FBgn0034245","log2FoldChange: -0.0229937288<br />-log10(padj): 1.079808e-02<br />log10(baseMean): 3.4609028<br />gene: FBgn0034246","log2FoldChange: -0.0908872222<br />-log10(padj): 5.290275e-02<br />log10(baseMean): 2.5009579<br />gene: FBgn0034248","log2FoldChange: 0.5084930401<br />-log10(padj): 2.520179e+00<br />log10(baseMean): 2.7381109<br />gene: FBgn0034249","log2FoldChange: -0.1345554220<br />-log10(padj): 1.388218e-01<br />log10(baseMean): 2.5309749<br />gene: FBgn0034253","log2FoldChange: 0.1741208167<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 2.6990601<br />gene: FBgn0034255","log2FoldChange: -0.0902088509<br />-log10(padj): 9.517988e-02<br />log10(baseMean): 4.0393501<br />gene: FBgn0034258","log2FoldChange: 0.3035309517<br />-log10(padj): 6.554094e-01<br />log10(baseMean): 2.9711758<br />gene: FBgn0034259","log2FoldChange: 0.2150007522<br />-log10(padj): 3.969022e-01<br />log10(baseMean): 2.4679058<br />gene: FBgn0034261","log2FoldChange: 0.3252492164<br />-log10(padj): 6.913758e-02<br />log10(baseMean): 1.0893435<br />gene: FBgn0034264","log2FoldChange: -0.0156276604<br />-log10(padj): 7.188587e-03<br />log10(baseMean): 2.4383432<br />gene: FBgn0034265","log2FoldChange: -0.3307514321<br />-log10(padj): 6.639638e-02<br />log10(baseMean): 1.1028565<br />gene: FBgn0034267","log2FoldChange: -0.1726556089<br />-log10(padj): 4.101715e-01<br />log10(baseMean): 2.9375238<br />gene: FBgn0034269","log2FoldChange: 0.3133452612<br />-log10(padj): 3.309824e-01<br />log10(baseMean): 2.1838728<br />gene: FBgn0034270","log2FoldChange: 0.0836164402<br />-log10(padj): 7.695746e-02<br />log10(baseMean): 2.6868216<br />gene: FBgn0034271","log2FoldChange: 0.1083487598<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 3.7427481<br />gene: FBgn0034277","log2FoldChange: 0.2544212845<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 1.1681862<br />gene: FBgn0034279","log2FoldChange: -0.2207869601<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.0490183<br />gene: FBgn0034281","log2FoldChange: 0.1534633404<br />-log10(padj): 1.236393e-01<br />log10(baseMean): 3.3763878<br />gene: FBgn0034282","log2FoldChange: -1.5029348685<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 1.1315167<br />gene: FBgn0034290","log2FoldChange: 0.0440792210<br />-log10(padj): 1.763922e-02<br />log10(baseMean): 2.3540362<br />gene: FBgn0034299","log2FoldChange: -0.0399168377<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.8865470<br />gene: FBgn0034300","log2FoldChange: 0.0535634924<br />-log10(padj): 4.355667e-02<br />log10(baseMean): 2.6712062<br />gene: FBgn0034304","log2FoldChange: -0.0099648917<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 2.3432747<br />gene: FBgn0034307","log2FoldChange: -0.1509772782<br />-log10(padj): 1.409127e-01<br />log10(baseMean): 2.8878341<br />gene: FBgn0034308","log2FoldChange: -0.0182744754<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.9117264<br />gene: FBgn0034310","log2FoldChange: 0.2514836947<br />-log10(padj): 6.092514e-01<br />log10(baseMean): 2.6620112<br />gene: FBgn0034312","log2FoldChange: 0.1455574485<br />-log10(padj): 2.167041e-01<br />log10(baseMean): 2.9683931<br />gene: FBgn0034313","log2FoldChange: -0.3651919261<br />-log10(padj): 3.665136e-01<br />log10(baseMean): 1.9548290<br />gene: FBgn0034314","log2FoldChange: 0.0322142528<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 3.0726152<br />gene: FBgn0034315","log2FoldChange: 0.8409444250<br />-log10(padj): 4.557988e-01<br />log10(baseMean): 1.4076729<br />gene: FBgn0034321","log2FoldChange: -0.0957132651<br />-log10(padj): 1.032380e-02<br />log10(baseMean): 1.2846005<br />gene: FBgn0034327","log2FoldChange: -0.0347639004<br />-log10(padj): 2.214450e-02<br />log10(baseMean): 3.1705890<br />gene: FBgn0034345","log2FoldChange: -0.1008945375<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.0668810<br />gene: FBgn0034346","log2FoldChange: 0.1736425158<br />-log10(padj): 1.778309e-01<br />log10(baseMean): 2.3840704<br />gene: FBgn0034351","log2FoldChange: 0.9547633027<br />-log10(padj): 4.858938e+00<br />log10(baseMean): 2.5712604<br />gene: FBgn0034354","log2FoldChange: 0.3647937467<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 1.0294692<br />gene: FBgn0034356","log2FoldChange: -0.1382047394<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.9307157<br />gene: FBgn0034360","log2FoldChange: 0.2490329799<br />-log10(padj): 1.873412e-01<br />log10(baseMean): 2.3408069<br />gene: FBgn0034361","log2FoldChange: 0.2145911560<br />-log10(padj): 3.618100e-01<br />log10(baseMean): 2.6802793<br />gene: FBgn0034362","log2FoldChange: -0.4751022459<br />-log10(padj): 2.171234e+00<br />log10(baseMean): 2.5724059<br />gene: FBgn0034365","log2FoldChange: 0.3074522030<br />-log10(padj): 4.386646e-01<br />log10(baseMean): 2.3774309<br />gene: FBgn0034366","log2FoldChange: -0.0324115349<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.8485697<br />gene: FBgn0034367","log2FoldChange: -0.0614072354<br />-log10(padj): 5.788090e-02<br />log10(baseMean): 3.1223066<br />gene: FBgn0034368","log2FoldChange: -0.1660986568<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 2.8723615<br />gene: FBgn0034371","log2FoldChange: -0.4969309350<br />-log10(padj): 1.765028e+00<br />log10(baseMean): 2.8420656<br />gene: FBgn0034376","log2FoldChange: -0.4838977497<br />-log10(padj): 2.147764e+00<br />log10(baseMean): 2.8728637<br />gene: FBgn0034378","log2FoldChange: 0.1751860552<br />-log10(padj): 1.467275e-01<br />log10(baseMean): 2.1691686<br />gene: FBgn0034379","log2FoldChange: 0.1809440924<br />-log10(padj): 1.882016e-01<br />log10(baseMean): 2.3458379<br />gene: FBgn0034380","log2FoldChange: -1.9215780275<br />-log10(padj): 1.113664e+01<br />log10(baseMean): 2.0443337<br />gene: FBgn0034389","log2FoldChange: 0.4364442646<br />-log10(padj): 1.287709e+00<br />log10(baseMean): 2.8956123<br />gene: FBgn0034390","log2FoldChange: 1.5609374747<br />-log10(padj): 7.174653e+00<br />log10(baseMean): 1.8137639<br />gene: FBgn0034391","log2FoldChange: -0.1674491461<br />-log10(padj): 2.959017e-01<br />log10(baseMean): 3.0732763<br />gene: FBgn0034392","log2FoldChange: -0.1471679798<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.4673491<br />gene: FBgn0034396","log2FoldChange: -0.5221157003<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 1.8064107<br />gene: FBgn0034397","log2FoldChange: -0.4051933739<br />-log10(padj): 2.793397e+00<br />log10(baseMean): 3.6845070<br />gene: FBgn0034398","log2FoldChange: 0.3056016034<br />-log10(padj): 4.211833e-01<br />log10(baseMean): 2.8510261<br />gene: FBgn0034399","log2FoldChange: -0.5419382748<br />-log10(padj): 5.387097e+00<br />log10(baseMean): 3.7737510<br />gene: FBgn0034400","log2FoldChange: -0.0132133529<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.4154895<br />gene: FBgn0034401","log2FoldChange: -0.2276172937<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.0039913<br />gene: FBgn0034402","log2FoldChange: 1.0502849074<br />-log10(padj): 2.638012e+00<br />log10(baseMean): 1.8327344<br />gene: FBgn0034403","log2FoldChange: 0.6565583242<br />-log10(padj): 1.031943e+01<br />log10(baseMean): 3.3599308<br />gene: FBgn0034405","log2FoldChange: 0.0347952861<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 1.4144510<br />gene: FBgn0034407","log2FoldChange: 0.5673864654<br />-log10(padj): 1.380347e+00<br />log10(baseMean): 2.2783118<br />gene: FBgn0034408","log2FoldChange: 0.1763997683<br />-log10(padj): 3.067249e-01<br />log10(baseMean): 2.7404484<br />gene: FBgn0034410","log2FoldChange: 0.1186521280<br />-log10(padj): 7.778007e-02<br />log10(baseMean): 2.2498482<br />gene: FBgn0034411","log2FoldChange: -0.4779097063<br />-log10(padj): 1.165532e+00<br />log10(baseMean): 2.2753212<br />gene: FBgn0034417","log2FoldChange: 0.3135919925<br />-log10(padj): 1.427539e+00<br />log10(baseMean): 3.0054249<br />gene: FBgn0034418","log2FoldChange: -0.1301087596<br />-log10(padj): 1.784549e-01<br />log10(baseMean): 2.6916907<br />gene: FBgn0034419","log2FoldChange: -0.1690374556<br />-log10(padj): 1.770380e-01<br />log10(baseMean): 3.5892597<br />gene: FBgn0034420","log2FoldChange: 0.1773577795<br />-log10(padj): 2.638586e-01<br />log10(baseMean): 3.3376430<br />gene: FBgn0034421","log2FoldChange: -0.1596994150<br />-log10(padj): 1.072175e-01<br />log10(baseMean): 2.2771801<br />gene: FBgn0034422","log2FoldChange: 0.0048363329<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.5579889<br />gene: FBgn0034425","log2FoldChange: 0.2343986894<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 2.6291843<br />gene: FBgn0034430","log2FoldChange: -0.2645838108<br />-log10(padj): 1.388937e+00<br />log10(baseMean): 3.1691130<br />gene: FBgn0034432","log2FoldChange: -0.0188959033<br />-log10(padj): 9.356238e-03<br />log10(baseMean): 2.6031142<br />gene: FBgn0034433","log2FoldChange: -3.6424792361<br />-log10(padj): 3.552143e+01<br />log10(baseMean): 2.0592731<br />gene: FBgn0034434","log2FoldChange: 0.1746498828<br />-log10(padj): 4.360557e-01<br />log10(baseMean): 3.2386882<br />gene: FBgn0034436","log2FoldChange: 1.9006257530<br />-log10(padj): 1.169940e+01<br />log10(baseMean): 1.8727126<br />gene: FBgn0034438","log2FoldChange: -0.0967273522<br />-log10(padj): 3.091232e-02<br />log10(baseMean): 1.7601925<br />gene: FBgn0034442","log2FoldChange: 0.0228631214<br />-log10(padj): 5.265523e-03<br />log10(baseMean): 2.3683853<br />gene: FBgn0034443","log2FoldChange: -0.3300458793<br />-log10(padj): 1.414546e+00<br />log10(baseMean): 3.0282200<br />gene: FBgn0034447","log2FoldChange: 0.1827357004<br />-log10(padj): 2.974095e-01<br />log10(baseMean): 2.8272761<br />gene: FBgn0034451","log2FoldChange: 0.2848106006<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 1.6800788<br />gene: FBgn0034452","log2FoldChange: -0.1950145349<br />-log10(padj): 4.334437e-01<br />log10(baseMean): 2.6770024<br />gene: FBgn0034455","log2FoldChange: -0.9385016891<br />-log10(padj): 3.342224e+00<br />log10(baseMean): 2.2074296<br />gene: FBgn0034476","log2FoldChange: 0.6822460140<br />-log10(padj): 2.303311e-01<br />log10(baseMean): 1.0434915<br />gene: FBgn0034479","log2FoldChange: -0.0134250235<br />-log10(padj): 5.418477e-03<br />log10(baseMean): 3.0667976<br />gene: FBgn0034488","log2FoldChange: 0.9330310312<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 1.1122588<br />gene: FBgn0034490","log2FoldChange: -0.2573373366<br />-log10(padj): 9.951660e-01<br />log10(baseMean): 3.0852958<br />gene: FBgn0034491","log2FoldChange: -0.2619406421<br />-log10(padj): 3.713583e-01<br />log10(baseMean): 2.5353358<br />gene: FBgn0034493","log2FoldChange: -0.3156672733<br />-log10(padj): 6.212189e-01<br />log10(baseMean): 3.4841356<br />gene: FBgn0034494","log2FoldChange: 0.0368923138<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.3081407<br />gene: FBgn0034495","log2FoldChange: 0.0421698702<br />-log10(padj): 3.537453e-02<br />log10(baseMean): 2.7081262<br />gene: FBgn0034496","log2FoldChange: -0.6858308541<br />-log10(padj): 2.685650e-01<br />log10(baseMean): 1.3969407<br />gene: FBgn0034497","log2FoldChange: 0.0075764656<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 3.0481377<br />gene: FBgn0034498","log2FoldChange: -0.1156538031<br />-log10(padj): 5.539985e-02<br />log10(baseMean): 2.3235904<br />gene: FBgn0034500","log2FoldChange: -0.4817918107<br />-log10(padj): 2.083457e+00<br />log10(baseMean): 3.4302876<br />gene: FBgn0034501","log2FoldChange: -0.0091716503<br />-log10(padj): 4.190849e-03<br />log10(baseMean): 2.4673634<br />gene: FBgn0034503","log2FoldChange: -0.0188678099<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.1613254<br />gene: FBgn0034504","log2FoldChange: 0.1116808953<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.7452726<br />gene: FBgn0034521","log2FoldChange: 0.4688481405<br />-log10(padj): 1.607725e+00<br />log10(baseMean): 2.4551198<br />gene: FBgn0034523","log2FoldChange: 0.0929619298<br />-log10(padj): 6.895106e-02<br />log10(baseMean): 2.6396837<br />gene: FBgn0034527","log2FoldChange: -0.1489143606<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.6698860<br />gene: FBgn0034528","log2FoldChange: 0.1712718434<br />-log10(padj): 1.117342e-01<br />log10(baseMean): 2.5117887<br />gene: FBgn0034529","log2FoldChange: -0.5117956160<br />-log10(padj): 9.543967e-01<br />log10(baseMean): 1.9703004<br />gene: FBgn0034530","log2FoldChange: -0.5798031951<br />-log10(padj): 4.015880e-01<br />log10(baseMean): 1.4254792<br />gene: FBgn0034532","log2FoldChange: -0.0940169509<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.7935813<br />gene: FBgn0034534","log2FoldChange: 0.0766497572<br />-log10(padj): 3.169064e-02<br />log10(baseMean): 2.1102903<br />gene: FBgn0034535","log2FoldChange: 0.1511129890<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 2.7674133<br />gene: FBgn0034537","log2FoldChange: 0.0062230929<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.7593982<br />gene: FBgn0034542","log2FoldChange: -0.0627713689<br />-log10(padj): 5.290275e-02<br />log10(baseMean): 2.7007915<br />gene: FBgn0034543","log2FoldChange: 0.4160831269<br />-log10(padj): 1.671556e+00<br />log10(baseMean): 2.5143972<br />gene: FBgn0034564","log2FoldChange: 0.4983880130<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 1.0612796<br />gene: FBgn0034568","log2FoldChange: 0.2652631448<br />-log10(padj): 7.217343e-01<br />log10(baseMean): 2.6185015<br />gene: FBgn0034569","log2FoldChange: -0.2167423327<br />-log10(padj): 2.368080e-01<br />log10(baseMean): 3.5061673<br />gene: FBgn0034570","log2FoldChange: -0.0486083024<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.7549960<br />gene: FBgn0034572","log2FoldChange: -0.1512018903<br />-log10(padj): 1.483407e-01<br />log10(baseMean): 3.0376102<br />gene: FBgn0034573","log2FoldChange: 0.1882567392<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 2.9614440<br />gene: FBgn0034576","log2FoldChange: -0.1864951457<br />-log10(padj): 5.141810e-01<br />log10(baseMean): 3.3697354<br />gene: FBgn0034577","log2FoldChange: 0.1922191405<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.6563277<br />gene: FBgn0034578","log2FoldChange: 0.4080576337<br />-log10(padj): 6.393582e-01<br />log10(baseMean): 2.0925464<br />gene: FBgn0034579","log2FoldChange: -0.4102769308<br />-log10(padj): 1.274073e+00<br />log10(baseMean): 3.2378503<br />gene: FBgn0034583","log2FoldChange: -0.0885130295<br />-log10(padj): 7.382405e-02<br />log10(baseMean): 2.8714899<br />gene: FBgn0034585","log2FoldChange: -0.1522026154<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.2128219<br />gene: FBgn0034590","log2FoldChange: 0.1906243447<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 1.0134955<br />gene: FBgn0034595","log2FoldChange: -0.0154392945<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.3565806<br />gene: FBgn0034598","log2FoldChange: -0.0826304371<br />-log10(padj): 5.636755e-02<br />log10(baseMean): 2.3985769<br />gene: FBgn0034599","log2FoldChange: -0.6814626548<br />-log10(padj): 1.980409e-01<br />log10(baseMean): 1.0144374<br />gene: FBgn0034602","log2FoldChange: 0.4424582978<br />-log10(padj): 1.548827e+00<br />log10(baseMean): 2.7643518<br />gene: FBgn0034603","log2FoldChange: -0.8735692623<br />-log10(padj): 2.696819e-01<br />log10(baseMean): 0.9630237<br />gene: FBgn0034605","log2FoldChange: 0.1255978182<br />-log10(padj): 1.117342e-01<br />log10(baseMean): 3.0923534<br />gene: FBgn0034611","log2FoldChange: 0.2282704286<br />-log10(padj): 4.685527e-02<br />log10(baseMean): 1.1367558<br />gene: FBgn0034612","log2FoldChange: -0.1486345791<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 2.5182042<br />gene: FBgn0034614","log2FoldChange: 0.3872840879<br />-log10(padj): 7.931230e-01<br />log10(baseMean): 2.3464227<br />gene: FBgn0034617","log2FoldChange: -0.0577654636<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.3174797<br />gene: FBgn0034618","log2FoldChange: -0.2844136817<br />-log10(padj): 7.042247e-01<br />log10(baseMean): 2.6190360<br />gene: FBgn0034626","log2FoldChange: 0.0495753357<br />-log10(padj): 1.889413e-02<br />log10(baseMean): 1.9867624<br />gene: FBgn0034627","log2FoldChange: -0.0103681364<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.7792668<br />gene: FBgn0034628","log2FoldChange: -0.0191843177<br />-log10(padj): 5.083832e-03<br />log10(baseMean): 1.8137256<br />gene: FBgn0034629","log2FoldChange: -0.1456050470<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.5050863<br />gene: FBgn0034631","log2FoldChange: 0.0722244704<br />-log10(padj): 2.499648e-02<br />log10(baseMean): 1.9277296<br />gene: FBgn0034634","log2FoldChange: -2.3604901815<br />-log10(padj): 2.516329e+00<br />log10(baseMean): 1.1808687<br />gene: FBgn0034636","log2FoldChange: -0.9119902815<br />-log10(padj): 1.171273e+01<br />log10(baseMean): 3.2211507<br />gene: FBgn0034638","log2FoldChange: 0.3360052624<br />-log10(padj): 8.973024e-02<br />log10(baseMean): 1.2519069<br />gene: FBgn0034639","log2FoldChange: -0.1021652378<br />-log10(padj): 1.564430e-01<br />log10(baseMean): 3.2903059<br />gene: FBgn0034641","log2FoldChange: -0.2678025174<br />-log10(padj): 8.409143e-01<br />log10(baseMean): 3.1227419<br />gene: FBgn0034643","log2FoldChange: -0.3086348199<br />-log10(padj): 8.563559e-01<br />log10(baseMean): 3.1813852<br />gene: FBgn0034644","log2FoldChange: 0.0963944195<br />-log10(padj): 6.229977e-02<br />log10(baseMean): 2.8925073<br />gene: FBgn0034645","log2FoldChange: -0.0128163676<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.8877928<br />gene: FBgn0034646","log2FoldChange: 0.9055509638<br />-log10(padj): 4.970633e-01<br />log10(baseMean): 1.1247555<br />gene: FBgn0034647","log2FoldChange: -0.0024620693<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 2.3954430<br />gene: FBgn0034650","log2FoldChange: -0.1225185197<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.4704193<br />gene: FBgn0034654","log2FoldChange: 0.0130709259<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.4772344<br />gene: FBgn0034655","log2FoldChange: -0.2078795545<br />-log10(padj): 7.555807e-02<br />log10(baseMean): 1.5541215<br />gene: FBgn0034656","log2FoldChange: 0.1673091200<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 2.9635724<br />gene: FBgn0034657","log2FoldChange: 0.7727371421<br />-log10(padj): 1.986652e-01<br />log10(baseMean): 0.9158904<br />gene: FBgn0034662","log2FoldChange: -0.1447802940<br />-log10(padj): 2.869327e-01<br />log10(baseMean): 2.9966603<br />gene: FBgn0034674","log2FoldChange: -0.0426840176<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 1.6536973<br />gene: FBgn0034684","log2FoldChange: 0.0171022474<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.0292686<br />gene: FBgn0034687","log2FoldChange: -0.3513342247<br />-log10(padj): 9.518441e-01<br />log10(baseMean): 2.4803072<br />gene: FBgn0034688","log2FoldChange: 0.0219427106<br />-log10(padj): 1.398029e-02<br />log10(baseMean): 2.5422150<br />gene: FBgn0034689","log2FoldChange: -0.1198166292<br />-log10(padj): 1.451405e-01<br />log10(baseMean): 3.3397425<br />gene: FBgn0034691","log2FoldChange: -0.6053697546<br />-log10(padj): 3.678384e+00<br />log10(baseMean): 2.6165653<br />gene: FBgn0034694","log2FoldChange: -0.1284740394<br />-log10(padj): 1.865095e-01<br />log10(baseMean): 3.0523799<br />gene: FBgn0034697","log2FoldChange: -0.1625251196<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.2655981<br />gene: FBgn0034703","log2FoldChange: 0.0966942778<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.4916945<br />gene: FBgn0034704","log2FoldChange: 0.0706900206<br />-log10(padj): 3.337508e-02<br />log10(baseMean): 2.2209925<br />gene: FBgn0034705","log2FoldChange: -0.1636412905<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 1.3717064<br />gene: FBgn0034706","log2FoldChange: 0.1413392481<br />-log10(padj): 1.563133e-01<br />log10(baseMean): 2.6164170<br />gene: FBgn0034707","log2FoldChange: 0.0735070989<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 3.2928821<br />gene: FBgn0034708","log2FoldChange: -0.2418108167<br />-log10(padj): 2.147990e-01<br />log10(baseMean): 3.0262722<br />gene: FBgn0034709","log2FoldChange: 1.3906957994<br />-log10(padj): 6.425902e+00<br />log10(baseMean): 2.6955201<br />gene: FBgn0034718","log2FoldChange: -0.1023742498<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.9565460<br />gene: FBgn0034722","log2FoldChange: 0.0220942649<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 1.1626932<br />gene: FBgn0034724","log2FoldChange: -0.3509085525<br />-log10(padj): 2.388750e-01<br />log10(baseMean): 2.2674167<br />gene: FBgn0034726","log2FoldChange: -0.0238882974<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 2.6827036<br />gene: FBgn0034727","log2FoldChange: -0.0960522560<br />-log10(padj): 7.166896e-02<br />log10(baseMean): 3.2714329<br />gene: FBgn0034728","log2FoldChange: 0.1014397759<br />-log10(padj): 3.802180e-02<br />log10(baseMean): 1.8096366<br />gene: FBgn0034729","log2FoldChange: 0.5356206314<br />-log10(padj): 4.196183e-01<br />log10(baseMean): 1.6106606<br />gene: FBgn0034733","log2FoldChange: -0.2368437874<br />-log10(padj): 4.079775e-01<br />log10(baseMean): 3.0746919<br />gene: FBgn0034734","log2FoldChange: -0.1731900849<br />-log10(padj): 3.135429e-01<br />log10(baseMean): 2.7980463<br />gene: FBgn0034735","log2FoldChange: -3.5113188756<br />-log10(padj): 5.628158e+01<br />log10(baseMean): 2.3538598<br />gene: FBgn0034736","log2FoldChange: 0.1889189250<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.5350950<br />gene: FBgn0034741","log2FoldChange: -0.0727886261<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.8829698<br />gene: FBgn0034742","log2FoldChange: -0.0536576705<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 4.2320300<br />gene: FBgn0034743","log2FoldChange: -0.1936931709<br />-log10(padj): 2.877323e-01<br />log10(baseMean): 2.9693757<br />gene: FBgn0034744","log2FoldChange: -0.3660666416<br />-log10(padj): 2.579744e-01<br />log10(baseMean): 1.6662665<br />gene: FBgn0034745","log2FoldChange: 0.0144267946<br />-log10(padj): 7.543257e-03<br />log10(baseMean): 2.5292270<br />gene: FBgn0034748","log2FoldChange: 0.2050158228<br />-log10(padj): 3.292870e-01<br />log10(baseMean): 2.8446356<br />gene: FBgn0034750","log2FoldChange: -1.0613823341<br />-log10(padj): 1.588573e+01<br />log10(baseMean): 4.2211754<br />gene: FBgn0034751","log2FoldChange: 0.3935147518<br />-log10(padj): 1.000222e+00<br />log10(baseMean): 3.8907155<br />gene: FBgn0034753","log2FoldChange: 0.2360512744<br />-log10(padj): 1.150920e-01<br />log10(baseMean): 1.7087645<br />gene: FBgn0034756","log2FoldChange: -0.3597912373<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 1.6425300<br />gene: FBgn0034758","log2FoldChange: -0.5919343998<br />-log10(padj): 1.269622e-01<br />log10(baseMean): 0.9073308<br />gene: FBgn0034759","log2FoldChange: 0.0182219128<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.0964708<br />gene: FBgn0034761","log2FoldChange: -0.1616051887<br />-log10(padj): 2.442868e-01<br />log10(baseMean): 2.6569119<br />gene: FBgn0034763","log2FoldChange: -0.2551672972<br />-log10(padj): 5.943739e-01<br />log10(baseMean): 2.7829716<br />gene: FBgn0034786","log2FoldChange: 0.7195461375<br />-log10(padj): 1.100856e-01<br />log10(baseMean): 0.9749201<br />gene: FBgn0034789","log2FoldChange: 0.0212769975<br />-log10(padj): 9.732012e-03<br />log10(baseMean): 2.6447503<br />gene: FBgn0034791","log2FoldChange: 0.0957522850<br />-log10(padj): 9.620880e-02<br />log10(baseMean): 3.2855581<br />gene: FBgn0034792","log2FoldChange: 0.6018819816<br />-log10(padj): 3.052884e+00<br />log10(baseMean): 2.7181627<br />gene: FBgn0034793","log2FoldChange: 0.1486385121<br />-log10(padj): 8.747108e-02<br />log10(baseMean): 2.1332040<br />gene: FBgn0034794","log2FoldChange: 0.4217903054<br />-log10(padj): 3.710694e-01<br />log10(baseMean): 1.9955421<br />gene: FBgn0034797","log2FoldChange: -0.2310708951<br />-log10(padj): 1.269622e-01<br />log10(baseMean): 1.8235439<br />gene: FBgn0034800","log2FoldChange: 0.1505965344<br />-log10(padj): 1.451671e-01<br />log10(baseMean): 3.8419169<br />gene: FBgn0034802","log2FoldChange: -0.2319735248<br />-log10(padj): 3.935528e-01<br />log10(baseMean): 2.4059812<br />gene: FBgn0034803","log2FoldChange: -1.5005251125<br />-log10(padj): 3.172190e+00<br />log10(baseMean): 1.5913090<br />gene: FBgn0034804","log2FoldChange: -0.0485603885<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 2.4244436<br />gene: FBgn0034814","log2FoldChange: 0.5242164652<br />-log10(padj): 4.505560e-01<br />log10(baseMean): 1.6707453<br />gene: FBgn0034816","log2FoldChange: -0.0504423534<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.7196321<br />gene: FBgn0034817","log2FoldChange: 0.2847053655<br />-log10(padj): 2.140120e-01<br />log10(baseMean): 2.5353378<br />gene: FBgn0034849","log2FoldChange: -0.1968881951<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 0.8112178<br />gene: FBgn0034850","log2FoldChange: -0.2071681687<br />-log10(padj): 5.648423e-01<br />log10(baseMean): 2.8414993<br />gene: FBgn0034853","log2FoldChange: -0.0268972539<br />-log10(padj): 2.155213e-02<br />log10(baseMean): 3.3680217<br />gene: FBgn0034854","log2FoldChange: -0.0763230896<br />-log10(padj): 2.214450e-02<br />log10(baseMean): 1.7517248<br />gene: FBgn0034856","log2FoldChange: 0.1629788969<br />-log10(padj): 1.588341e-01<br />log10(baseMean): 3.1634318<br />gene: FBgn0034858","log2FoldChange: 0.2373917865<br />-log10(padj): 3.628149e-01<br />log10(baseMean): 2.7640884<br />gene: FBgn0034859","log2FoldChange: -0.1307331873<br />-log10(padj): 1.926605e-01<br />log10(baseMean): 2.7718273<br />gene: FBgn0034873","log2FoldChange: 0.0268460794<br />-log10(padj): 1.548214e-02<br />log10(baseMean): 2.8276615<br />gene: FBgn0034876","log2FoldChange: 0.2276837406<br />-log10(padj): 7.894877e-01<br />log10(baseMean): 3.1495566<br />gene: FBgn0034877","log2FoldChange: 0.0183883403<br />-log10(padj): 9.830631e-03<br />log10(baseMean): 3.1317074<br />gene: FBgn0034878","log2FoldChange: 0.1100202701<br />-log10(padj): 9.252481e-02<br />log10(baseMean): 2.7332692<br />gene: FBgn0034879","log2FoldChange: 0.6102023516<br />-log10(padj): 3.818032e-01<br />log10(baseMean): 1.3516601<br />gene: FBgn0034880","log2FoldChange: 1.6334763593<br />-log10(padj): 1.033066e+01<br />log10(baseMean): 2.2055435<br />gene: FBgn0034885","log2FoldChange: -0.3699978136<br />-log10(padj): 1.482539e+00<br />log10(baseMean): 3.7830069<br />gene: FBgn0034886","log2FoldChange: 0.1251074835<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 1.6111923<br />gene: FBgn0034887","log2FoldChange: -0.0910370562<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 2.2981728<br />gene: FBgn0034893","log2FoldChange: -0.5240798911<br />-log10(padj): 2.623011e+00<br />log10(baseMean): 2.7458548<br />gene: FBgn0034894","log2FoldChange: -1.9213947372<br />-log10(padj): 3.019798e+01<br />log10(baseMean): 2.9506010<br />gene: FBgn0034897","log2FoldChange: -2.6012271385<br />-log10(padj): 1.402750e+00<br />log10(baseMean): 0.8248976<br />gene: FBgn0034898","log2FoldChange: 0.1726126993<br />-log10(padj): 1.616856e-01<br />log10(baseMean): 2.5628514<br />gene: FBgn0034902","log2FoldChange: 0.5750299208<br />-log10(padj): 1.571543e-01<br />log10(baseMean): 1.1250799<br />gene: FBgn0034906","log2FoldChange: -0.0951957777<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 2.6636192<br />gene: FBgn0034908","log2FoldChange: 0.0137315576<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.2629209<br />gene: FBgn0034909","log2FoldChange: 1.7127618745<br />-log10(padj): 8.561056e-01<br />log10(baseMean): 0.8393736<br />gene: FBgn0034911","log2FoldChange: 0.0346515919<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 2.7762067<br />gene: FBgn0034913","log2FoldChange: 0.2998576253<br />-log10(padj): 1.210635e+00<br />log10(baseMean): 3.4914450<br />gene: FBgn0034914","log2FoldChange: 0.0564970832<br />-log10(padj): 3.646846e-02<br />log10(baseMean): 3.0221071<br />gene: FBgn0034915","log2FoldChange: 0.0020018934<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 2.3319336<br />gene: FBgn0034918","log2FoldChange: -0.1499873037<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 2.2010790<br />gene: FBgn0034919","log2FoldChange: -0.1153118890<br />-log10(padj): 9.077430e-02<br />log10(baseMean): 2.5988937<br />gene: FBgn0034921","log2FoldChange: -0.1929360533<br />-log10(padj): 3.492311e-01<br />log10(baseMean): 2.8586236<br />gene: FBgn0034922","log2FoldChange: 0.1310997945<br />-log10(padj): 1.297141e-01<br />log10(baseMean): 2.6117330<br />gene: FBgn0034923","log2FoldChange: 0.0478566126<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 1.1347811<br />gene: FBgn0034924","log2FoldChange: -0.0336478831<br />-log10(padj): 1.685188e-02<br />log10(baseMean): 2.5189820<br />gene: FBgn0034925","log2FoldChange: 0.1149719217<br />-log10(padj): 1.326801e-01<br />log10(baseMean): 2.8291668<br />gene: FBgn0034926","log2FoldChange: -0.0445548181<br />-log10(padj): 5.235416e-03<br />log10(baseMean): 1.0578332<br />gene: FBgn0034928","log2FoldChange: -0.0932276053<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 0.9898688<br />gene: FBgn0034931","log2FoldChange: -0.3407781748<br />-log10(padj): 5.092879e-01<br />log10(baseMean): 2.4271429<br />gene: FBgn0034933","log2FoldChange: 0.1198885564<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.8389396<br />gene: FBgn0034936","log2FoldChange: 0.2312680242<br />-log10(padj): 6.765428e-02<br />log10(baseMean): 1.4045555<br />gene: FBgn0034937","log2FoldChange: 0.3261009572<br />-log10(padj): 6.440008e-01<br />log10(baseMean): 2.3545380<br />gene: FBgn0034938","log2FoldChange: -0.0196079786<br />-log10(padj): 1.186237e-02<br />log10(baseMean): 2.7253660<br />gene: FBgn0034939","log2FoldChange: 0.1647072531<br />-log10(padj): 1.766420e-01<br />log10(baseMean): 2.4040526<br />gene: FBgn0034940","log2FoldChange: 0.0694554572<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.1721308<br />gene: FBgn0034945","log2FoldChange: 0.1221934699<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 2.8406940<br />gene: FBgn0034946","log2FoldChange: 0.0513313263<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.0795635<br />gene: FBgn0034948","log2FoldChange: 0.8641112744<br />-log10(padj): 2.771046e+00<br />log10(baseMean): 1.9814427<br />gene: FBgn0034950","log2FoldChange: -0.2382719303<br />-log10(padj): 3.278556e-01<br />log10(baseMean): 2.4843316<br />gene: FBgn0034951","log2FoldChange: 0.2285302341<br />-log10(padj): 9.164097e-02<br />log10(baseMean): 1.6113849<br />gene: FBgn0034956","log2FoldChange: -0.0425413859<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.9489873<br />gene: FBgn0034958","log2FoldChange: 0.1258444691<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.5311909<br />gene: FBgn0034959","log2FoldChange: 0.0106056719<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.3423401<br />gene: FBgn0034961","log2FoldChange: 0.1001380199<br />-log10(padj): 1.100856e-01<br />log10(baseMean): 2.8476477<br />gene: FBgn0034962","log2FoldChange: 0.0663627645<br />-log10(padj): 2.482922e-02<br />log10(baseMean): 2.1824663<br />gene: FBgn0034963","log2FoldChange: -0.2104814557<br />-log10(padj): 6.121250e-01<br />log10(baseMean): 3.3191454<br />gene: FBgn0034964","log2FoldChange: 0.1286645443<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 1.8605353<br />gene: FBgn0034965","log2FoldChange: 0.0701599665<br />-log10(padj): 2.227551e-02<br />log10(baseMean): 1.7839330<br />gene: FBgn0034966","log2FoldChange: -0.0948701908<br />-log10(padj): 6.926412e-02<br />log10(baseMean): 4.0209540<br />gene: FBgn0034967","log2FoldChange: -0.1214510610<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 4.3698322<br />gene: FBgn0034968","log2FoldChange: -0.3505803517<br />-log10(padj): 1.947530e+00<br />log10(baseMean): 3.0980856<br />gene: FBgn0034970","log2FoldChange: -0.0064990869<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.2774209<br />gene: FBgn0034971","log2FoldChange: -0.1275306177<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 3.3140795<br />gene: FBgn0034975","log2FoldChange: -0.0831900209<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.6774886<br />gene: FBgn0034976","log2FoldChange: 0.0624825024<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.8569955<br />gene: FBgn0034982","log2FoldChange: -0.2634608217<br />-log10(padj): 2.823653e-01<br />log10(baseMean): 2.5090846<br />gene: FBgn0034985","log2FoldChange: 0.0612788491<br />-log10(padj): 4.675296e-02<br />log10(baseMean): 2.5032117<br />gene: FBgn0034986","log2FoldChange: -0.0701615291<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 3.1274060<br />gene: FBgn0034987","log2FoldChange: 0.0687235175<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.3623991<br />gene: FBgn0034988","log2FoldChange: -0.0714624288<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 3.1772996<br />gene: FBgn0034989","log2FoldChange: 0.2483824115<br />-log10(padj): 9.122208e-01<br />log10(baseMean): 3.7091738<br />gene: FBgn0034997","log2FoldChange: 0.4466491608<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 0.9584000<br />gene: FBgn0034999","log2FoldChange: -0.4264340873<br />-log10(padj): 3.566237e+00<br />log10(baseMean): 3.9150039<br />gene: FBgn0035001","log2FoldChange: -0.0258575712<br />-log10(padj): 1.500149e-02<br />log10(baseMean): 2.5680127<br />gene: FBgn0035002","log2FoldChange: -0.2575754894<br />-log10(padj): 5.766076e-01<br />log10(baseMean): 3.1132590<br />gene: FBgn0035016","log2FoldChange: -0.7776195784<br />-log10(padj): 3.591048e+00<br />log10(baseMean): 2.4514320<br />gene: FBgn0035020","log2FoldChange: 0.0096275400<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.4750878<br />gene: FBgn0035021","log2FoldChange: -0.4193957391<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 3.0563918<br />gene: FBgn0035023","log2FoldChange: -0.0362234334<br />-log10(padj): 2.945997e-02<br />log10(baseMean): 3.0542795<br />gene: FBgn0035024","log2FoldChange: 0.0581053338<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 2.6354531<br />gene: FBgn0035025","log2FoldChange: -0.0555789183<br />-log10(padj): 4.686228e-02<br />log10(baseMean): 2.7335270<br />gene: FBgn0035026","log2FoldChange: -0.0628951322<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.6175009<br />gene: FBgn0035027","log2FoldChange: -0.0611347535<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 2.6495855<br />gene: FBgn0035028","log2FoldChange: -0.0491976968<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 3.1720731<br />gene: FBgn0035032","log2FoldChange: 0.0071799537<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 2.7221714<br />gene: FBgn0035033","log2FoldChange: -0.0210418405<br />-log10(padj): 1.124756e-02<br />log10(baseMean): 2.6307965<br />gene: FBgn0035035","log2FoldChange: 0.1494502060<br />-log10(padj): 1.613646e-01<br />log10(baseMean): 2.4533904<br />gene: FBgn0035036","log2FoldChange: 0.0880037428<br />-log10(padj): 7.795537e-02<br />log10(baseMean): 2.5274591<br />gene: FBgn0035039","log2FoldChange: 0.3550087970<br />-log10(padj): 3.810279e-01<br />log10(baseMean): 2.1541500<br />gene: FBgn0035044","log2FoldChange: 0.0915264649<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.6841077<br />gene: FBgn0035046","log2FoldChange: 0.0311917528<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.3723663<br />gene: FBgn0035047","log2FoldChange: -0.0218516788<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.3629478<br />gene: FBgn0035049","log2FoldChange: -0.0184868498<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 2.4652333<br />gene: FBgn0035059","log2FoldChange: -0.0732811279<br />-log10(padj): 8.465078e-02<br />log10(baseMean): 3.3981571<br />gene: FBgn0035060","log2FoldChange: -0.0628798655<br />-log10(padj): 3.829192e-02<br />log10(baseMean): 2.3692712<br />gene: FBgn0035063","log2FoldChange: -0.0264814364<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.4614626<br />gene: FBgn0035064","log2FoldChange: -0.1868592629<br />-log10(padj): 2.028787e-01<br />log10(baseMean): 2.4712191<br />gene: FBgn0035065","log2FoldChange: 0.3243318712<br />-log10(padj): 1.261286e+00<br />log10(baseMean): 2.6881581<br />gene: FBgn0035073","log2FoldChange: 1.3783294622<br />-log10(padj): 2.518353e+00<br />log10(baseMean): 1.4887286<br />gene: FBgn0035076","log2FoldChange: 0.3721967669<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 1.1089714<br />gene: FBgn0035077","log2FoldChange: 0.0322283708<br />-log10(padj): 4.054271e-03<br />log10(baseMean): 0.8947726<br />gene: FBgn0035078","log2FoldChange: 0.0542167595<br />-log10(padj): 2.525758e-02<br />log10(baseMean): 2.4028877<br />gene: FBgn0035082","log2FoldChange: -0.3646921536<br />-log10(padj): 1.500904e+00<br />log10(baseMean): 3.3697486<br />gene: FBgn0035083","log2FoldChange: -0.9275653886<br />-log10(padj): 3.322332e+00<br />log10(baseMean): 2.0742101<br />gene: FBgn0035084","log2FoldChange: -2.5602423187<br />-log10(padj): 7.308828e+01<br />log10(baseMean): 2.8049700<br />gene: FBgn0035085","log2FoldChange: -0.3317037370<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 1.5707077<br />gene: FBgn0035086","log2FoldChange: -0.0248284114<br />-log10(padj): 1.349861e-02<br />log10(baseMean): 3.5701942<br />gene: FBgn0035087","log2FoldChange: 0.0127359258<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 2.7661975<br />gene: FBgn0035088","log2FoldChange: 0.2592045047<br />-log10(padj): 3.802180e-02<br />log10(baseMean): 0.8878334<br />gene: FBgn0035089","log2FoldChange: 0.4907221560<br />-log10(padj): 2.241023e+00<br />log10(baseMean): 2.6289399<br />gene: FBgn0035091","log2FoldChange: 0.2729843974<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 1.0409592<br />gene: FBgn0035097","log2FoldChange: 0.0684754835<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.9052219<br />gene: FBgn0035099","log2FoldChange: -0.5884713732<br />-log10(padj): 1.699820e-01<br />log10(baseMean): 1.0896516<br />gene: FBgn0035100","log2FoldChange: 0.1214113666<br />-log10(padj): 1.873412e-01<br />log10(baseMean): 3.1357713<br />gene: FBgn0035101","log2FoldChange: 0.1686567813<br />-log10(padj): 3.329052e-01<br />log10(baseMean): 2.7474038<br />gene: FBgn0035102","log2FoldChange: -0.2472912626<br />-log10(padj): 5.470330e-01<br />log10(baseMean): 3.4507632<br />gene: FBgn0035106","log2FoldChange: 0.0988839567<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.9473882<br />gene: FBgn0035107","log2FoldChange: 0.3089905282<br />-log10(padj): 4.329783e-01<br />log10(baseMean): 2.1750316<br />gene: FBgn0035109","log2FoldChange: 0.0818332768<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.5207917<br />gene: FBgn0035110","log2FoldChange: -0.0592296358<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.9773880<br />gene: FBgn0035111","log2FoldChange: 0.7861372205<br />-log10(padj): 2.908581e+00<br />log10(baseMean): 2.0197047<br />gene: FBgn0035113","log2FoldChange: 0.2840870051<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 2.5751787<br />gene: FBgn0035120","log2FoldChange: -0.0438594485<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 3.8297481<br />gene: FBgn0035121","log2FoldChange: -0.0121832492<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 2.5992372<br />gene: FBgn0035122","log2FoldChange: 0.1409293935<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.4869993<br />gene: FBgn0035131","log2FoldChange: -0.3612299636<br />-log10(padj): 1.274073e+00<br />log10(baseMean): 3.0148030<br />gene: FBgn0035132","log2FoldChange: 0.0950687392<br />-log10(padj): 9.412115e-02<br />log10(baseMean): 3.2988189<br />gene: FBgn0035133","log2FoldChange: 0.4548873290<br />-log10(padj): 7.954962e-02<br />log10(baseMean): 0.8629140<br />gene: FBgn0035134","log2FoldChange: 0.0169402133<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.0738476<br />gene: FBgn0035136","log2FoldChange: -0.2655358986<br />-log10(padj): 1.037479e+00<br />log10(baseMean): 3.0291807<br />gene: FBgn0035137","log2FoldChange: 0.0610669163<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.2312797<br />gene: FBgn0035140","log2FoldChange: -0.1716176941<br />-log10(padj): 2.397553e-01<br />log10(baseMean): 2.7162853<br />gene: FBgn0035141","log2FoldChange: -0.1983291288<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 4.2755304<br />gene: FBgn0035142","log2FoldChange: 0.6572781809<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 0.9858550<br />gene: FBgn0035143","log2FoldChange: -0.2532173424<br />-log10(padj): 3.067409e-01<br />log10(baseMean): 2.4452520<br />gene: FBgn0035144","log2FoldChange: -0.0495204993<br />-log10(padj): 5.128138e-02<br />log10(baseMean): 3.3109236<br />gene: FBgn0035145","log2FoldChange: 0.0610616814<br />-log10(padj): 5.469498e-02<br />log10(baseMean): 2.9064987<br />gene: FBgn0035146","log2FoldChange: -1.1038276092<br />-log10(padj): 8.513773e+00<br />log10(baseMean): 4.2860678<br />gene: FBgn0035147","log2FoldChange: 0.3093096258<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 2.9973205<br />gene: FBgn0035148","log2FoldChange: 0.2039764887<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 3.3021533<br />gene: FBgn0035149","log2FoldChange: 0.5650728605<br />-log10(padj): 6.274764e+00<br />log10(baseMean): 3.4159789<br />gene: FBgn0035150","log2FoldChange: 0.6074509470<br />-log10(padj): 8.815624e-01<br />log10(baseMean): 1.9351258<br />gene: FBgn0035151","log2FoldChange: -0.2488172129<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 2.3363064<br />gene: FBgn0035152","log2FoldChange: -0.0713963892<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 3.2075360<br />gene: FBgn0035153","log2FoldChange: 0.5505184822<br />-log10(padj): 1.375293e+00<br />log10(baseMean): 2.0033580<br />gene: FBgn0035154","log2FoldChange: 0.1211940152<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.1095987<br />gene: FBgn0035155","log2FoldChange: 0.4300179154<br />-log10(padj): 1.499167e-01<br />log10(baseMean): 1.3009378<br />gene: FBgn0035157","log2FoldChange: 0.2432748915<br />-log10(padj): 1.007882e+00<br />log10(baseMean): 2.9627189<br />gene: FBgn0035158","log2FoldChange: 0.1917063918<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 2.3365332<br />gene: FBgn0035159","log2FoldChange: 0.2758738027<br />-log10(padj): 2.054522e-01<br />log10(baseMean): 1.9872597<br />gene: FBgn0035160","log2FoldChange: -0.4183410233<br />-log10(padj): 2.579744e-01<br />log10(baseMean): 1.8573682<br />gene: FBgn0035161","log2FoldChange: -0.0462704196<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 4.1725733<br />gene: FBgn0035162","log2FoldChange: 0.2207726648<br />-log10(padj): 3.896990e-01<br />log10(baseMean): 2.5829059<br />gene: FBgn0035164","log2FoldChange: 0.3492577439<br />-log10(padj): 2.418834e+00<br />log10(baseMean): 3.5922644<br />gene: FBgn0035165","log2FoldChange: 0.1785711681<br />-log10(padj): 2.238635e-01<br />log10(baseMean): 2.5507237<br />gene: FBgn0035166","log2FoldChange: 0.6883686911<br />-log10(padj): 1.246909e+00<br />log10(baseMean): 1.9693940<br />gene: FBgn0035167","log2FoldChange: -0.0653726001<br />-log10(padj): 1.164540e-02<br />log10(baseMean): 1.3932504<br />gene: FBgn0035168","log2FoldChange: 0.2369380411<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.6345811<br />gene: FBgn0035169","log2FoldChange: 1.6912007331<br />-log10(padj): 1.086229e+00<br />log10(baseMean): 1.2242132<br />gene: FBgn0035170","log2FoldChange: -1.3549280318<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 0.9596204<br />gene: FBgn0035171","log2FoldChange: 0.5323091176<br />-log10(padj): 3.623814e+00<br />log10(baseMean): 2.7148082<br />gene: FBgn0035173","log2FoldChange: -0.1094764527<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.1193147<br />gene: FBgn0035181","log2FoldChange: 2.9751089699<br />-log10(padj): 2.869229e+01<br />log10(baseMean): 2.3337594<br />gene: FBgn0035189","log2FoldChange: -0.2418157245<br />-log10(padj): 2.424491e-01<br />log10(baseMean): 2.4418271<br />gene: FBgn0035194","log2FoldChange: 0.0410263625<br />-log10(padj): 2.774356e-02<br />log10(baseMean): 3.2707722<br />gene: FBgn0035195","log2FoldChange: -0.0122837178<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.9556772<br />gene: FBgn0035202","log2FoldChange: 0.4370312144<br />-log10(padj): 1.277061e+00<br />log10(baseMean): 2.9131370<br />gene: FBgn0035203","log2FoldChange: -0.3735437835<br />-log10(padj): 1.207338e+00<br />log10(baseMean): 2.7505503<br />gene: FBgn0035204","log2FoldChange: -0.0400289976<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 3.4294264<br />gene: FBgn0035205","log2FoldChange: -0.1245622876<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 2.8784012<br />gene: FBgn0035206","log2FoldChange: 0.0834082887<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.4677379<br />gene: FBgn0035207","log2FoldChange: -0.1492213548<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 1.5183284<br />gene: FBgn0035208","log2FoldChange: 0.1314654528<br />-log10(padj): 1.030332e-01<br />log10(baseMean): 2.5007826<br />gene: FBgn0035209","log2FoldChange: 0.3393730424<br />-log10(padj): 9.631074e-01<br />log10(baseMean): 2.5212284<br />gene: FBgn0035210","log2FoldChange: 0.3539947008<br />-log10(padj): 4.413021e-01<br />log10(baseMean): 2.0459590<br />gene: FBgn0035211","log2FoldChange: -0.2414475027<br />-log10(padj): 5.181536e-01<br />log10(baseMean): 3.0229702<br />gene: FBgn0035213","log2FoldChange: -0.1299053383<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 1.4155831<br />gene: FBgn0035216","log2FoldChange: -0.1745430463<br />-log10(padj): 3.321162e-01<br />log10(baseMean): 3.1993805<br />gene: FBgn0035227","log2FoldChange: -0.1418474038<br />-log10(padj): 1.727186e-01<br />log10(baseMean): 3.2773134<br />gene: FBgn0035228","log2FoldChange: 0.0325042908<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.1317473<br />gene: FBgn0035229","log2FoldChange: -0.8021573789<br />-log10(padj): 1.589409e+00<br />log10(baseMean): 1.7220616<br />gene: FBgn0035231","log2FoldChange: -0.5353870795<br />-log10(padj): 5.187476e+00<br />log10(baseMean): 3.2367613<br />gene: FBgn0035232","log2FoldChange: -0.1671131895<br />-log10(padj): 1.658154e-01<br />log10(baseMean): 2.2812788<br />gene: FBgn0035233","log2FoldChange: 0.3470719155<br />-log10(padj): 1.222172e-01<br />log10(baseMean): 1.3859485<br />gene: FBgn0035234","log2FoldChange: -0.2063049095<br />-log10(padj): 3.713583e-01<br />log10(baseMean): 2.8247879<br />gene: FBgn0035235","log2FoldChange: -0.2695348305<br />-log10(padj): 6.813664e-01<br />log10(baseMean): 3.3215119<br />gene: FBgn0035236","log2FoldChange: 0.2177694037<br />-log10(padj): 2.262589e-01<br />log10(baseMean): 2.9758473<br />gene: FBgn0035237","log2FoldChange: 0.0473027700<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.8775365<br />gene: FBgn0035238","log2FoldChange: -0.6416522250<br />-log10(padj): 6.741894e-01<br />log10(baseMean): 1.6715729<br />gene: FBgn0035241","log2FoldChange: 0.1315374440<br />-log10(padj): 1.084218e-01<br />log10(baseMean): 2.5407715<br />gene: FBgn0035243","log2FoldChange: -0.1518848119<br />-log10(padj): 1.677254e-01<br />log10(baseMean): 3.2028005<br />gene: FBgn0035244","log2FoldChange: 0.1355384381<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 1.7543216<br />gene: FBgn0035246","log2FoldChange: -0.2614362941<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 2.7769942<br />gene: FBgn0035247","log2FoldChange: -0.0516655647<br />-log10(padj): 1.609026e-02<br />log10(baseMean): 1.7796263<br />gene: FBgn0035248","log2FoldChange: -0.1256693406<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.4202556<br />gene: FBgn0035249","log2FoldChange: -0.1387502776<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 3.1107511<br />gene: FBgn0035251","log2FoldChange: -0.0730183499<br />-log10(padj): 5.701360e-02<br />log10(baseMean): 2.7777260<br />gene: FBgn0035252","log2FoldChange: 0.0638454455<br />-log10(padj): 2.434834e-02<br />log10(baseMean): 3.5174504<br />gene: FBgn0035253","log2FoldChange: -0.0781109560<br />-log10(padj): 4.371691e-02<br />log10(baseMean): 2.2385771<br />gene: FBgn0035254","log2FoldChange: -1.1494010171<br />-log10(padj): 4.353194e-01<br />log10(baseMean): 0.8980016<br />gene: FBgn0035260","log2FoldChange: -0.4987381719<br />-log10(padj): 1.192336e+00<br />log10(baseMean): 2.5331252<br />gene: FBgn0035262","log2FoldChange: -0.3124869647<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 1.5126165<br />gene: FBgn0035263","log2FoldChange: -0.2126207570<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 1.6076959<br />gene: FBgn0035264","log2FoldChange: 1.0752919406<br />-log10(padj): 6.729740e+00<br />log10(baseMean): 2.5837605<br />gene: FBgn0035266","log2FoldChange: 0.9985840313<br />-log10(padj): 1.264161e+00<br />log10(baseMean): 1.5468876<br />gene: FBgn0035267","log2FoldChange: -0.0930359067<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 2.9132543<br />gene: FBgn0035268","log2FoldChange: -0.1660645659<br />-log10(padj): 2.764490e-01<br />log10(baseMean): 2.7530939<br />gene: FBgn0035270","log2FoldChange: 0.1968863854<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 2.0413118<br />gene: FBgn0035271","log2FoldChange: -0.0261897493<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 2.3569981<br />gene: FBgn0035272","log2FoldChange: -0.2566944793<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.5314402<br />gene: FBgn0035283","log2FoldChange: 0.4127227100<br />-log10(padj): 2.355266e+00<br />log10(baseMean): 2.8519422<br />gene: FBgn0035285","log2FoldChange: -0.1312762066<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 1.6067668<br />gene: FBgn0035286","log2FoldChange: -0.7864107547<br />-log10(padj): 1.535085e+00<br />log10(baseMean): 1.7373550<br />gene: FBgn0035287","log2FoldChange: -0.1023677648<br />-log10(padj): 7.765385e-02<br />log10(baseMean): 3.0561135<br />gene: FBgn0035294","log2FoldChange: 0.3620175562<br />-log10(padj): 1.567645e+00<br />log10(baseMean): 2.7270725<br />gene: FBgn0035295","log2FoldChange: -0.2672444587<br />-log10(padj): 7.592377e-01<br />log10(baseMean): 3.0218131<br />gene: FBgn0035296","log2FoldChange: -0.2570661845<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 2.5035308<br />gene: FBgn0035297","log2FoldChange: 0.1070420410<br />-log10(padj): 1.136128e-01<br />log10(baseMean): 2.8350265<br />gene: FBgn0035298","log2FoldChange: 0.1992428143<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 1.3702077<br />gene: FBgn0035308","log2FoldChange: -0.1966954240<br />-log10(padj): 1.572749e-01<br />log10(baseMean): 2.2649642<br />gene: FBgn0035309","log2FoldChange: -0.4364926676<br />-log10(padj): 1.751247e-01<br />log10(baseMean): 1.4829417<br />gene: FBgn0035311","log2FoldChange: -0.5959042103<br />-log10(padj): 2.496249e+00<br />log10(baseMean): 2.3434836<br />gene: FBgn0035312","log2FoldChange: -0.0145055286<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.1770725<br />gene: FBgn0035317","log2FoldChange: 0.0973559574<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.7357336<br />gene: FBgn0035318","log2FoldChange: -0.1630676057<br />-log10(padj): 1.437531e-01<br />log10(baseMean): 3.1074644<br />gene: FBgn0035321","log2FoldChange: 0.1587208725<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 2.4242035<br />gene: FBgn0035323","log2FoldChange: -0.0581237739<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 3.3290650<br />gene: FBgn0035333","log2FoldChange: -0.0801686594<br />-log10(padj): 6.106439e-02<br />log10(baseMean): 2.7723571<br />gene: FBgn0035334","log2FoldChange: 0.0728409962<br />-log10(padj): 3.645199e-02<br />log10(baseMean): 2.4035143<br />gene: FBgn0035335","log2FoldChange: -0.0425164804<br />-log10(padj): 2.499648e-02<br />log10(baseMean): 2.5886999<br />gene: FBgn0035336","log2FoldChange: -0.0546918792<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.3988146<br />gene: FBgn0035337","log2FoldChange: -0.4322358579<br />-log10(padj): 9.909494e-01<br />log10(baseMean): 2.9377720<br />gene: FBgn0035338","log2FoldChange: -1.6470504254<br />-log10(padj): 9.587960e+00<br />log10(baseMean): 1.8886789<br />gene: FBgn0035344","log2FoldChange: 0.2961810864<br />-log10(padj): 1.269015e+00<br />log10(baseMean): 2.9075723<br />gene: FBgn0035346","log2FoldChange: -0.3992407973<br />-log10(padj): 1.949249e+00<br />log10(baseMean): 2.7740329<br />gene: FBgn0035347","log2FoldChange: -0.2808586178<br />-log10(padj): 5.584344e-01<br />log10(baseMean): 2.7936514<br />gene: FBgn0035348","log2FoldChange: -0.0222717923<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.0273150<br />gene: FBgn0035355","log2FoldChange: 0.1262854032<br />-log10(padj): 4.842354e-02<br />log10(baseMean): 1.9822179<br />gene: FBgn0035356","log2FoldChange: -0.1063972049<br />-log10(padj): 2.280743e-02<br />log10(baseMean): 1.9500799<br />gene: FBgn0035357","log2FoldChange: 0.1627301029<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 1.5801216<br />gene: FBgn0035358","log2FoldChange: -0.2044623014<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.7513037<br />gene: FBgn0035359","log2FoldChange: 0.1129617486<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 1.3041754<br />gene: FBgn0035360","log2FoldChange: 0.0875957945<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 0.8927142<br />gene: FBgn0035367","log2FoldChange: 0.0480422976<br />-log10(padj): 4.913937e-02<br />log10(baseMean): 3.4254033<br />gene: FBgn0035370","log2FoldChange: -0.0271007055<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 3.2722049<br />gene: FBgn0035371","log2FoldChange: -0.0310596905<br />-log10(padj): 2.009957e-02<br />log10(baseMean): 2.9617003<br />gene: FBgn0035372","log2FoldChange: 0.1475057749<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 2.8114041<br />gene: FBgn0035374","log2FoldChange: -0.0782937592<br />-log10(padj): 7.401100e-02<br />log10(baseMean): 3.0037810<br />gene: FBgn0035375","log2FoldChange: -0.3254014476<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 3.6721945<br />gene: FBgn0035376","log2FoldChange: -0.3203614465<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 0.8792831<br />gene: FBgn0035377","log2FoldChange: 0.0780565759<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.5841915<br />gene: FBgn0035378","log2FoldChange: -0.0779562840<br />-log10(padj): 9.294115e-02<br />log10(baseMean): 2.8464823<br />gene: FBgn0035383","log2FoldChange: -0.1390283185<br />-log10(padj): 3.247082e-01<br />log10(baseMean): 3.1566700<br />gene: FBgn0035388","log2FoldChange: 0.4090628356<br />-log10(padj): 2.559329e+00<br />log10(baseMean): 3.0513968<br />gene: FBgn0035390","log2FoldChange: 0.2023419120<br />-log10(padj): 1.115369e-01<br />log10(baseMean): 1.8278937<br />gene: FBgn0035392","log2FoldChange: 0.1839153740<br />-log10(padj): 1.408800e-01<br />log10(baseMean): 2.4916629<br />gene: FBgn0035393","log2FoldChange: -0.0415592528<br />-log10(padj): 2.185367e-02<br />log10(baseMean): 3.4554437<br />gene: FBgn0035397","log2FoldChange: -0.3911371731<br />-log10(padj): 6.389853e-02<br />log10(baseMean): 0.8200967<br />gene: FBgn0035399","log2FoldChange: -0.1431991851<br />-log10(padj): 2.331077e-01<br />log10(baseMean): 3.1879363<br />gene: FBgn0035400","log2FoldChange: -0.0984093425<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.9891101<br />gene: FBgn0035401","log2FoldChange: 0.1530916018<br />-log10(padj): 2.381656e-01<br />log10(baseMean): 2.8898225<br />gene: FBgn0035402","log2FoldChange: -0.9529893071<br />-log10(padj): 6.935639e+00<br />log10(baseMean): 2.6957743<br />gene: FBgn0035403","log2FoldChange: 0.1032507140<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.0109763<br />gene: FBgn0035404","log2FoldChange: 0.1862215457<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 2.7357480<br />gene: FBgn0035405","log2FoldChange: -0.1152246567<br />-log10(padj): 4.949434e-02<br />log10(baseMean): 2.1077644<br />gene: FBgn0035407","log2FoldChange: -0.5511966003<br />-log10(padj): 1.782891e-01<br />log10(baseMean): 1.2280665<br />gene: FBgn0035409","log2FoldChange: 0.3527374317<br />-log10(padj): 6.871396e-01<br />log10(baseMean): 2.5176500<br />gene: FBgn0035410","log2FoldChange: 0.0812330226<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 3.1244017<br />gene: FBgn0035411","log2FoldChange: 0.1465404341<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.2585055<br />gene: FBgn0035414","log2FoldChange: -0.0781768388<br />-log10(padj): 2.847533e-02<br />log10(baseMean): 2.3373459<br />gene: FBgn0035415","log2FoldChange: -0.0610938386<br />-log10(padj): 7.406088e-02<br />log10(baseMean): 3.3039709<br />gene: FBgn0035416","log2FoldChange: -0.0444039247<br />-log10(padj): 2.830218e-02<br />log10(baseMean): 3.3230481<br />gene: FBgn0035420","log2FoldChange: 0.2203662336<br />-log10(padj): 4.328319e-01<br />log10(baseMean): 2.4940596<br />gene: FBgn0035421","log2FoldChange: -0.1316702357<br />-log10(padj): 1.331873e-01<br />log10(baseMean): 4.3880720<br />gene: FBgn0035422","log2FoldChange: 0.0427036422<br />-log10(padj): 3.645199e-02<br />log10(baseMean): 3.3261410<br />gene: FBgn0035423","log2FoldChange: -0.0766539973<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 3.6303576<br />gene: FBgn0035424","log2FoldChange: -0.1471003542<br />-log10(padj): 1.674937e-01<br />log10(baseMean): 3.0803745<br />gene: FBgn0035425","log2FoldChange: -0.1329810373<br />-log10(padj): 1.235517e-01<br />log10(baseMean): 3.5830961<br />gene: FBgn0035432","log2FoldChange: -0.0941727818<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 1.9929268<br />gene: FBgn0035435","log2FoldChange: -0.0937298695<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 3.4441559<br />gene: FBgn0035437","log2FoldChange: 0.2288063341<br />-log10(padj): 8.243211e-01<br />log10(baseMean): 3.5779929<br />gene: FBgn0035438","log2FoldChange: 0.2091601612<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 2.6954295<br />gene: FBgn0035439","log2FoldChange: 0.0396298393<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 3.0586697<br />gene: FBgn0035440","log2FoldChange: 0.2255653253<br />-log10(padj): 2.466627e-01<br />log10(baseMean): 2.1864023<br />gene: FBgn0035443","log2FoldChange: -0.0734957121<br />-log10(padj): 6.642066e-02<br />log10(baseMean): 2.7279975<br />gene: FBgn0035444","log2FoldChange: 0.3044732585<br />-log10(padj): 1.522534e-01<br />log10(baseMean): 1.9012145<br />gene: FBgn0035445","log2FoldChange: -0.0012525746<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 2.8833470<br />gene: FBgn0035449","log2FoldChange: 0.1182549501<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.6696176<br />gene: FBgn0035452","log2FoldChange: 0.0921676801<br />-log10(padj): 3.661190e-02<br />log10(baseMean): 2.0501713<br />gene: FBgn0035461","log2FoldChange: 0.1622635771<br />-log10(padj): 1.176136e-01<br />log10(baseMean): 2.2860897<br />gene: FBgn0035462","log2FoldChange: 0.1421040902<br />-log10(padj): 2.058877e-01<br />log10(baseMean): 3.0492188<br />gene: FBgn0035464","log2FoldChange: -0.0454327184<br />-log10(padj): 2.125730e-02<br />log10(baseMean): 2.3493018<br />gene: FBgn0035469","log2FoldChange: -0.1522248690<br />-log10(padj): 9.869851e-02<br />log10(baseMean): 2.1142736<br />gene: FBgn0035470","log2FoldChange: 0.1599850849<br />-log10(padj): 2.255230e-01<br />log10(baseMean): 3.0854076<br />gene: FBgn0035471","log2FoldChange: -0.1182939263<br />-log10(padj): 1.782943e-01<br />log10(baseMean): 2.9608966<br />gene: FBgn0035473","log2FoldChange: -0.0440361478<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.9787890<br />gene: FBgn0035475","log2FoldChange: -0.0380748254<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 1.2030328<br />gene: FBgn0035476","log2FoldChange: -0.0484760563<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 2.1526780<br />gene: FBgn0035477","log2FoldChange: -0.1848037331<br />-log10(padj): 1.235517e-01<br />log10(baseMean): 2.2631246<br />gene: FBgn0035480","log2FoldChange: 0.0374486142<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.3117948<br />gene: FBgn0035482","log2FoldChange: 0.1823543473<br />-log10(padj): 1.284897e-01<br />log10(baseMean): 2.2147170<br />gene: FBgn0035483","log2FoldChange: -0.0835669235<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.4050747<br />gene: FBgn0035484","log2FoldChange: -0.1710211095<br />-log10(padj): 2.260508e-01<br />log10(baseMean): 2.7326858<br />gene: FBgn0035488","log2FoldChange: 0.0154811400<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 3.0381737<br />gene: FBgn0035489","log2FoldChange: -0.6472341103<br />-log10(padj): 2.167378e+00<br />log10(baseMean): 2.3873124<br />gene: FBgn0035496","log2FoldChange: -0.1269016250<br />-log10(padj): 8.940060e-02<br />log10(baseMean): 2.6704309<br />gene: FBgn0035497","log2FoldChange: -0.0382826425<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 3.0052120<br />gene: FBgn0035498","log2FoldChange: 0.1156605195<br />-log10(padj): 9.150092e-02<br />log10(baseMean): 3.8671173<br />gene: FBgn0035499","log2FoldChange: -0.5656832495<br />-log10(padj): 4.129762e+00<br />log10(baseMean): 3.6388112<br />gene: FBgn0035500","log2FoldChange: -0.3888645916<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 1.0839898<br />gene: FBgn0035501","log2FoldChange: -0.2849063427<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 2.8961116<br />gene: FBgn0035507","log2FoldChange: -0.3953241688<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 1.7742313<br />gene: FBgn0035509","log2FoldChange: 0.7275466661<br />-log10(padj): 2.666288e-01<br />log10(baseMean): 1.1871342<br />gene: FBgn0035512","log2FoldChange: -0.4607544775<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 1.1577360<br />gene: FBgn0035513","log2FoldChange: -0.2485942746<br />-log10(padj): 3.116702e-01<br />log10(baseMean): 2.8390667<br />gene: FBgn0035514","log2FoldChange: 0.3779369667<br />-log10(padj): 1.219243e+00<br />log10(baseMean): 2.3743415<br />gene: FBgn0035515","log2FoldChange: 0.3681216260<br />-log10(padj): 5.491635e-01<br />log10(baseMean): 2.4848254<br />gene: FBgn0035517","log2FoldChange: 0.1414026629<br />-log10(padj): 1.228884e-01<br />log10(baseMean): 2.5708290<br />gene: FBgn0035518","log2FoldChange: 0.0168884164<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.7872416<br />gene: FBgn0035519","log2FoldChange: 0.1516432657<br />-log10(padj): 1.084626e-01<br />log10(baseMean): 2.3190221<br />gene: FBgn0035520","log2FoldChange: 1.5482833752<br />-log10(padj): 3.051554e+00<br />log10(baseMean): 1.6803789<br />gene: FBgn0035523","log2FoldChange: -0.2835403385<br />-log10(padj): 5.743118e-01<br />log10(baseMean): 2.7391717<br />gene: FBgn0035524","log2FoldChange: -0.1961248778<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 3.1365437<br />gene: FBgn0035526","log2FoldChange: -0.1779943435<br />-log10(padj): 1.382248e-01<br />log10(baseMean): 3.0924342<br />gene: FBgn0035528","log2FoldChange: -0.2081104755<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 2.2984268<br />gene: FBgn0035529","log2FoldChange: 0.0172806606<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.3260099<br />gene: FBgn0035532","log2FoldChange: -0.2508850920<br />-log10(padj): 1.110254e+00<br />log10(baseMean): 3.5173583<br />gene: FBgn0035533","log2FoldChange: 0.0357889521<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.0162802<br />gene: FBgn0035534","log2FoldChange: 0.5905559553<br />-log10(padj): 3.592539e-01<br />log10(baseMean): 1.3997356<br />gene: FBgn0035537","log2FoldChange: 1.5715505759<br />-log10(padj): 1.792584e+00<br />log10(baseMean): 1.3113690<br />gene: FBgn0035539","log2FoldChange: -0.3333457708<br />-log10(padj): 6.749513e-01<br />log10(baseMean): 2.3506129<br />gene: FBgn0035540","log2FoldChange: -0.3604640640<br />-log10(padj): 5.517127e-01<br />log10(baseMean): 2.7099879<br />gene: FBgn0035541","log2FoldChange: -0.9475381087<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 2.7339109<br />gene: FBgn0035542","log2FoldChange: -0.3034519673<br />-log10(padj): 8.815624e-01<br />log10(baseMean): 3.7383183<br />gene: FBgn0035558","log2FoldChange: 0.5289647841<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.0061360<br />gene: FBgn0035577","log2FoldChange: 0.4606314896<br />-log10(padj): 1.084915e-01<br />log10(baseMean): 1.0893410<br />gene: FBgn0035581","log2FoldChange: -0.0773512888<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.8325950<br />gene: FBgn0035586","log2FoldChange: 0.2159313819<br />-log10(padj): 3.069340e-02<br />log10(baseMean): 0.9989497<br />gene: FBgn0035587","log2FoldChange: 0.1929296620<br />-log10(padj): 1.344421e-01<br />log10(baseMean): 2.3886440<br />gene: FBgn0035588","log2FoldChange: -0.2426540580<br />-log10(padj): 3.862570e-01<br />log10(baseMean): 2.5638104<br />gene: FBgn0035589","log2FoldChange: 0.4085638531<br />-log10(padj): 1.021690e+00<br />log10(baseMean): 2.3113067<br />gene: FBgn0035590","log2FoldChange: -0.0019405649<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 2.4170644<br />gene: FBgn0035591","log2FoldChange: 0.1095497504<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.4469992<br />gene: FBgn0035592","log2FoldChange: 0.1856809816<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.6496557<br />gene: FBgn0035593","log2FoldChange: -0.1533549755<br />-log10(padj): 2.136474e-01<br />log10(baseMean): 3.4867777<br />gene: FBgn0035600","log2FoldChange: -0.0219988028<br />-log10(padj): 9.670303e-03<br />log10(baseMean): 3.4185990<br />gene: FBgn0035601","log2FoldChange: 0.3873921314<br />-log10(padj): 1.777973e+00<br />log10(baseMean): 2.7357440<br />gene: FBgn0035603","log2FoldChange: 0.3900953524<br />-log10(padj): 8.086463e-01<br />log10(baseMean): 3.5477191<br />gene: FBgn0035608","log2FoldChange: -0.4588722505<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.1214646<br />gene: FBgn0035612","log2FoldChange: -0.0595637854<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.0790263<br />gene: FBgn0035617","log2FoldChange: 0.0769426938<br />-log10(padj): 5.455075e-02<br />log10(baseMean): 3.2468289<br />gene: FBgn0035622","log2FoldChange: 0.2190396584<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.5831157<br />gene: FBgn0035623","log2FoldChange: 0.1866006527<br />-log10(padj): 1.331016e-01<br />log10(baseMean): 2.0474313<br />gene: FBgn0035624","log2FoldChange: -0.0300196293<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.9940867<br />gene: FBgn0035626","log2FoldChange: 0.0136099512<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.2241284<br />gene: FBgn0035627","log2FoldChange: -0.1439660286<br />-log10(padj): 2.761376e-01<br />log10(baseMean): 3.6302384<br />gene: FBgn0035630","log2FoldChange: 0.0059162134<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.9304784<br />gene: FBgn0035631","log2FoldChange: -0.1147491998<br />-log10(padj): 7.279571e-02<br />log10(baseMean): 2.4397441<br />gene: FBgn0035632","log2FoldChange: -0.1561497182<br />-log10(padj): 1.148200e-01<br />log10(baseMean): 2.8072685<br />gene: FBgn0035639","log2FoldChange: -0.0767849601<br />-log10(padj): 4.614145e-02<br />log10(baseMean): 2.6340517<br />gene: FBgn0035640","log2FoldChange: 0.1334636291<br />-log10(padj): 6.340796e-02<br />log10(baseMean): 2.4184989<br />gene: FBgn0035641","log2FoldChange: 0.1074734325<br />-log10(padj): 6.633703e-02<br />log10(baseMean): 2.2992705<br />gene: FBgn0035642","log2FoldChange: -0.4844383475<br />-log10(padj): 2.025520e+00<br />log10(baseMean): 2.5978424<br />gene: FBgn0035644","log2FoldChange: -0.0648681004<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 1.7771370<br />gene: FBgn0035670","log2FoldChange: 0.3220616239<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 1.5416878<br />gene: FBgn0035673","log2FoldChange: 0.1254157348<br />-log10(padj): 9.620880e-02<br />log10(baseMean): 2.4972995<br />gene: FBgn0035674","log2FoldChange: -0.1504932141<br />-log10(padj): 7.555807e-02<br />log10(baseMean): 2.1838782<br />gene: FBgn0035675","log2FoldChange: -0.6928305150<br />-log10(padj): 3.621771e-01<br />log10(baseMean): 1.2794022<br />gene: FBgn0035676","log2FoldChange: -0.4129277447<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 1.9938408<br />gene: FBgn0035678","log2FoldChange: -0.2318458680<br />-log10(padj): 4.925073e-01<br />log10(baseMean): 2.7176166<br />gene: FBgn0035679","log2FoldChange: -0.0786465739<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 3.2686633<br />gene: FBgn0035688","log2FoldChange: 0.2421136978<br />-log10(padj): 3.019354e-01<br />log10(baseMean): 2.5055666<br />gene: FBgn0035689","log2FoldChange: 0.0119488981<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.7179745<br />gene: FBgn0035690","log2FoldChange: -0.0025237753<br />-log10(padj): 1.175523e-03<br />log10(baseMean): 1.6554112<br />gene: FBgn0035691","log2FoldChange: 0.0643709691<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.7232279<br />gene: FBgn0035692","log2FoldChange: 0.5214122065<br />-log10(padj): 1.903028e-01<br />log10(baseMean): 1.4555659<br />gene: FBgn0035693","log2FoldChange: -0.2204688837<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 1.7884186<br />gene: FBgn0035702","log2FoldChange: -0.0761860294<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.1834123<br />gene: FBgn0035703","log2FoldChange: -0.0915887493<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 2.7247617<br />gene: FBgn0035704","log2FoldChange: -0.1859056707<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 2.9578353<br />gene: FBgn0035707","log2FoldChange: -0.4266592587<br />-log10(padj): 2.253174e+00<br />log10(baseMean): 3.0777208<br />gene: FBgn0035708","log2FoldChange: 2.3071960406<br />-log10(padj): 3.497362e+00<br />log10(baseMean): 1.2392917<br />gene: FBgn0035710","log2FoldChange: -0.0109348513<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.4463084<br />gene: FBgn0035713","log2FoldChange: 0.1778058633<br />-log10(padj): 2.362813e-01<br />log10(baseMean): 2.6268417<br />gene: FBgn0035714","log2FoldChange: -0.1534873020<br />-log10(padj): 1.959070e-01<br />log10(baseMean): 3.1162659<br />gene: FBgn0035715","log2FoldChange: 0.1363560342<br />-log10(padj): 6.237762e-02<br />log10(baseMean): 1.8784434<br />gene: FBgn0035719","log2FoldChange: -0.1322362238<br />-log10(padj): 2.286833e-01<br />log10(baseMean): 3.7481162<br />gene: FBgn0035720","log2FoldChange: 0.0311644898<br />-log10(padj): 9.014029e-03<br />log10(baseMean): 2.2681206<br />gene: FBgn0035721","log2FoldChange: -0.1765971078<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 2.1168012<br />gene: FBgn0035722","log2FoldChange: -0.6019600582<br />-log10(padj): 3.566542e-01<br />log10(baseMean): 1.4574120<br />gene: FBgn0035724","log2FoldChange: 0.1084904059<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 2.3938605<br />gene: FBgn0035725","log2FoldChange: 0.0598819103<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.6911473<br />gene: FBgn0035726","log2FoldChange: 1.5889123034<br />-log10(padj): 8.592062e+00<br />log10(baseMean): 1.8145073<br />gene: FBgn0035727","log2FoldChange: -0.1701357308<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.6592167<br />gene: FBgn0035751","log2FoldChange: -0.1365371984<br />-log10(padj): 1.141528e-01<br />log10(baseMean): 4.2353175<br />gene: FBgn0035753","log2FoldChange: -0.0361701288<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.2752603<br />gene: FBgn0035754","log2FoldChange: 0.4108625923<br />-log10(padj): 2.666288e-01<br />log10(baseMean): 1.7390221<br />gene: FBgn0035760","log2FoldChange: 0.7394028671<br />-log10(padj): 8.306237e+00<br />log10(baseMean): 2.7665949<br />gene: FBgn0035761","log2FoldChange: -0.0035333732<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.6349675<br />gene: FBgn0035762","log2FoldChange: -0.1138016988<br />-log10(padj): 2.024417e-01<br />log10(baseMean): 3.1641520<br />gene: FBgn0035763","log2FoldChange: 0.9902457204<br />-log10(padj): 1.058082e+01<br />log10(baseMean): 2.7455165<br />gene: FBgn0035765","log2FoldChange: -0.3594707558<br />-log10(padj): 2.187385e+00<br />log10(baseMean): 3.2218518<br />gene: FBgn0035766","log2FoldChange: 0.0620260375<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.8553246<br />gene: FBgn0035767","log2FoldChange: 0.3277206033<br />-log10(padj): 6.187511e-02<br />log10(baseMean): 1.0673002<br />gene: FBgn0035768","log2FoldChange: -0.3150439290<br />-log10(padj): 1.566219e+00<br />log10(baseMean): 2.9113623<br />gene: FBgn0035769","log2FoldChange: -0.1408036134<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 2.5134609<br />gene: FBgn0035770","log2FoldChange: -0.1094288064<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 3.5678139<br />gene: FBgn0035771","log2FoldChange: 0.1816989251<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 3.6664874<br />gene: FBgn0035772","log2FoldChange: 0.2517370959<br />-log10(padj): 3.237581e-02<br />log10(baseMean): 0.8331932<br />gene: FBgn0035777","log2FoldChange: -2.7247164746<br />-log10(padj): 1.875051e+00<br />log10(baseMean): 1.1340206<br />gene: FBgn0035789","log2FoldChange: -0.1882233331<br />-log10(padj): 4.992606e-01<br />log10(baseMean): 3.4808603<br />gene: FBgn0035793","log2FoldChange: -0.6230461255<br />-log10(padj): 4.393303e-01<br />log10(baseMean): 1.8948450<br />gene: FBgn0035802","log2FoldChange: 0.1953999377<br />-log10(padj): 1.858855e-01<br />log10(baseMean): 2.3751196<br />gene: FBgn0035805","log2FoldChange: -0.8636108304<br />-log10(padj): 2.556949e-01<br />log10(baseMean): 0.9746544<br />gene: FBgn0035806","log2FoldChange: -0.0627319843<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.8569788<br />gene: FBgn0035807","log2FoldChange: 0.8817914801<br />-log10(padj): 5.748602e+00<br />log10(baseMean): 3.0152389<br />gene: FBgn0035811","log2FoldChange: 0.0293159856<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.8980372<br />gene: FBgn0035812","log2FoldChange: -0.0013806817<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 2.1728577<br />gene: FBgn0035824","log2FoldChange: 0.0741812766<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.6964391<br />gene: FBgn0035825","log2FoldChange: 0.0649811540<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.4565163<br />gene: FBgn0035827","log2FoldChange: 0.2563266907<br />-log10(padj): 5.293197e-01<br />log10(baseMean): 2.7462281<br />gene: FBgn0035829","log2FoldChange: 0.1013717717<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 3.0777528<br />gene: FBgn0035830","log2FoldChange: -0.1089986210<br />-log10(padj): 6.470652e-02<br />log10(baseMean): 2.3074588<br />gene: FBgn0035831","log2FoldChange: -0.2913792682<br />-log10(padj): 1.382248e-01<br />log10(baseMean): 1.9944419<br />gene: FBgn0035832","log2FoldChange: 0.0788453158<br />-log10(padj): 7.716672e-02<br />log10(baseMean): 2.7901682<br />gene: FBgn0035833","log2FoldChange: 0.0828552292<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 2.1490260<br />gene: FBgn0035838","log2FoldChange: 0.0539447850<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.7481537<br />gene: FBgn0035839","log2FoldChange: -0.0365222348<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 2.6265002<br />gene: FBgn0035842","log2FoldChange: -0.2059365705<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 1.5855481<br />gene: FBgn0035844","log2FoldChange: -0.2359364523<br />-log10(padj): 4.603572e-01<br />log10(baseMean): 2.5195913<br />gene: FBgn0035848","log2FoldChange: 0.0190055054<br />-log10(padj): 1.194862e-02<br />log10(baseMean): 2.6738600<br />gene: FBgn0035849","log2FoldChange: -0.3265350386<br />-log10(padj): 8.034442e-01<br />log10(baseMean): 3.1136016<br />gene: FBgn0035850","log2FoldChange: 0.1020620120<br />-log10(padj): 1.217867e-01<br />log10(baseMean): 2.8386510<br />gene: FBgn0035851","log2FoldChange: 0.4228271356<br />-log10(padj): 8.373395e-02<br />log10(baseMean): 1.0182603<br />gene: FBgn0035852","log2FoldChange: 0.1689694674<br />-log10(padj): 1.618824e-01<br />log10(baseMean): 2.8613391<br />gene: FBgn0035853","log2FoldChange: -0.0453397710<br />-log10(padj): 2.332938e-02<br />log10(baseMean): 2.2694208<br />gene: FBgn0035854","log2FoldChange: -0.4414301075<br />-log10(padj): 1.240841e-01<br />log10(baseMean): 1.1815596<br />gene: FBgn0035855","log2FoldChange: 0.0083840048<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 0.7964879<br />gene: FBgn0035856","log2FoldChange: -0.0929512962<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.4028269<br />gene: FBgn0035866","log2FoldChange: 0.0169017222<br />-log10(padj): 9.732012e-03<br />log10(baseMean): 2.9563832<br />gene: FBgn0035867","log2FoldChange: 0.0457265857<br />-log10(padj): 1.349861e-02<br />log10(baseMean): 1.9650998<br />gene: FBgn0035868","log2FoldChange: -0.0171874655<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.2210891<br />gene: FBgn0035871","log2FoldChange: -0.0708993262<br />-log10(padj): 5.702578e-02<br />log10(baseMean): 3.5121945<br />gene: FBgn0035872","log2FoldChange: -0.3006189951<br />-log10(padj): 2.840521e-01<br />log10(baseMean): 2.1349841<br />gene: FBgn0035876","log2FoldChange: -0.3648363538<br />-log10(padj): 6.929979e-01<br />log10(baseMean): 2.4831518<br />gene: FBgn0035877","log2FoldChange: -0.1134508920<br />-log10(padj): 1.141528e-01<br />log10(baseMean): 3.1894956<br />gene: FBgn0035878","log2FoldChange: -0.4847509243<br />-log10(padj): 3.858341e+00<br />log10(baseMean): 3.1378710<br />gene: FBgn0035879","log2FoldChange: 0.0996120872<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.0968576<br />gene: FBgn0035880","log2FoldChange: -0.1714420022<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.2278911<br />gene: FBgn0035888","log2FoldChange: 0.1620563377<br />-log10(padj): 1.553941e-01<br />log10(baseMean): 2.3400494<br />gene: FBgn0035890","log2FoldChange: 0.5185878544<br />-log10(padj): 9.122208e-01<br />log10(baseMean): 2.1196638<br />gene: FBgn0035891","log2FoldChange: -0.0405473711<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 2.7125360<br />gene: FBgn0035892","log2FoldChange: 0.0349881761<br />-log10(padj): 2.209430e-02<br />log10(baseMean): 3.6191460<br />gene: FBgn0035895","log2FoldChange: 0.0749445152<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.5439819<br />gene: FBgn0035896","log2FoldChange: -0.3227890571<br />-log10(padj): 8.258184e-02<br />log10(baseMean): 1.4065337<br />gene: FBgn0035898","log2FoldChange: 0.0919240067<br />-log10(padj): 6.787207e-02<br />log10(baseMean): 2.3225252<br />gene: FBgn0035900","log2FoldChange: 0.1852693654<br />-log10(padj): 2.202089e-01<br />log10(baseMean): 2.3939781<br />gene: FBgn0035901","log2FoldChange: 0.1440889225<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 1.7515519<br />gene: FBgn0035902","log2FoldChange: -0.7779533998<br />-log10(padj): 2.647498e+00<br />log10(baseMean): 2.0183863<br />gene: FBgn0035903","log2FoldChange: -0.1113532649<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 3.2700954<br />gene: FBgn0035904","log2FoldChange: -0.1640559583<br />-log10(padj): 2.395095e-01<br />log10(baseMean): 2.9373539<br />gene: FBgn0035906","log2FoldChange: 0.1816656428<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 1.9501973<br />gene: FBgn0035907","log2FoldChange: -0.0466711458<br />-log10(padj): 4.985877e-02<br />log10(baseMean): 3.5814269<br />gene: FBgn0035909","log2FoldChange: -0.1912940216<br />-log10(padj): 2.666288e-01<br />log10(baseMean): 2.5752282<br />gene: FBgn0035911","log2FoldChange: 0.0338877018<br />-log10(padj): 1.393747e-02<br />log10(baseMean): 2.4577356<br />gene: FBgn0035916","log2FoldChange: 0.0782211111<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.5862832<br />gene: FBgn0035918","log2FoldChange: -0.1918592588<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.7657406<br />gene: FBgn0035923","log2FoldChange: 1.5337751253<br />-log10(padj): 2.802954e+00<br />log10(baseMean): 1.4376438<br />gene: FBgn0035932","log2FoldChange: 1.3619923649<br />-log10(padj): 9.518441e-01<br />log10(baseMean): 1.0232108<br />gene: FBgn0035933","log2FoldChange: 0.0944442652<br />-log10(padj): 2.274835e-02<br />log10(baseMean): 1.4441942<br />gene: FBgn0035936","log2FoldChange: 0.5408890376<br />-log10(padj): 4.586956e+00<br />log10(baseMean): 2.9554641<br />gene: FBgn0035937","log2FoldChange: -0.0920143252<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 2.7334183<br />gene: FBgn0035938","log2FoldChange: 0.4428443404<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 1.0969776<br />gene: FBgn0035941","log2FoldChange: -0.0852153225<br />-log10(padj): 4.993802e-02<br />log10(baseMean): 2.1872831<br />gene: FBgn0035942","log2FoldChange: -0.0960524285<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 2.9072317<br />gene: FBgn0035944","log2FoldChange: -0.0572270566<br />-log10(padj): 5.729797e-02<br />log10(baseMean): 2.8640320<br />gene: FBgn0035945","log2FoldChange: 0.0726301684<br />-log10(padj): 7.339738e-02<br />log10(baseMean): 3.0377463<br />gene: FBgn0035947","log2FoldChange: 0.0598176983<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 0.9938912<br />gene: FBgn0035948","log2FoldChange: -0.0310643331<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.4859351<br />gene: FBgn0035949","log2FoldChange: 0.2409939728<br />-log10(padj): 5.228430e-01<br />log10(baseMean): 2.8059216<br />gene: FBgn0035950","log2FoldChange: 0.0721368517<br />-log10(padj): 6.793828e-02<br />log10(baseMean): 2.7879196<br />gene: FBgn0035951","log2FoldChange: -0.0723963922<br />-log10(padj): 7.894813e-02<br />log10(baseMean): 2.8428511<br />gene: FBgn0035953","log2FoldChange: 1.7338904195<br />-log10(padj): 1.727599e+00<br />log10(baseMean): 1.1525639<br />gene: FBgn0035957","log2FoldChange: -0.1874464907<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.7204657<br />gene: FBgn0035959","log2FoldChange: 0.2683345922<br />-log10(padj): 1.084626e-01<br />log10(baseMean): 1.6403593<br />gene: FBgn0035960","log2FoldChange: 0.0736442118<br />-log10(padj): 5.408986e-02<br />log10(baseMean): 2.5014267<br />gene: FBgn0035964","log2FoldChange: 0.2424018210<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.8617522<br />gene: FBgn0035965","log2FoldChange: 1.1073446819<br />-log10(padj): 1.290959e+01<br />log10(baseMean): 2.4699437<br />gene: FBgn0035968","log2FoldChange: 1.7634393829<br />-log10(padj): 1.650052e+00<br />log10(baseMean): 1.1110878<br />gene: FBgn0035969","log2FoldChange: 0.3664835443<br />-log10(padj): 6.017031e-01<br />log10(baseMean): 2.1131856<br />gene: FBgn0035975","log2FoldChange: 0.0954387343<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 2.4981209<br />gene: FBgn0035976","log2FoldChange: -0.2185198608<br />-log10(padj): 1.792343e-01<br />log10(baseMean): 2.1693994<br />gene: FBgn0035977","log2FoldChange: 0.1522678001<br />-log10(padj): 2.737850e-01<br />log10(baseMean): 2.8948449<br />gene: FBgn0035978","log2FoldChange: -0.1625669081<br />-log10(padj): 5.977921e-02<br />log10(baseMean): 1.6966386<br />gene: FBgn0035980","log2FoldChange: -0.0572040642<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.6604524<br />gene: FBgn0035981","log2FoldChange: -0.1271922837<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.3864065<br />gene: FBgn0035983","log2FoldChange: 0.3797386610<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 1.2020892<br />gene: FBgn0035985","log2FoldChange: 0.2250977682<br />-log10(padj): 6.838419e-01<br />log10(baseMean): 2.8201639<br />gene: FBgn0035986","log2FoldChange: -0.0238447424<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.9237520<br />gene: FBgn0035987","log2FoldChange: -0.4219533521<br />-log10(padj): 1.099102e-01<br />log10(baseMean): 1.1763865<br />gene: FBgn0035988","log2FoldChange: -0.1323588742<br />-log10(padj): 1.236393e-01<br />log10(baseMean): 2.5193543<br />gene: FBgn0035989","log2FoldChange: 0.0965337268<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.6359425<br />gene: FBgn0035993","log2FoldChange: 0.2031395973<br />-log10(padj): 2.401727e-01<br />log10(baseMean): 2.7374120<br />gene: FBgn0035995","log2FoldChange: 0.2151930789<br />-log10(padj): 1.115845e-01<br />log10(baseMean): 1.9393170<br />gene: FBgn0035996","log2FoldChange: -0.2340173381<br />-log10(padj): 5.014606e-01<br />log10(baseMean): 2.8908246<br />gene: FBgn0035997","log2FoldChange: 0.0481495473<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 1.7982024<br />gene: FBgn0035998","log2FoldChange: -0.0213313553<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.0094232<br />gene: FBgn0035999","log2FoldChange: -0.0141909619<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.4303467<br />gene: FBgn0036000","log2FoldChange: -0.2378524723<br />-log10(padj): 4.798402e-01<br />log10(baseMean): 3.2962565<br />gene: FBgn0036004","log2FoldChange: -0.0523001734<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.2554935<br />gene: FBgn0036005","log2FoldChange: 0.7925622936<br />-log10(padj): 6.385785e+00<br />log10(baseMean): 3.1013186<br />gene: FBgn0036007","log2FoldChange: -0.2149543861<br />-log10(padj): 1.222402e-01<br />log10(baseMean): 2.1241763<br />gene: FBgn0036008","log2FoldChange: 0.0030728385<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.1886450<br />gene: FBgn0036017","log2FoldChange: -0.2611864792<br />-log10(padj): 4.203212e-01<br />log10(baseMean): 2.5970323<br />gene: FBgn0036018","log2FoldChange: 0.5326192506<br />-log10(padj): 1.552426e-01<br />log10(baseMean): 1.2842155<br />gene: FBgn0036019","log2FoldChange: 0.0005307642<br />-log10(padj): 6.303468e-04<br />log10(baseMean): 2.4468005<br />gene: FBgn0036020","log2FoldChange: 0.1778635600<br />-log10(padj): 1.476243e-01<br />log10(baseMean): 2.1269102<br />gene: FBgn0036028","log2FoldChange: -0.0962189105<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.6622513<br />gene: FBgn0036030","log2FoldChange: -0.1318809100<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 2.8907699<br />gene: FBgn0036032","log2FoldChange: -0.1391175926<br />-log10(padj): 1.279068e-01<br />log10(baseMean): 2.5943618<br />gene: FBgn0036035","log2FoldChange: 0.2479936405<br />-log10(padj): 1.303420e-01<br />log10(baseMean): 1.8783346<br />gene: FBgn0036036","log2FoldChange: 0.0036171018<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 2.7069173<br />gene: FBgn0036038","log2FoldChange: -0.0752877493<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 2.4650272<br />gene: FBgn0036039","log2FoldChange: -0.1189373200<br />-log10(padj): 1.483407e-01<br />log10(baseMean): 3.3665560<br />gene: FBgn0036043","log2FoldChange: 0.1208745303<br />-log10(padj): 1.103685e-01<br />log10(baseMean): 2.4530724<br />gene: FBgn0036052","log2FoldChange: -0.3587273612<br />-log10(padj): 1.427539e+00<br />log10(baseMean): 3.3999337<br />gene: FBgn0036053","log2FoldChange: -0.0165496405<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 3.1676452<br />gene: FBgn0036058","log2FoldChange: -0.5037066102<br />-log10(padj): 1.833931e+00<br />log10(baseMean): 2.9002587<br />gene: FBgn0036059","log2FoldChange: 0.5823831952<br />-log10(padj): 4.139797e-01<br />log10(baseMean): 1.5135630<br />gene: FBgn0036062","log2FoldChange: -0.1273926967<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 1.9386162<br />gene: FBgn0036063","log2FoldChange: -0.2720684677<br />-log10(padj): 6.787207e-02<br />log10(baseMean): 1.2814157<br />gene: FBgn0036068","log2FoldChange: 0.2173493230<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 1.1377191<br />gene: FBgn0036082","log2FoldChange: -0.7455869227<br />-log10(padj): 1.311218e-01<br />log10(baseMean): 1.3371453<br />gene: FBgn0036083","log2FoldChange: -0.0667608211<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 2.2927928<br />gene: FBgn0036090","log2FoldChange: -0.1644973682<br />-log10(padj): 1.687510e-02<br />log10(baseMean): 0.7949761<br />gene: FBgn0036093","log2FoldChange: -0.2805131335<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 0.9201628<br />gene: FBgn0036094","log2FoldChange: 0.1874951980<br />-log10(padj): 3.660934e-01<br />log10(baseMean): 2.9886944<br />gene: FBgn0036096","log2FoldChange: -0.0052080891<br />-log10(padj): 3.088847e-03<br />log10(baseMean): 2.9466663<br />gene: FBgn0036099","log2FoldChange: -0.4973476800<br />-log10(padj): 8.522667e-01<br />log10(baseMean): 2.3200954<br />gene: FBgn0036101","log2FoldChange: 0.0672852464<br />-log10(padj): 7.142732e-02<br />log10(baseMean): 2.8007468<br />gene: FBgn0036104","log2FoldChange: 0.3074147646<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.7305112<br />gene: FBgn0036105","log2FoldChange: 0.0113473743<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.1206285<br />gene: FBgn0036107","log2FoldChange: -0.7290239497<br />-log10(padj): 7.894619e-01<br />log10(baseMean): 1.7542804<br />gene: FBgn0036110","log2FoldChange: -0.0334720118<br />-log10(padj): 3.120938e-02<br />log10(baseMean): 3.3938630<br />gene: FBgn0036111","log2FoldChange: -0.7973670194<br />-log10(padj): 6.813664e-01<br />log10(baseMean): 1.4459597<br />gene: FBgn0036116","log2FoldChange: -0.0354790750<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.2021487<br />gene: FBgn0036117","log2FoldChange: -0.0570558863<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 1.8717360<br />gene: FBgn0036118","log2FoldChange: -0.0223629245<br />-log10(padj): 1.242995e-02<br />log10(baseMean): 3.1699815<br />gene: FBgn0036121","log2FoldChange: 0.0273482536<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.4422232<br />gene: FBgn0036122","log2FoldChange: -0.2328186802<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 2.8572139<br />gene: FBgn0036124","log2FoldChange: 0.2252084305<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 2.1140194<br />gene: FBgn0036126","log2FoldChange: -0.1016861544<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 2.8239162<br />gene: FBgn0036133","log2FoldChange: -0.1623357589<br />-log10(padj): 3.259289e-01<br />log10(baseMean): 3.0574934<br />gene: FBgn0036134","log2FoldChange: 0.1297055675<br />-log10(padj): 7.848184e-02<br />log10(baseMean): 2.5953104<br />gene: FBgn0036135","log2FoldChange: 0.1370514970<br />-log10(padj): 1.798294e-01<br />log10(baseMean): 2.6869910<br />gene: FBgn0036136","log2FoldChange: 0.1125697392<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 3.0813709<br />gene: FBgn0036141","log2FoldChange: -0.0809183075<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 1.7701296<br />gene: FBgn0036142","log2FoldChange: 0.0114964620<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.3076404<br />gene: FBgn0036144","log2FoldChange: -0.9167573260<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 1.3703974<br />gene: FBgn0036145","log2FoldChange: -0.3088757851<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.5221953<br />gene: FBgn0036146","log2FoldChange: -0.3017249875<br />-log10(padj): 1.852747e+00<br />log10(baseMean): 3.2179190<br />gene: FBgn0036147","log2FoldChange: 0.4481684219<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.1158194<br />gene: FBgn0036150","log2FoldChange: -0.1626025794<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.2836061<br />gene: FBgn0036152","log2FoldChange: -0.3179167100<br />-log10(padj): 1.061206e-01<br />log10(baseMean): 1.4650348<br />gene: FBgn0036153","log2FoldChange: -0.1992693782<br />-log10(padj): 4.497479e-02<br />log10(baseMean): 1.3423643<br />gene: FBgn0036154","log2FoldChange: -0.5842402926<br />-log10(padj): 3.019219e-01<br />log10(baseMean): 1.8825509<br />gene: FBgn0036155","log2FoldChange: 0.1042513135<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.5319813<br />gene: FBgn0036159","log2FoldChange: -0.0255408931<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 0.7900112<br />gene: FBgn0036162","log2FoldChange: -0.0585284548<br />-log10(padj): 1.897739e-02<br />log10(baseMean): 2.6600629<br />gene: FBgn0036165","log2FoldChange: 0.1750636771<br />-log10(padj): 1.201659e-01<br />log10(baseMean): 2.6524029<br />gene: FBgn0036173","log2FoldChange: -0.0151128291<br />-log10(padj): 9.724218e-03<br />log10(baseMean): 2.9146838<br />gene: FBgn0036180","log2FoldChange: -0.8685653067<br />-log10(padj): 2.488595e-01<br />log10(baseMean): 1.9515933<br />gene: FBgn0036181","log2FoldChange: 0.0544597408<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.3378748<br />gene: FBgn0036184","log2FoldChange: -0.0390900158<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.0680647<br />gene: FBgn0036186","log2FoldChange: -0.2291694213<br />-log10(padj): 4.614889e-01<br />log10(baseMean): 2.5619913<br />gene: FBgn0036187","log2FoldChange: 0.1748122774<br />-log10(padj): 2.222556e-01<br />log10(baseMean): 2.5050889<br />gene: FBgn0036188","log2FoldChange: -0.2250586977<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.8086212<br />gene: FBgn0036191","log2FoldChange: -0.1194555263<br />-log10(padj): 2.540586e-02<br />log10(baseMean): 1.4359400<br />gene: FBgn0036192","log2FoldChange: 0.0396039825<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.5264129<br />gene: FBgn0036193","log2FoldChange: -0.3114438717<br />-log10(padj): 8.273230e-01<br />log10(baseMean): 2.9616059<br />gene: FBgn0036194","log2FoldChange: -0.6153351759<br />-log10(padj): 1.511357e+00<br />log10(baseMean): 2.2162419<br />gene: FBgn0036196","log2FoldChange: 0.5153922520<br />-log10(padj): 7.592377e-01<br />log10(baseMean): 2.0195785<br />gene: FBgn0036198","log2FoldChange: -0.3274490912<br />-log10(padj): 1.127608e+00<br />log10(baseMean): 2.6953743<br />gene: FBgn0036199","log2FoldChange: 0.1564670825<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 2.3526639<br />gene: FBgn0036207","log2FoldChange: -0.1371597382<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.9479768<br />gene: FBgn0036210","log2FoldChange: -0.2401316602<br />-log10(padj): 5.969497e-01<br />log10(baseMean): 2.8576687<br />gene: FBgn0036211","log2FoldChange: 0.3996576270<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 1.3193703<br />gene: FBgn0036212","log2FoldChange: -0.0027468954<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 4.2830131<br />gene: FBgn0036213","log2FoldChange: -0.4050673290<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.2448083<br />gene: FBgn0036214","log2FoldChange: 0.0231455893<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.0788913<br />gene: FBgn0036237","log2FoldChange: -0.1143922356<br />-log10(padj): 1.602229e-01<br />log10(baseMean): 3.0981620<br />gene: FBgn0036239","log2FoldChange: 0.1460986536<br />-log10(padj): 1.163594e-01<br />log10(baseMean): 2.4372172<br />gene: FBgn0036240","log2FoldChange: -0.0737936831<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 2.4602555<br />gene: FBgn0036248","log2FoldChange: 0.0431346066<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.1711246<br />gene: FBgn0036249","log2FoldChange: -0.3448267407<br />-log10(padj): 5.344507e-01<br />log10(baseMean): 2.5786578<br />gene: FBgn0036254","log2FoldChange: -0.2096379904<br />-log10(padj): 5.112058e-02<br />log10(baseMean): 1.6600886<br />gene: FBgn0036255","log2FoldChange: -0.3527224988<br />-log10(padj): 2.246697e+00<br />log10(baseMean): 3.0349957<br />gene: FBgn0036257","log2FoldChange: 0.0572156537<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 3.6131534<br />gene: FBgn0036258","log2FoldChange: 0.1121967026<br />-log10(padj): 2.499648e-02<br />log10(baseMean): 1.4278357<br />gene: FBgn0036260","log2FoldChange: -0.1238054789<br />-log10(padj): 1.609026e-02<br />log10(baseMean): 0.9826697<br />gene: FBgn0036262","log2FoldChange: 0.0330886584<br />-log10(padj): 1.708130e-02<br />log10(baseMean): 2.5334048<br />gene: FBgn0036263","log2FoldChange: 0.0860877494<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 1.8542829<br />gene: FBgn0036266","log2FoldChange: 0.1391464332<br />-log10(padj): 1.268350e-01<br />log10(baseMean): 2.6910500<br />gene: FBgn0036271","log2FoldChange: 0.3549334114<br />-log10(padj): 1.819569e+00<br />log10(baseMean): 2.9418421<br />gene: FBgn0036272","log2FoldChange: -0.1221301938<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.1307283<br />gene: FBgn0036273","log2FoldChange: 0.3321260307<br />-log10(padj): 6.964581e-01<br />log10(baseMean): 2.5036902<br />gene: FBgn0036277","log2FoldChange: -0.4233712912<br />-log10(padj): 1.668830e+00<br />log10(baseMean): 2.9645608<br />gene: FBgn0036279","log2FoldChange: 0.7065666272<br />-log10(padj): 3.182794e-01<br />log10(baseMean): 1.1791112<br />gene: FBgn0036282","log2FoldChange: -0.0152511278<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.7754793<br />gene: FBgn0036286","log2FoldChange: 1.5239037596<br />-log10(padj): 1.163612e+00<br />log10(baseMean): 1.1517336<br />gene: FBgn0036287","log2FoldChange: -0.1862062827<br />-log10(padj): 1.230588e-01<br />log10(baseMean): 1.9887099<br />gene: FBgn0036288","log2FoldChange: 0.3154873792<br />-log10(padj): 2.427348e-01<br />log10(baseMean): 1.7906126<br />gene: FBgn0036289","log2FoldChange: 0.3974018902<br />-log10(padj): 1.828944e+00<br />log10(baseMean): 3.0691863<br />gene: FBgn0036290","log2FoldChange: 0.0432108895<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 1.9663686<br />gene: FBgn0036291","log2FoldChange: 0.0739896499<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 2.2509093<br />gene: FBgn0036292","log2FoldChange: -0.0165213020<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.8708714<br />gene: FBgn0036298","log2FoldChange: 0.9397715307<br />-log10(padj): 7.646732e+00<br />log10(baseMean): 2.8163275<br />gene: FBgn0036299","log2FoldChange: 0.1066044436<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.5541199<br />gene: FBgn0036300","log2FoldChange: -0.1789028795<br />-log10(padj): 2.007831e-01<br />log10(baseMean): 2.4842086<br />gene: FBgn0036301","log2FoldChange: -0.1294417460<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 3.1270058<br />gene: FBgn0036302","log2FoldChange: -0.0491086907<br />-log10(padj): 2.950840e-02<br />log10(baseMean): 2.6787175<br />gene: FBgn0036305","log2FoldChange: -0.1189962880<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 2.4608130<br />gene: FBgn0036306","log2FoldChange: -0.1227885976<br />-log10(padj): 6.237762e-02<br />log10(baseMean): 3.0742545<br />gene: FBgn0036309","log2FoldChange: -0.2947130254<br />-log10(padj): 5.108891e-01<br />log10(baseMean): 2.7002969<br />gene: FBgn0036312","log2FoldChange: 0.0536727134<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.8212826<br />gene: FBgn0036314","log2FoldChange: 0.3426157666<br />-log10(padj): 7.332208e-01<br />log10(baseMean): 3.6161559<br />gene: FBgn0036316","log2FoldChange: 0.3306179239<br />-log10(padj): 1.414546e+00<br />log10(baseMean): 2.7262660<br />gene: FBgn0036317","log2FoldChange: -0.2170317419<br />-log10(padj): 7.724669e-01<br />log10(baseMean): 3.2321404<br />gene: FBgn0036318","log2FoldChange: 0.1219184982<br />-log10(padj): 1.619278e-02<br />log10(baseMean): 0.9083707<br />gene: FBgn0036319","log2FoldChange: 0.2950297345<br />-log10(padj): 4.613482e-02<br />log10(baseMean): 1.0749624<br />gene: FBgn0036324","log2FoldChange: -0.0032911839<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.7393573<br />gene: FBgn0036331","log2FoldChange: -0.2516217752<br />-log10(padj): 1.238949e-01<br />log10(baseMean): 2.7276930<br />gene: FBgn0036332","log2FoldChange: 0.0366610701<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.9653772<br />gene: FBgn0036333","log2FoldChange: -0.1434533879<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 3.0820342<br />gene: FBgn0036334","log2FoldChange: 0.3109864386<br />-log10(padj): 3.982016e-01<br />log10(baseMean): 2.1232888<br />gene: FBgn0036335","log2FoldChange: 0.0243841564<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.4974884<br />gene: FBgn0036336","log2FoldChange: -0.2693652323<br />-log10(padj): 8.389397e-01<br />log10(baseMean): 2.8441902<br />gene: FBgn0036337","log2FoldChange: -0.0778331482<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.3464970<br />gene: FBgn0036338","log2FoldChange: 0.0659481513<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 3.2487199<br />gene: FBgn0036340","log2FoldChange: -0.1575741724<br />-log10(padj): 2.426927e-01<br />log10(baseMean): 2.8632500<br />gene: FBgn0036341","log2FoldChange: -0.1511297621<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.2162578<br />gene: FBgn0036342","log2FoldChange: 0.3220097080<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 2.6100782<br />gene: FBgn0036353","log2FoldChange: -0.3462553850<br />-log10(padj): 8.111198e-01<br />log10(baseMean): 3.0477825<br />gene: FBgn0036354","log2FoldChange: 0.3178390304<br />-log10(padj): 4.125902e-01<br />log10(baseMean): 2.1004787<br />gene: FBgn0036356","log2FoldChange: 0.3014759512<br />-log10(padj): 9.833063e-01<br />log10(baseMean): 3.0034787<br />gene: FBgn0036365","log2FoldChange: 0.0853003612<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.2811008<br />gene: FBgn0036366","log2FoldChange: 0.1923413558<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 1.7952782<br />gene: FBgn0036368","log2FoldChange: 0.0183170731<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.2911357<br />gene: FBgn0036369","log2FoldChange: 0.2385353781<br />-log10(padj): 7.133393e-01<br />log10(baseMean): 3.1204367<br />gene: FBgn0036372","log2FoldChange: -0.9787105654<br />-log10(padj): 3.829703e+00<br />log10(baseMean): 3.5864792<br />gene: FBgn0036373","log2FoldChange: -0.0769608507<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.0985111<br />gene: FBgn0036374","log2FoldChange: -0.1357687983<br />-log10(padj): 3.396391e-01<br />log10(baseMean): 3.3463988<br />gene: FBgn0036376","log2FoldChange: -1.3866729215<br />-log10(padj): 5.631643e-01<br />log10(baseMean): 0.8403371<br />gene: FBgn0036379","log2FoldChange: 0.0734114704<br />-log10(padj): 2.373932e-02<br />log10(baseMean): 2.1361930<br />gene: FBgn0036381","log2FoldChange: 0.1906135262<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 1.5146492<br />gene: FBgn0036382","log2FoldChange: 0.0112957716<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 2.4152036<br />gene: FBgn0036386","log2FoldChange: 0.2576550915<br />-log10(padj): 1.068433e+00<br />log10(baseMean): 3.3699330<br />gene: FBgn0036389","log2FoldChange: -0.0404440589<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.8019313<br />gene: FBgn0036395","log2FoldChange: 0.0123536292<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 1.9856306<br />gene: FBgn0036396","log2FoldChange: 0.2404437721<br />-log10(padj): 2.388550e-01<br />log10(baseMean): 2.3782170<br />gene: FBgn0036397","log2FoldChange: -0.2752034505<br />-log10(padj): 5.279329e-01<br />log10(baseMean): 3.6804635<br />gene: FBgn0036398","log2FoldChange: 0.0157519390<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.1997398<br />gene: FBgn0036402","log2FoldChange: 0.1173283352<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.4841626<br />gene: FBgn0036405","log2FoldChange: -0.0673454687<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 1.9751673<br />gene: FBgn0036406","log2FoldChange: 0.0346949617<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.2395379<br />gene: FBgn0036410","log2FoldChange: -1.1709957240<br />-log10(padj): 4.071180e-01<br />log10(baseMean): 0.8245537<br />gene: FBgn0036419","log2FoldChange: 0.1381390143<br />-log10(padj): 6.811621e-02<br />log10(baseMean): 1.9226879<br />gene: FBgn0036423","log2FoldChange: -0.3114671011<br />-log10(padj): 2.426927e-01<br />log10(baseMean): 2.1889804<br />gene: FBgn0036428","log2FoldChange: -0.1634598701<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.5711170<br />gene: FBgn0036433","log2FoldChange: 0.1004658040<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.7166644<br />gene: FBgn0036446","log2FoldChange: -0.0372464767<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.5154124<br />gene: FBgn0036448","log2FoldChange: 0.3924985757<br />-log10(padj): 2.356496e+00<br />log10(baseMean): 3.1978092<br />gene: FBgn0036449","log2FoldChange: 0.0350674092<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 3.0166225<br />gene: FBgn0036450","log2FoldChange: -0.0410004127<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 3.6061797<br />gene: FBgn0036451","log2FoldChange: -0.2088598835<br />-log10(padj): 5.490546e-02<br />log10(baseMean): 2.8633326<br />gene: FBgn0036454","log2FoldChange: -0.3158354351<br />-log10(padj): 1.074738e+00<br />log10(baseMean): 2.8164077<br />gene: FBgn0036460","log2FoldChange: 0.0172397387<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 0.9976126<br />gene: FBgn0036461","log2FoldChange: 0.1870463078<br />-log10(padj): 3.352636e-01<br />log10(baseMean): 2.7860802<br />gene: FBgn0036462","log2FoldChange: 0.2115832692<br />-log10(padj): 3.996402e-01<br />log10(baseMean): 2.7960023<br />gene: FBgn0036463","log2FoldChange: -0.0138637112<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.4300998<br />gene: FBgn0036476","log2FoldChange: -0.2535258093<br />-log10(padj): 9.833063e-01<br />log10(baseMean): 3.0561534<br />gene: FBgn0036478","log2FoldChange: 0.1964377524<br />-log10(padj): 4.740326e-01<br />log10(baseMean): 2.7794102<br />gene: FBgn0036480","log2FoldChange: 0.6939225472<br />-log10(padj): 3.075858e-01<br />log10(baseMean): 1.1280120<br />gene: FBgn0036481","log2FoldChange: 0.0258860911<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.5794260<br />gene: FBgn0036483","log2FoldChange: 0.0010055937<br />-log10(padj): 1.175523e-03<br />log10(baseMean): 2.9352689<br />gene: FBgn0036484","log2FoldChange: -0.0022609550<br />-log10(padj): 1.883530e-03<br />log10(baseMean): 3.0574888<br />gene: FBgn0036486","log2FoldChange: 0.1432383029<br />-log10(padj): 2.605771e-01<br />log10(baseMean): 2.9144948<br />gene: FBgn0036487","log2FoldChange: 0.2246271349<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 2.6005477<br />gene: FBgn0036488","log2FoldChange: -0.0969818344<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.5904315<br />gene: FBgn0036489","log2FoldChange: -0.0636613599<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 2.1184437<br />gene: FBgn0036493","log2FoldChange: -0.0209699046<br />-log10(padj): 1.186237e-02<br />log10(baseMean): 2.6046441<br />gene: FBgn0036500","log2FoldChange: -0.2124163161<br />-log10(padj): 3.492311e-01<br />log10(baseMean): 2.7080226<br />gene: FBgn0036501","log2FoldChange: -0.0275411317<br />-log10(padj): 1.083072e-02<br />log10(baseMean): 2.1148023<br />gene: FBgn0036502","log2FoldChange: -0.1282899738<br />-log10(padj): 1.245608e-01<br />log10(baseMean): 3.1236267<br />gene: FBgn0036505","log2FoldChange: 0.0508737197<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 3.0958594<br />gene: FBgn0036509","log2FoldChange: 0.0195579358<br />-log10(padj): 9.200321e-03<br />log10(baseMean): 2.6659010<br />gene: FBgn0036510","log2FoldChange: -0.1181803347<br />-log10(padj): 1.399139e-01<br />log10(baseMean): 3.3253285<br />gene: FBgn0036511","log2FoldChange: 0.0372865137<br />-log10(padj): 1.952727e-02<br />log10(baseMean): 2.6267648<br />gene: FBgn0036512","log2FoldChange: -0.0854737817<br />-log10(padj): 5.452627e-02<br />log10(baseMean): 2.7554742<br />gene: FBgn0036514","log2FoldChange: 0.1413082141<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 2.8438181<br />gene: FBgn0036515","log2FoldChange: 0.0876729161<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 2.9766761<br />gene: FBgn0036516","log2FoldChange: -0.1850865693<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 3.5085617<br />gene: FBgn0036518","log2FoldChange: -0.0973177641<br />-log10(padj): 7.679941e-02<br />log10(baseMean): 2.4448376<br />gene: FBgn0036519","log2FoldChange: -0.4472036748<br />-log10(padj): 1.032628e-01<br />log10(baseMean): 1.0517351<br />gene: FBgn0036520","log2FoldChange: 0.0513844483<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 2.6611698<br />gene: FBgn0036522","log2FoldChange: 0.0777747240<br />-log10(padj): 8.535439e-02<br />log10(baseMean): 2.9201905<br />gene: FBgn0036533","log2FoldChange: -0.0141233944<br />-log10(padj): 7.543257e-03<br />log10(baseMean): 3.3897853<br />gene: FBgn0036534","log2FoldChange: -0.2776733778<br />-log10(padj): 3.982645e-01<br />log10(baseMean): 2.1765654<br />gene: FBgn0036536","log2FoldChange: -0.0379547192<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.5979112<br />gene: FBgn0036537","log2FoldChange: 0.0423138968<br />-log10(padj): 2.075232e-02<br />log10(baseMean): 2.4925895<br />gene: FBgn0036538","log2FoldChange: 0.5902912871<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 0.8030368<br />gene: FBgn0036544","log2FoldChange: 0.0969869648<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.5228393<br />gene: FBgn0036545","log2FoldChange: 0.3686974853<br />-log10(padj): 9.062172e-01<br />log10(baseMean): 2.3319318<br />gene: FBgn0036546","log2FoldChange: 0.2303950896<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.4121880<br />gene: FBgn0036547","log2FoldChange: 0.0598768103<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.6689920<br />gene: FBgn0036548","log2FoldChange: -0.2746430545<br />-log10(padj): 4.107652e-01<br />log10(baseMean): 2.3061927<br />gene: FBgn0036549","log2FoldChange: -0.0169985191<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.0058760<br />gene: FBgn0036551","log2FoldChange: -0.3594174577<br />-log10(padj): 6.008733e-02<br />log10(baseMean): 0.9116353<br />gene: FBgn0036552","log2FoldChange: -0.1487664300<br />-log10(padj): 7.443250e-02<br />log10(baseMean): 2.9150118<br />gene: FBgn0036556","log2FoldChange: 0.0016282807<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 2.6573821<br />gene: FBgn0036557","log2FoldChange: 0.2088563896<br />-log10(padj): 2.182740e-01<br />log10(baseMean): 3.5693302<br />gene: FBgn0036558","log2FoldChange: 2.1472273788<br />-log10(padj): 3.527306e+00<br />log10(baseMean): 1.2763458<br />gene: FBgn0036560","log2FoldChange: 0.1879585061<br />-log10(padj): 2.697490e-01<br />log10(baseMean): 2.5998036<br />gene: FBgn0036564","log2FoldChange: 0.2339179217<br />-log10(padj): 1.748230e-01<br />log10(baseMean): 3.0037800<br />gene: FBgn0036565","log2FoldChange: -0.0190052960<br />-log10(padj): 1.224480e-02<br />log10(baseMean): 3.4697604<br />gene: FBgn0036566","log2FoldChange: 0.1215922760<br />-log10(padj): 1.672272e-01<br />log10(baseMean): 2.8860378<br />gene: FBgn0036567","log2FoldChange: 0.1960207750<br />-log10(padj): 2.522953e-01<br />log10(baseMean): 2.5621424<br />gene: FBgn0036568","log2FoldChange: -0.1037048376<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.2873045<br />gene: FBgn0036569","log2FoldChange: 0.1652330431<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 2.5991156<br />gene: FBgn0036570","log2FoldChange: -0.0277844912<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.4912291<br />gene: FBgn0036571","log2FoldChange: 0.0178311340<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 1.9399633<br />gene: FBgn0036573","log2FoldChange: -0.1975937553<br />-log10(padj): 2.817945e-01<br />log10(baseMean): 2.8803587<br />gene: FBgn0036574","log2FoldChange: 0.1326223712<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 1.2677116<br />gene: FBgn0036575","log2FoldChange: -1.0328772277<br />-log10(padj): 4.282691e+00<br />log10(baseMean): 3.2493519<br />gene: FBgn0036576","log2FoldChange: -0.1773799980<br />-log10(padj): 1.578071e-01<br />log10(baseMean): 2.4421727<br />gene: FBgn0036578","log2FoldChange: 0.0693112538<br />-log10(padj): 5.091132e-02<br />log10(baseMean): 2.4549783<br />gene: FBgn0036579","log2FoldChange: 0.1130341965<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 2.5213196<br />gene: FBgn0036580","log2FoldChange: 0.0322771255<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.4409741<br />gene: FBgn0036581","log2FoldChange: -0.0985455257<br />-log10(padj): 6.895106e-02<br />log10(baseMean): 2.4351061<br />gene: FBgn0036614","log2FoldChange: 0.2702381789<br />-log10(padj): 3.182794e-01<br />log10(baseMean): 2.2793724<br />gene: FBgn0036615","log2FoldChange: -0.8546941499<br />-log10(padj): 2.802954e+00<br />log10(baseMean): 2.0679453<br />gene: FBgn0036616","log2FoldChange: 0.7511016302<br />-log10(padj): 9.152652e-01<br />log10(baseMean): 1.6105058<br />gene: FBgn0036620","log2FoldChange: -0.2431930941<br />-log10(padj): 9.527824e-01<br />log10(baseMean): 3.2253433<br />gene: FBgn0036621","log2FoldChange: -0.1459871207<br />-log10(padj): 6.482097e-02<br />log10(baseMean): 1.8483551<br />gene: FBgn0036622","log2FoldChange: 0.0451894308<br />-log10(padj): 5.164392e-02<br />log10(baseMean): 3.3897836<br />gene: FBgn0036623","log2FoldChange: -0.1239421417<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.8593408<br />gene: FBgn0036624","log2FoldChange: -0.0964906306<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 1.8837257<br />gene: FBgn0036629","log2FoldChange: 0.2975792281<br />-log10(padj): 6.520807e-01<br />log10(baseMean): 2.4268369<br />gene: FBgn0036640","log2FoldChange: 0.7385880967<br />-log10(padj): 3.690924e+00<br />log10(baseMean): 2.3486584<br />gene: FBgn0036641","log2FoldChange: -0.2857454072<br />-log10(padj): 3.844517e-01<br />log10(baseMean): 2.4183199<br />gene: FBgn0036643","log2FoldChange: 0.4591959512<br />-log10(padj): 2.666288e-01<br />log10(baseMean): 2.1938067<br />gene: FBgn0036646","log2FoldChange: 0.0594698960<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.2852962<br />gene: FBgn0036648","log2FoldChange: 0.1248022256<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 0.8664641<br />gene: FBgn0036652","log2FoldChange: 0.4392213711<br />-log10(padj): 8.334717e-02<br />log10(baseMean): 0.9966379<br />gene: FBgn0036654","log2FoldChange: 0.3411029234<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 1.2908901<br />gene: FBgn0036655","log2FoldChange: 0.1974477622<br />-log10(padj): 2.962956e-01<br />log10(baseMean): 2.3693803<br />gene: FBgn0036660","log2FoldChange: -0.0839190654<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 3.2229228<br />gene: FBgn0036661","log2FoldChange: -0.1066671568<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.5541559<br />gene: FBgn0036662","log2FoldChange: -0.6630520588<br />-log10(padj): 4.885920e+00<br />log10(baseMean): 3.8854143<br />gene: FBgn0036663","log2FoldChange: -0.1476301252<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 3.2084617<br />gene: FBgn0036666","log2FoldChange: 0.1014702504<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 2.2889735<br />gene: FBgn0036667","log2FoldChange: -0.1287113558<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.2860557<br />gene: FBgn0036668","log2FoldChange: 0.2041953222<br />-log10(padj): 3.425529e-01<br />log10(baseMean): 2.6719892<br />gene: FBgn0036671","log2FoldChange: -0.8795386393<br />-log10(padj): 1.141165e+01<br />log10(baseMean): 2.9582060<br />gene: FBgn0036684","log2FoldChange: 0.1045369020<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 3.1893136<br />gene: FBgn0036685","log2FoldChange: -0.2185966742<br />-log10(padj): 8.639578e-01<br />log10(baseMean): 3.1711248<br />gene: FBgn0036686","log2FoldChange: 0.6234896883<br />-log10(padj): 2.883483e+00<br />log10(baseMean): 2.5741092<br />gene: FBgn0036688","log2FoldChange: -0.0002289573<br />-log10(padj): 3.176531e-04<br />log10(baseMean): 2.0702605<br />gene: FBgn0036689","log2FoldChange: -0.0087549205<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 2.4390647<br />gene: FBgn0036691","log2FoldChange: -0.0910890884<br />-log10(padj): 7.066721e-02<br />log10(baseMean): 2.5396021<br />gene: FBgn0036695","log2FoldChange: 0.2752640529<br />-log10(padj): 3.982645e-01<br />log10(baseMean): 2.1750975<br />gene: FBgn0036696","log2FoldChange: 0.0600349462<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 2.7092483<br />gene: FBgn0036697","log2FoldChange: 0.2938433010<br />-log10(padj): 1.160183e+00<br />log10(baseMean): 3.4926740<br />gene: FBgn0036702","log2FoldChange: -0.0547464070<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.8839788<br />gene: FBgn0036710","log2FoldChange: 0.0385176465<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.5953487<br />gene: FBgn0036714","log2FoldChange: 0.0884264449<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.5825614<br />gene: FBgn0036723","log2FoldChange: -0.0215143508<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.6760527<br />gene: FBgn0036726","log2FoldChange: 0.1948528052<br />-log10(padj): 2.399073e-01<br />log10(baseMean): 3.1683486<br />gene: FBgn0036728","log2FoldChange: -0.5601851136<br />-log10(padj): 3.912562e+00<br />log10(baseMean): 3.3885401<br />gene: FBgn0036732","log2FoldChange: 0.0567252985<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.6894771<br />gene: FBgn0036733","log2FoldChange: -0.2000301320<br />-log10(padj): 2.167041e-01<br />log10(baseMean): 3.0135554<br />gene: FBgn0036734","log2FoldChange: -0.1337781552<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.9680773<br />gene: FBgn0036735","log2FoldChange: 0.0614612616<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.9818939<br />gene: FBgn0036740","log2FoldChange: -0.2235771202<br />-log10(padj): 3.966458e-01<br />log10(baseMean): 3.0208232<br />gene: FBgn0036741","log2FoldChange: 0.0274303137<br />-log10(padj): 4.547574e-03<br />log10(baseMean): 1.4280066<br />gene: FBgn0036742","log2FoldChange: 0.1139800743<br />-log10(padj): 1.032218e-01<br />log10(baseMean): 2.9728962<br />gene: FBgn0036745","log2FoldChange: -0.4591716485<br />-log10(padj): 9.992028e-01<br />log10(baseMean): 3.0830023<br />gene: FBgn0036746","log2FoldChange: -0.4065245629<br />-log10(padj): 3.134901e-01<br />log10(baseMean): 1.6277555<br />gene: FBgn0036749","log2FoldChange: 0.4559880294<br />-log10(padj): 1.703374e+00<br />log10(baseMean): 3.7688105<br />gene: FBgn0036752","log2FoldChange: -0.0951514464<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.3180748<br />gene: FBgn0036754","log2FoldChange: -0.1282246634<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 1.5789930<br />gene: FBgn0036759","log2FoldChange: -0.0528745420<br />-log10(padj): 2.986811e-02<br />log10(baseMean): 2.3617457<br />gene: FBgn0036760","log2FoldChange: -0.0760837505<br />-log10(padj): 8.670466e-02<br />log10(baseMean): 2.9626400<br />gene: FBgn0036761","log2FoldChange: -0.2523629916<br />-log10(padj): 4.334437e-01<br />log10(baseMean): 3.4717109<br />gene: FBgn0036762","log2FoldChange: 0.0267413100<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.6988615<br />gene: FBgn0036763","log2FoldChange: 0.4147577514<br />-log10(padj): 1.675065e+00<br />log10(baseMean): 3.5626554<br />gene: FBgn0036764","log2FoldChange: 0.3088535625<br />-log10(padj): 4.255595e-01<br />log10(baseMean): 2.3576690<br />gene: FBgn0036765","log2FoldChange: 0.5419840165<br />-log10(padj): 8.626754e-02<br />log10(baseMean): 0.7937381<br />gene: FBgn0036769","log2FoldChange: -0.4025707386<br />-log10(padj): 2.142626e+00<br />log10(baseMean): 3.2294178<br />gene: FBgn0036770","log2FoldChange: 0.2748888666<br />-log10(padj): 4.125902e-01<br />log10(baseMean): 2.2675832<br />gene: FBgn0036771","log2FoldChange: -0.2362940675<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.9159168<br />gene: FBgn0036772","log2FoldChange: -0.0178991934<br />-log10(padj): 9.060734e-03<br />log10(baseMean): 2.6751308<br />gene: FBgn0036774","log2FoldChange: -0.2261155884<br />-log10(padj): 3.042328e-01<br />log10(baseMean): 2.6790376<br />gene: FBgn0036775","log2FoldChange: -0.2676151600<br />-log10(padj): 1.576016e-01<br />log10(baseMean): 1.7967213<br />gene: FBgn0036777","log2FoldChange: -1.0220815597<br />-log10(padj): 1.467019e+00<br />log10(baseMean): 1.5305261<br />gene: FBgn0036801","log2FoldChange: 0.3477946646<br />-log10(padj): 5.991162e-01<br />log10(baseMean): 2.2522519<br />gene: FBgn0036804","log2FoldChange: -0.1407552114<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 2.8783723<br />gene: FBgn0036805","log2FoldChange: -0.0611915904<br />-log10(padj): 5.091132e-02<br />log10(baseMean): 2.6091491<br />gene: FBgn0036806","log2FoldChange: 0.4816087331<br />-log10(padj): 2.869327e-01<br />log10(baseMean): 1.4760756<br />gene: FBgn0036809","log2FoldChange: 0.2762233131<br />-log10(padj): 3.027591e-01<br />log10(baseMean): 2.2414196<br />gene: FBgn0036811","log2FoldChange: -0.0441685709<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 1.9028248<br />gene: FBgn0036812","log2FoldChange: -0.1319795054<br />-log10(padj): 1.397724e-01<br />log10(baseMean): 2.7109073<br />gene: FBgn0036813","log2FoldChange: -0.1985040341<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.2283554<br />gene: FBgn0036814","log2FoldChange: -0.1526619237<br />-log10(padj): 2.035081e-01<br />log10(baseMean): 2.6841794<br />gene: FBgn0036815","log2FoldChange: -0.1365176371<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 3.5740675<br />gene: FBgn0036816","log2FoldChange: 0.2878993182<br />-log10(padj): 1.062820e-01<br />log10(baseMean): 1.5062671<br />gene: FBgn0036817","log2FoldChange: 0.0406765345<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.7089013<br />gene: FBgn0036818","log2FoldChange: 0.0376506876<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 1.5988117<br />gene: FBgn0036819","log2FoldChange: -0.0347202462<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.8142653<br />gene: FBgn0036820","log2FoldChange: -1.7432882071<br />-log10(padj): 2.952011e+00<br />log10(baseMean): 2.6194161<br />gene: FBgn0036821","log2FoldChange: -0.6120364149<br />-log10(padj): 3.248076e+00<br />log10(baseMean): 3.4771408<br />gene: FBgn0036824","log2FoldChange: -0.1951360587<br />-log10(padj): 1.622799e-01<br />log10(baseMean): 4.4781455<br />gene: FBgn0036825","log2FoldChange: 0.0499123653<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.1898171<br />gene: FBgn0036826","log2FoldChange: 0.0382283970<br />-log10(padj): 1.897739e-02<br />log10(baseMean): 2.4309495<br />gene: FBgn0036827","log2FoldChange: -0.0818758416<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.7311967<br />gene: FBgn0036828","log2FoldChange: -0.2492564966<br />-log10(padj): 1.026860e-01<br />log10(baseMean): 1.8317094<br />gene: FBgn0036837","log2FoldChange: 0.2094298700<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 2.1433316<br />gene: FBgn0036838","log2FoldChange: -0.0013853835<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 3.0693760<br />gene: FBgn0036842","log2FoldChange: 0.2408749588<br />-log10(padj): 1.468821e-01<br />log10(baseMean): 2.3400404<br />gene: FBgn0036843","log2FoldChange: 0.0757705027<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.1834154<br />gene: FBgn0036844","log2FoldChange: -0.1804509238<br />-log10(padj): 3.188201e-01<br />log10(baseMean): 2.6533977<br />gene: FBgn0036846","log2FoldChange: 0.1270436588<br />-log10(padj): 1.179979e-01<br />log10(baseMean): 2.9534566<br />gene: FBgn0036847","log2FoldChange: 0.1925672951<br />-log10(padj): 2.053470e-01<br />log10(baseMean): 2.6042844<br />gene: FBgn0036848","log2FoldChange: -0.0185058720<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 1.6361465<br />gene: FBgn0036849","log2FoldChange: 0.3037563187<br />-log10(padj): 4.076629e-01<br />log10(baseMean): 2.1734985<br />gene: FBgn0036850","log2FoldChange: -0.0099782716<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.1766275<br />gene: FBgn0036853","log2FoldChange: -0.3908786890<br />-log10(padj): 1.842568e+00<br />log10(baseMean): 2.6631028<br />gene: FBgn0036857","log2FoldChange: 1.0875666828<br />-log10(padj): 3.052884e+00<br />log10(baseMean): 2.4167328<br />gene: FBgn0036862","log2FoldChange: 0.5018719806<br />-log10(padj): 8.338134e-02<br />log10(baseMean): 0.8118252<br />gene: FBgn0036875","log2FoldChange: -0.3814264128<br />-log10(padj): 1.618824e-01<br />log10(baseMean): 1.4757420<br />gene: FBgn0036882","log2FoldChange: -0.1979711864<br />-log10(padj): 2.778236e-01<br />log10(baseMean): 2.4762552<br />gene: FBgn0036886","log2FoldChange: -0.0167108065<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.2585358<br />gene: FBgn0036887","log2FoldChange: -0.0074262397<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.2069104<br />gene: FBgn0036888","log2FoldChange: 0.1377101877<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.0854177<br />gene: FBgn0036889","log2FoldChange: -0.0076293416<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 3.4263975<br />gene: FBgn0036891","log2FoldChange: 0.0419823755<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 2.9857673<br />gene: FBgn0036892","log2FoldChange: 0.5414206070<br />-log10(padj): 2.101963e+00<br />log10(baseMean): 2.2287675<br />gene: FBgn0036893","log2FoldChange: -0.8600201983<br />-log10(padj): 5.930380e+00<br />log10(baseMean): 2.8087095<br />gene: FBgn0036896","log2FoldChange: 0.0951342046<br />-log10(padj): 7.339738e-02<br />log10(baseMean): 2.4058955<br />gene: FBgn0036897","log2FoldChange: -0.1364374718<br />-log10(padj): 1.797085e-01<br />log10(baseMean): 2.6783509<br />gene: FBgn0036900","log2FoldChange: 0.0116981260<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.6775459<br />gene: FBgn0036906","log2FoldChange: -0.0939203448<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.5831281<br />gene: FBgn0036908","log2FoldChange: 0.3882538621<br />-log10(padj): 2.857419e-01<br />log10(baseMean): 1.9437647<br />gene: FBgn0036909","log2FoldChange: 0.1207000587<br />-log10(padj): 8.619087e-02<br />log10(baseMean): 2.2374342<br />gene: FBgn0036911","log2FoldChange: -0.2395518552<br />-log10(padj): 6.199465e-01<br />log10(baseMean): 3.2649653<br />gene: FBgn0036913","log2FoldChange: -0.1778286220<br />-log10(padj): 3.633488e-01<br />log10(baseMean): 2.8249558<br />gene: FBgn0036915","log2FoldChange: 0.0778095784<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.3166042<br />gene: FBgn0036916","log2FoldChange: -0.0382132128<br />-log10(padj): 2.525758e-02<br />log10(baseMean): 2.7656127<br />gene: FBgn0036918","log2FoldChange: -0.0512658321<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.9477638<br />gene: FBgn0036919","log2FoldChange: 0.1136848576<br />-log10(padj): 8.738510e-02<br />log10(baseMean): 2.3732389<br />gene: FBgn0036920","log2FoldChange: -0.2490239072<br />-log10(padj): 3.324575e-01<br />log10(baseMean): 3.3922049<br />gene: FBgn0036921","log2FoldChange: 0.1917449311<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 1.7481677<br />gene: FBgn0036922","log2FoldChange: -0.1214915202<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 1.6587559<br />gene: FBgn0036926","log2FoldChange: -0.3826591887<br />-log10(padj): 3.449925e-01<br />log10(baseMean): 1.8925863<br />gene: FBgn0036927","log2FoldChange: 0.1001266070<br />-log10(padj): 9.544956e-02<br />log10(baseMean): 3.0396900<br />gene: FBgn0036928","log2FoldChange: 0.1042298031<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.7027811<br />gene: FBgn0036929","log2FoldChange: 0.2982111733<br />-log10(padj): 1.723919e-01<br />log10(baseMean): 1.7162444<br />gene: FBgn0036931","log2FoldChange: -0.0956615711<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.3750958<br />gene: FBgn0036932","log2FoldChange: 0.0755605799<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 1.3502251<br />gene: FBgn0036942","log2FoldChange: 0.3536808073<br />-log10(padj): 2.944630e-01<br />log10(baseMean): 1.8539547<br />gene: FBgn0036945","log2FoldChange: -0.1558918077<br />-log10(padj): 1.693695e-01<br />log10(baseMean): 3.2086942<br />gene: FBgn0036958","log2FoldChange: -0.3738764613<br />-log10(padj): 1.637484e-01<br />log10(baseMean): 1.4737370<br />gene: FBgn0036959","log2FoldChange: 0.2267630423<br />-log10(padj): 2.737850e-01<br />log10(baseMean): 2.2752605<br />gene: FBgn0036964","log2FoldChange: -0.0353102122<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.6849920<br />gene: FBgn0036967","log2FoldChange: -0.9756221155<br />-log10(padj): 1.412460e+01<br />log10(baseMean): 2.8671147<br />gene: FBgn0036968","log2FoldChange: -0.2231626255<br />-log10(padj): 2.919246e-01<br />log10(baseMean): 2.3212250<br />gene: FBgn0036973","log2FoldChange: -0.1979617181<br />-log10(padj): 6.692284e-01<br />log10(baseMean): 3.3401084<br />gene: FBgn0036974","log2FoldChange: -0.1858699765<br />-log10(padj): 1.669556e-01<br />log10(baseMean): 2.2242958<br />gene: FBgn0036975","log2FoldChange: -0.3195340080<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 0.9492230<br />gene: FBgn0036978","log2FoldChange: -0.0062270457<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 2.8923475<br />gene: FBgn0036980","log2FoldChange: 0.6261270729<br />-log10(padj): 1.439937e+00<br />log10(baseMean): 1.9910866<br />gene: FBgn0036984","log2FoldChange: -1.3481424388<br />-log10(padj): 1.226822e+01<br />log10(baseMean): 3.2861742<br />gene: FBgn0036985","log2FoldChange: -0.0547785715<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.2387674<br />gene: FBgn0036987","log2FoldChange: 0.0605982307<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.2382011<br />gene: FBgn0036988","log2FoldChange: -0.0337398139<br />-log10(padj): 1.356040e-02<br />log10(baseMean): 2.3487602<br />gene: FBgn0036990","log2FoldChange: -0.0323909375<br />-log10(padj): 9.732012e-03<br />log10(baseMean): 2.1067677<br />gene: FBgn0036991","log2FoldChange: 0.2443033165<br />-log10(padj): 7.229956e-02<br />log10(baseMean): 1.3463276<br />gene: FBgn0036992","log2FoldChange: 0.1071258752<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.3428941<br />gene: FBgn0036994","log2FoldChange: -0.1279480227<br />-log10(padj): 1.789957e-02<br />log10(baseMean): 1.0892254<br />gene: FBgn0036995","log2FoldChange: 0.1984527462<br />-log10(padj): 9.551590e-02<br />log10(baseMean): 1.7687178<br />gene: FBgn0036997","log2FoldChange: 0.0975336523<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.1995360<br />gene: FBgn0036998","log2FoldChange: 0.1143895511<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.3524234<br />gene: FBgn0036999","log2FoldChange: 0.1483127849<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 1.7968060<br />gene: FBgn0037000","log2FoldChange: -0.0489541930<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.2655833<br />gene: FBgn0037001","log2FoldChange: 0.0230916468<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 0.8572241<br />gene: FBgn0037003","log2FoldChange: 0.8154495246<br />-log10(padj): 1.619966e-01<br />log10(baseMean): 0.8636781<br />gene: FBgn0037004","log2FoldChange: 0.1584108511<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 1.0397179<br />gene: FBgn0037005","log2FoldChange: -0.1542893518<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 2.9103035<br />gene: FBgn0037007","log2FoldChange: 0.0700161576<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 1.9869612<br />gene: FBgn0037008","log2FoldChange: -0.0184396884<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.6167211<br />gene: FBgn0037009","log2FoldChange: -0.0908979836<br />-log10(padj): 1.125061e-01<br />log10(baseMean): 2.9272856<br />gene: FBgn0037010","log2FoldChange: 0.2995700009<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.4490300<br />gene: FBgn0037011","log2FoldChange: -0.0462197175<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.1515081<br />gene: FBgn0037012","log2FoldChange: -0.1656641839<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.0107651<br />gene: FBgn0037013","log2FoldChange: 0.3045289127<br />-log10(padj): 3.418896e-01<br />log10(baseMean): 2.4912524<br />gene: FBgn0037016","log2FoldChange: -0.0844736577<br />-log10(padj): 1.426905e-02<br />log10(baseMean): 1.3940001<br />gene: FBgn0037017","log2FoldChange: 0.0853019889<br />-log10(padj): 6.229977e-02<br />log10(baseMean): 2.2696742<br />gene: FBgn0037018","log2FoldChange: 0.1036015016<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.2196023<br />gene: FBgn0037019","log2FoldChange: -0.0735616093<br />-log10(padj): 6.536412e-02<br />log10(baseMean): 2.5539566<br />gene: FBgn0037020","log2FoldChange: -0.2589927318<br />-log10(padj): 9.064644e-01<br />log10(baseMean): 2.7792195<br />gene: FBgn0037021","log2FoldChange: -0.0524658399<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.0105958<br />gene: FBgn0037022","log2FoldChange: -0.0454842357<br />-log10(padj): 1.940444e-02<br />log10(baseMean): 2.4406366<br />gene: FBgn0037024","log2FoldChange: 0.1455992359<br />-log10(padj): 8.492934e-02<br />log10(baseMean): 2.9242952<br />gene: FBgn0037025","log2FoldChange: 0.0711949940<br />-log10(padj): 7.111451e-02<br />log10(baseMean): 2.9971569<br />gene: FBgn0037026","log2FoldChange: 0.0295988124<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.9769681<br />gene: FBgn0037027","log2FoldChange: 0.5372425195<br />-log10(padj): 1.729550e+00<br />log10(baseMean): 2.5108756<br />gene: FBgn0037028","log2FoldChange: 0.1569782734<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.8869854<br />gene: FBgn0037031","log2FoldChange: 0.2436243350<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 0.7913883<br />gene: FBgn0037037","log2FoldChange: 0.1255484236<br />-log10(padj): 1.082707e-01<br />log10(baseMean): 2.5645154<br />gene: FBgn0037044","log2FoldChange: 0.3258454345<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 1.9458511<br />gene: FBgn0037045","log2FoldChange: 1.5141189373<br />-log10(padj): 2.117400e+00<br />log10(baseMean): 1.3949173<br />gene: FBgn0037046","log2FoldChange: -0.1515008543<br />-log10(padj): 8.598498e-02<br />log10(baseMean): 2.1794562<br />gene: FBgn0037050","log2FoldChange: -0.0723694772<br />-log10(padj): 7.986683e-02<br />log10(baseMean): 3.4410557<br />gene: FBgn0037051","log2FoldChange: -0.3454892755<br />-log10(padj): 7.470616e-01<br />log10(baseMean): 2.5280962<br />gene: FBgn0037057","log2FoldChange: 0.0003149364<br />-log10(padj): 3.176531e-04<br />log10(baseMean): 2.1606736<br />gene: FBgn0037061","log2FoldChange: 0.1569892835<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 2.6986899<br />gene: FBgn0037063","log2FoldChange: 0.0872113273<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 1.1852162<br />gene: FBgn0037064","log2FoldChange: -0.7513823879<br />-log10(padj): 1.757891e-01<br />log10(baseMean): 0.8525976<br />gene: FBgn0037065","log2FoldChange: -0.1444565291<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 2.3313553<br />gene: FBgn0037070","log2FoldChange: 0.0304738121<br />-log10(padj): 1.356040e-02<br />log10(baseMean): 2.3328761<br />gene: FBgn0037071","log2FoldChange: -0.2521460041<br />-log10(padj): 1.000222e+00<br />log10(baseMean): 2.9881582<br />gene: FBgn0037073","log2FoldChange: -0.2987365109<br />-log10(padj): 1.842354e+00<br />log10(baseMean): 3.4484688<br />gene: FBgn0037074","log2FoldChange: -0.1754026246<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 2.2145875<br />gene: FBgn0037076","log2FoldChange: 0.1577666203<br />-log10(padj): 3.727518e-02<br />log10(baseMean): 1.4190563<br />gene: FBgn0037078","log2FoldChange: -0.1151602161<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 2.7951181<br />gene: FBgn0037081","log2FoldChange: 0.0601203110<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.4490122<br />gene: FBgn0037082","log2FoldChange: -0.1896083715<br />-log10(padj): 1.707210e-01<br />log10(baseMean): 2.2610784<br />gene: FBgn0037084","log2FoldChange: 0.1582413555<br />-log10(padj): 1.487293e-01<br />log10(baseMean): 2.2891457<br />gene: FBgn0037087","log2FoldChange: 0.3224601955<br />-log10(padj): 3.352636e-01<br />log10(baseMean): 2.1298717<br />gene: FBgn0037092","log2FoldChange: -0.1779100413<br />-log10(padj): 5.490360e-01<br />log10(baseMean): 3.1430774<br />gene: FBgn0037093","log2FoldChange: -0.0993391771<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 3.1309709<br />gene: FBgn0037094","log2FoldChange: -0.0715746976<br />-log10(padj): 3.150226e-02<br />log10(baseMean): 2.8639994<br />gene: FBgn0037097","log2FoldChange: -0.2917526094<br />-log10(padj): 7.234091e-01<br />log10(baseMean): 3.1858504<br />gene: FBgn0037098","log2FoldChange: 0.0319175696<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.4925871<br />gene: FBgn0037102","log2FoldChange: -0.0691441298<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.4601443<br />gene: FBgn0037105","log2FoldChange: 0.1319527769<br />-log10(padj): 6.843950e-02<br />log10(baseMean): 2.0603385<br />gene: FBgn0037106","log2FoldChange: -0.1343719175<br />-log10(padj): 1.653947e-01<br />log10(baseMean): 3.0292738<br />gene: FBgn0037108","log2FoldChange: 0.0354075421<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 3.0454845<br />gene: FBgn0037109","log2FoldChange: -0.3101655103<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.3415083<br />gene: FBgn0037110","log2FoldChange: -0.3085539711<br />-log10(padj): 1.439937e+00<br />log10(baseMean): 2.9050817<br />gene: FBgn0037116","log2FoldChange: -0.1872207847<br />-log10(padj): 1.202652e-01<br />log10(baseMean): 2.2437592<br />gene: FBgn0037117","log2FoldChange: -0.0522588201<br />-log10(padj): 4.426756e-02<br />log10(baseMean): 2.7710530<br />gene: FBgn0037120","log2FoldChange: -0.0139134972<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.4377223<br />gene: FBgn0037121","log2FoldChange: 0.1733423365<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 1.5621626<br />gene: FBgn0037126","log2FoldChange: 0.1237618545<br />-log10(padj): 1.222172e-01<br />log10(baseMean): 2.6550862<br />gene: FBgn0037130","log2FoldChange: -0.1533813710<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.3838248<br />gene: FBgn0037133","log2FoldChange: 0.2074859212<br />-log10(padj): 1.181581e-01<br />log10(baseMean): 1.9980772<br />gene: FBgn0037134","log2FoldChange: 0.0348872098<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 3.6719154<br />gene: FBgn0037135","log2FoldChange: -0.1255161932<br />-log10(padj): 1.809581e-01<br />log10(baseMean): 3.4098598<br />gene: FBgn0037137","log2FoldChange: 0.1243130951<br />-log10(padj): 2.004441e-01<br />log10(baseMean): 2.9879376<br />gene: FBgn0037138","log2FoldChange: 0.2528469977<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 1.0628656<br />gene: FBgn0037140","log2FoldChange: 0.0570328755<br />-log10(padj): 3.185575e-02<br />log10(baseMean): 2.5593031<br />gene: FBgn0037141","log2FoldChange: 0.0497589548<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 2.6476514<br />gene: FBgn0037142","log2FoldChange: -0.4041681940<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 1.7473472<br />gene: FBgn0037144","log2FoldChange: 0.3260974691<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 2.3438434<br />gene: FBgn0037149","log2FoldChange: -0.0581503515<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.3369937<br />gene: FBgn0037150","log2FoldChange: 0.2447502961<br />-log10(padj): 2.543856e-01<br />log10(baseMean): 2.1339876<br />gene: FBgn0037151","log2FoldChange: 1.8412706590<br />-log10(padj): 1.513759e+01<br />log10(baseMean): 2.0134278<br />gene: FBgn0037153","log2FoldChange: 0.1014744583<br />-log10(padj): 5.388257e-02<br />log10(baseMean): 2.2687162<br />gene: FBgn0037156","log2FoldChange: -0.1092948431<br />-log10(padj): 1.727186e-01<br />log10(baseMean): 3.1638022<br />gene: FBgn0037182","log2FoldChange: -0.2187157516<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 1.8326838<br />gene: FBgn0037183","log2FoldChange: 0.0608673545<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 2.0867158<br />gene: FBgn0037184","log2FoldChange: -0.1353423269<br />-log10(padj): 1.009511e-01<br />log10(baseMean): 2.2129826<br />gene: FBgn0037185","log2FoldChange: -0.2238595693<br />-log10(padj): 3.977426e-02<br />log10(baseMean): 1.1047631<br />gene: FBgn0037186","log2FoldChange: -0.0363319234<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.5216016<br />gene: FBgn0037188","log2FoldChange: -0.2432118110<br />-log10(padj): 2.303742e-01<br />log10(baseMean): 2.4003660<br />gene: FBgn0037199","log2FoldChange: 0.0888878427<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.3565575<br />gene: FBgn0037200","log2FoldChange: 0.2671274267<br />-log10(padj): 4.957971e-01<br />log10(baseMean): 2.5273142<br />gene: FBgn0037202","log2FoldChange: 0.3003803368<br />-log10(padj): 1.322521e-01<br />log10(baseMean): 1.5816528<br />gene: FBgn0037203","log2FoldChange: -0.1062151960<br />-log10(padj): 9.844763e-02<br />log10(baseMean): 2.7279619<br />gene: FBgn0037205","log2FoldChange: 0.1405073328<br />-log10(padj): 1.551123e-01<br />log10(baseMean): 2.7282831<br />gene: FBgn0037206","log2FoldChange: -0.1571666312<br />-log10(padj): 4.406148e-01<br />log10(baseMean): 3.5483756<br />gene: FBgn0037207","log2FoldChange: -0.0848062990<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 1.5467442<br />gene: FBgn0037212","log2FoldChange: 0.4610354003<br />-log10(padj): 1.776853e+00<br />log10(baseMean): 2.7169220<br />gene: FBgn0037213","log2FoldChange: -0.2054691981<br />-log10(padj): 8.237388e-01<br />log10(baseMean): 3.6735515<br />gene: FBgn0037215","log2FoldChange: -0.1785544378<br />-log10(padj): 6.588253e-02<br />log10(baseMean): 1.7161101<br />gene: FBgn0037217","log2FoldChange: -0.0226051716<br />-log10(padj): 1.901726e-02<br />log10(baseMean): 3.3321865<br />gene: FBgn0037218","log2FoldChange: -0.0740178342<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 3.1832517<br />gene: FBgn0037220","log2FoldChange: -0.6333530069<br />-log10(padj): 2.797356e+00<br />log10(baseMean): 2.4850838<br />gene: FBgn0037222","log2FoldChange: -2.6265624197<br />-log10(padj): 6.416928e+00<br />log10(baseMean): 1.3977268<br />gene: FBgn0037223","log2FoldChange: 0.1919525552<br />-log10(padj): 9.252481e-02<br />log10(baseMean): 1.8461485<br />gene: FBgn0037225","log2FoldChange: 0.2065578543<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 4.0925469<br />gene: FBgn0037228","log2FoldChange: -0.0192118928<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.6066253<br />gene: FBgn0037229","log2FoldChange: 0.1774562137<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 1.0747404<br />gene: FBgn0037230","log2FoldChange: -0.1333277930<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 2.6919977<br />gene: FBgn0037231","log2FoldChange: -0.1941617983<br />-log10(padj): 1.766045e-01<br />log10(baseMean): 2.4000818<br />gene: FBgn0037232","log2FoldChange: 0.1097457649<br />-log10(padj): 9.517988e-02<br />log10(baseMean): 2.7905899<br />gene: FBgn0037234","log2FoldChange: 0.0434941281<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 2.7708592<br />gene: FBgn0037235","log2FoldChange: -0.0574850387<br />-log10(padj): 4.944644e-02<br />log10(baseMean): 2.9922507<br />gene: FBgn0037236","log2FoldChange: -0.9358191231<br />-log10(padj): 6.738837e-01<br />log10(baseMean): 1.3136798<br />gene: FBgn0037238","log2FoldChange: -0.8754310051<br />-log10(padj): 1.431525e+01<br />log10(baseMean): 2.8737063<br />gene: FBgn0037239","log2FoldChange: 0.5303515919<br />-log10(padj): 2.587527e+00<br />log10(baseMean): 3.2349090<br />gene: FBgn0037240","log2FoldChange: 0.0605101681<br />-log10(padj): 4.503165e-02<br />log10(baseMean): 2.8013426<br />gene: FBgn0037241","log2FoldChange: 0.0731550940<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.7622403<br />gene: FBgn0037242","log2FoldChange: -0.1253181465<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.4246078<br />gene: FBgn0037244","log2FoldChange: 0.0946125536<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 4.0423801<br />gene: FBgn0037245","log2FoldChange: -0.2755583044<br />-log10(padj): 4.918229e-01<br />log10(baseMean): 3.2503326<br />gene: FBgn0037248","log2FoldChange: -0.0719705417<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 3.9781574<br />gene: FBgn0037249","log2FoldChange: 0.0329474056<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.3659599<br />gene: FBgn0037250","log2FoldChange: 0.3046651401<br />-log10(padj): 3.886434e-01<br />log10(baseMean): 2.3452829<br />gene: FBgn0037251","log2FoldChange: -0.0130151088<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.2497125<br />gene: FBgn0037252","log2FoldChange: 0.0048237384<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.0783724<br />gene: FBgn0037255","log2FoldChange: 0.3643800077<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.0878860<br />gene: FBgn0037260","log2FoldChange: 0.0175853797<br />-log10(padj): 1.347426e-02<br />log10(baseMean): 3.1637030<br />gene: FBgn0037261","log2FoldChange: 0.1438625015<br />-log10(padj): 1.469827e-01<br />log10(baseMean): 2.7624784<br />gene: FBgn0037262","log2FoldChange: -0.0293591138<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.0448093<br />gene: FBgn0037263","log2FoldChange: -0.1275897570<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 2.1875456<br />gene: FBgn0037265","log2FoldChange: -0.0307261351<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.7416968<br />gene: FBgn0037270","log2FoldChange: 0.4592711431<br />-log10(padj): 3.759752e-02<br />log10(baseMean): 0.8530362<br />gene: FBgn0037275","log2FoldChange: -0.1525979022<br />-log10(padj): 1.426320e-01<br />log10(baseMean): 3.0096480<br />gene: FBgn0037279","log2FoldChange: -0.0258099638<br />-log10(padj): 1.889413e-02<br />log10(baseMean): 2.9264782<br />gene: FBgn0037282","log2FoldChange: 3.0106884736<br />-log10(padj): 1.697915e+01<br />log10(baseMean): 1.8267213<br />gene: FBgn0037290","log2FoldChange: -0.1380066322<br />-log10(padj): 2.168900e-01<br />log10(baseMean): 2.8516069<br />gene: FBgn0037293","log2FoldChange: -0.0066892110<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.4831812<br />gene: FBgn0037297","log2FoldChange: 0.1729018774<br />-log10(padj): 3.205229e-01<br />log10(baseMean): 3.1189764<br />gene: FBgn0037298","log2FoldChange: -0.0482614373<br />-log10(padj): 3.436600e-02<br />log10(baseMean): 2.6651404<br />gene: FBgn0037299","log2FoldChange: 0.1501132382<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 2.9599137<br />gene: FBgn0037301","log2FoldChange: 0.0132113565<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 0.8884815<br />gene: FBgn0037304","log2FoldChange: 0.2246061859<br />-log10(padj): 3.027591e-01<br />log10(baseMean): 2.7476849<br />gene: FBgn0037305","log2FoldChange: 0.5404711393<br />-log10(padj): 4.084043e-01<br />log10(baseMean): 1.5292441<br />gene: FBgn0037309","log2FoldChange: -0.0936957324<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.2235882<br />gene: FBgn0037312","log2FoldChange: -0.1865229862<br />-log10(padj): 1.868422e-01<br />log10(baseMean): 3.1119926<br />gene: FBgn0037313","log2FoldChange: 0.0086906391<br />-log10(padj): 4.341892e-03<br />log10(baseMean): 3.2969607<br />gene: FBgn0037315","log2FoldChange: -0.0217045020<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.2021754<br />gene: FBgn0037317","log2FoldChange: -0.1514442083<br />-log10(padj): 2.968951e-01<br />log10(baseMean): 2.9683287<br />gene: FBgn0037321","log2FoldChange: 0.4719599373<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 0.7958305<br />gene: FBgn0037322","log2FoldChange: -0.0964570096<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.5919481<br />gene: FBgn0037327","log2FoldChange: -0.1824218395<br />-log10(padj): 1.224566e-01<br />log10(baseMean): 4.2527836<br />gene: FBgn0037328","log2FoldChange: -0.1666100558<br />-log10(padj): 1.638218e-01<br />log10(baseMean): 2.7429754<br />gene: FBgn0037329","log2FoldChange: 0.2041162522<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 2.5840333<br />gene: FBgn0037330","log2FoldChange: 0.0170538462<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.6307661<br />gene: FBgn0037332","log2FoldChange: -0.1265339176<br />-log10(padj): 2.036902e-01<br />log10(baseMean): 3.1366736<br />gene: FBgn0037336","log2FoldChange: 0.0232869518<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.3893275<br />gene: FBgn0037338","log2FoldChange: 0.1160579639<br />-log10(padj): 1.778309e-01<br />log10(baseMean): 3.0443609<br />gene: FBgn0037339","log2FoldChange: 0.2577814984<br />-log10(padj): 3.320113e-01<br />log10(baseMean): 2.2198322<br />gene: FBgn0037340","log2FoldChange: -0.2469375441<br />-log10(padj): 3.886434e-01<br />log10(baseMean): 2.3900477<br />gene: FBgn0037341","log2FoldChange: -0.0489057804<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 2.5816070<br />gene: FBgn0037342","log2FoldChange: -0.1049612763<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 3.5847307<br />gene: FBgn0037344","log2FoldChange: 0.2696596203<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.1136393<br />gene: FBgn0037345","log2FoldChange: -0.0383650790<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 2.4198458<br />gene: FBgn0037347","log2FoldChange: -0.4157663114<br />-log10(padj): 5.044849e-01<br />log10(baseMean): 2.0214718<br />gene: FBgn0037350","log2FoldChange: -0.1234337008<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 4.5611522<br />gene: FBgn0037351","log2FoldChange: 0.1278673494<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 0.8944883<br />gene: FBgn0037352","log2FoldChange: -0.0948184481<br />-log10(padj): 7.084153e-02<br />log10(baseMean): 2.6461241<br />gene: FBgn0037354","log2FoldChange: 0.3785077408<br />-log10(padj): 7.534971e-01<br />log10(baseMean): 2.3633089<br />gene: FBgn0037356","log2FoldChange: 0.0099913297<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.5757423<br />gene: FBgn0037357","log2FoldChange: -0.0026857600<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 2.8570672<br />gene: FBgn0037358","log2FoldChange: 0.2310287030<br />-log10(padj): 1.808983e-01<br />log10(baseMean): 2.2166348<br />gene: FBgn0037359","log2FoldChange: -0.0017051312<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 3.2534392<br />gene: FBgn0037360","log2FoldChange: -0.2775141085<br />-log10(padj): 1.011330e+00<br />log10(baseMean): 3.2277960<br />gene: FBgn0037363","log2FoldChange: 0.1475495446<br />-log10(padj): 1.410758e-01<br />log10(baseMean): 2.5840857<br />gene: FBgn0037364","log2FoldChange: 0.0711096324<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 1.8208342<br />gene: FBgn0037368","log2FoldChange: 0.4294336116<br />-log10(padj): 1.484047e+00<br />log10(baseMean): 2.4544055<br />gene: FBgn0037369","log2FoldChange: 0.1901724150<br />-log10(padj): 4.353194e-01<br />log10(baseMean): 2.7317381<br />gene: FBgn0037370","log2FoldChange: 0.0634416574<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 2.9303322<br />gene: FBgn0037371","log2FoldChange: 0.0230134461<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.3557922<br />gene: FBgn0037372","log2FoldChange: -0.0434983168<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 3.0223616<br />gene: FBgn0037374","log2FoldChange: -0.6057840189<br />-log10(padj): 4.603572e-01<br />log10(baseMean): 1.5951166<br />gene: FBgn0037375","log2FoldChange: -0.1216441482<br />-log10(padj): 1.588975e-01<br />log10(baseMean): 3.0173568<br />gene: FBgn0037376","log2FoldChange: -0.2138052615<br />-log10(padj): 3.957597e-01<br />log10(baseMean): 2.5672420<br />gene: FBgn0037377","log2FoldChange: 0.0748050607<br />-log10(padj): 5.611394e-02<br />log10(baseMean): 2.5985451<br />gene: FBgn0037378","log2FoldChange: -0.1119031075<br />-log10(padj): 1.324204e-01<br />log10(baseMean): 2.9450739<br />gene: FBgn0037379","log2FoldChange: 0.1203783628<br />-log10(padj): 1.277414e-01<br />log10(baseMean): 2.7613246<br />gene: FBgn0037382","log2FoldChange: -0.0538873534<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 2.2677493<br />gene: FBgn0037383","log2FoldChange: 0.1764840406<br />-log10(padj): 1.419664e-01<br />log10(baseMean): 2.3178717<br />gene: FBgn0037384","log2FoldChange: -0.2855938749<br />-log10(padj): 7.323648e-02<br />log10(baseMean): 1.3106163<br />gene: FBgn0037386","log2FoldChange: -0.5924851982<br />-log10(padj): 1.873412e-01<br />log10(baseMean): 1.1771924<br />gene: FBgn0037387","log2FoldChange: 0.0273232224<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.0999568<br />gene: FBgn0037391","log2FoldChange: 0.3460248909<br />-log10(padj): 1.511863e-01<br />log10(baseMean): 1.6190496<br />gene: FBgn0037405","log2FoldChange: 0.0535389746<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 2.0938314<br />gene: FBgn0037429","log2FoldChange: -0.5576701538<br />-log10(padj): 9.782572e-02<br />log10(baseMean): 0.8134628<br />gene: FBgn0037432","log2FoldChange: 0.0378317210<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.3073481<br />gene: FBgn0037433","log2FoldChange: 0.2971661374<br />-log10(padj): 9.549102e-01<br />log10(baseMean): 3.0046410<br />gene: FBgn0037434","log2FoldChange: 0.2014042234<br />-log10(padj): 1.512365e-01<br />log10(baseMean): 2.0507857<br />gene: FBgn0037435","log2FoldChange: -0.0940131275<br />-log10(padj): 9.738478e-02<br />log10(baseMean): 3.0200400<br />gene: FBgn0037439","log2FoldChange: 0.0697535297<br />-log10(padj): 4.426756e-02<br />log10(baseMean): 2.3769805<br />gene: FBgn0037440","log2FoldChange: -0.0883723125<br />-log10(padj): 1.339331e-01<br />log10(baseMean): 3.3858532<br />gene: FBgn0037442","log2FoldChange: -0.0709383914<br />-log10(padj): 3.560999e-02<br />log10(baseMean): 3.1597758<br />gene: FBgn0037443","log2FoldChange: -0.4496356460<br />-log10(padj): 1.864802e+00<br />log10(baseMean): 3.0990837<br />gene: FBgn0037445","log2FoldChange: -0.1052469101<br />-log10(padj): 1.024462e-01<br />log10(baseMean): 2.9412703<br />gene: FBgn0037446","log2FoldChange: -0.3579426936<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 0.8100525<br />gene: FBgn0037448","log2FoldChange: -0.0764400089<br />-log10(padj): 1.375785e-02<br />log10(baseMean): 1.2932553<br />gene: FBgn0037449","log2FoldChange: 0.1660033448<br />-log10(padj): 1.640969e-01<br />log10(baseMean): 2.6461379<br />gene: FBgn0037465","log2FoldChange: -0.1591466799<br />-log10(padj): 2.358852e-01<br />log10(baseMean): 2.7271509<br />gene: FBgn0037466","log2FoldChange: 0.0888541898<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.8690407<br />gene: FBgn0037467","log2FoldChange: 0.8965191934<br />-log10(padj): 5.092477e+00<br />log10(baseMean): 3.4978543<br />gene: FBgn0037468","log2FoldChange: 0.1196495036<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 2.2514932<br />gene: FBgn0037469","log2FoldChange: 0.2286630642<br />-log10(padj): 6.066753e-01<br />log10(baseMean): 2.9449106<br />gene: FBgn0037470","log2FoldChange: 0.0948128072<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.3788349<br />gene: FBgn0037472","log2FoldChange: -0.0653570196<br />-log10(padj): 3.852149e-02<br />log10(baseMean): 2.3631404<br />gene: FBgn0037473","log2FoldChange: 0.0605134191<br />-log10(padj): 3.580449e-02<br />log10(baseMean): 2.4207512<br />gene: FBgn0037478","log2FoldChange: -0.0042685748<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 2.2603870<br />gene: FBgn0037481","log2FoldChange: -0.1008301386<br />-log10(padj): 1.094620e-01<br />log10(baseMean): 2.9092442<br />gene: FBgn0037482","log2FoldChange: -0.3463421464<br />-log10(padj): 5.796379e-02<br />log10(baseMean): 0.9320482<br />gene: FBgn0037483","log2FoldChange: -0.0502280839<br />-log10(padj): 3.635191e-02<br />log10(baseMean): 2.8397598<br />gene: FBgn0037489","log2FoldChange: 0.5655193111<br />-log10(padj): 6.032900e-01<br />log10(baseMean): 1.7522254<br />gene: FBgn0037490","log2FoldChange: -0.1761954946<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 2.1590918<br />gene: FBgn0037491","log2FoldChange: -0.2398099182<br />-log10(padj): 1.166027e-01<br />log10(baseMean): 1.7177649<br />gene: FBgn0037492","log2FoldChange: 0.0076673130<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.4265587<br />gene: FBgn0037504","log2FoldChange: -0.4508484636<br />-log10(padj): 3.761526e-01<br />log10(baseMean): 1.6890339<br />gene: FBgn0037513","log2FoldChange: -0.0812121249<br />-log10(padj): 6.273245e-02<br />log10(baseMean): 2.8379144<br />gene: FBgn0037515","log2FoldChange: 0.3107657377<br />-log10(padj): 5.933883e-01<br />log10(baseMean): 2.3057451<br />gene: FBgn0037518","log2FoldChange: 0.4946670722<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 1.1611664<br />gene: FBgn0037525","log2FoldChange: -0.0455165696<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.2040124<br />gene: FBgn0037526","log2FoldChange: 0.0689137628<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.7851696<br />gene: FBgn0037529","log2FoldChange: -0.2298021616<br />-log10(padj): 5.566336e-01<br />log10(baseMean): 3.4770676<br />gene: FBgn0037530","log2FoldChange: 0.2641716139<br />-log10(padj): 3.382657e-01<br />log10(baseMean): 2.4165801<br />gene: FBgn0037531","log2FoldChange: 0.0198112893<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 3.6308666<br />gene: FBgn0037533","log2FoldChange: -0.1614278919<br />-log10(padj): 8.199836e-02<br />log10(baseMean): 2.0652186<br />gene: FBgn0037535","log2FoldChange: 0.0055985925<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.9735703<br />gene: FBgn0037536","log2FoldChange: 0.1075370066<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8780792<br />gene: FBgn0037537","log2FoldChange: 0.0191453861<br />-log10(padj): 9.356238e-03<br />log10(baseMean): 2.6983138<br />gene: FBgn0037538","log2FoldChange: -0.4024999678<br />-log10(padj): 1.666099e+00<br />log10(baseMean): 2.7487219<br />gene: FBgn0037539","log2FoldChange: 0.0487221065<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 2.0776283<br />gene: FBgn0037540","log2FoldChange: 0.0553949677<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 3.3290557<br />gene: FBgn0037541","log2FoldChange: -0.2096004357<br />-log10(padj): 2.427348e-01<br />log10(baseMean): 2.2520311<br />gene: FBgn0037543","log2FoldChange: -0.2543493105<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 1.8503903<br />gene: FBgn0037544","log2FoldChange: -0.4549984088<br />-log10(padj): 6.606335e-01<br />log10(baseMean): 2.1450371<br />gene: FBgn0037548","log2FoldChange: 0.0351898014<br />-log10(padj): 2.950840e-02<br />log10(baseMean): 3.3358206<br />gene: FBgn0037549","log2FoldChange: 0.1313753065<br />-log10(padj): 1.123756e-01<br />log10(baseMean): 2.3726522<br />gene: FBgn0037550","log2FoldChange: -0.1472998611<br />-log10(padj): 1.588723e-01<br />log10(baseMean): 2.9318414<br />gene: FBgn0037551","log2FoldChange: -0.2816596462<br />-log10(padj): 2.213831e-01<br />log10(baseMean): 3.3012060<br />gene: FBgn0037552","log2FoldChange: -0.5582923041<br />-log10(padj): 9.080726e-01<br />log10(baseMean): 2.4181123<br />gene: FBgn0037553","log2FoldChange: 0.3115862753<br />-log10(padj): 1.295310e+00<br />log10(baseMean): 2.8311917<br />gene: FBgn0037554","log2FoldChange: -0.0062655949<br />-log10(padj): 3.945119e-03<br />log10(baseMean): 2.7959522<br />gene: FBgn0037555","log2FoldChange: 0.0286255150<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.7747544<br />gene: FBgn0037556","log2FoldChange: 0.0105432244<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.7272938<br />gene: FBgn0037560","log2FoldChange: -0.1711926536<br />-log10(padj): 1.653947e-01<br />log10(baseMean): 2.4982006<br />gene: FBgn0037561","log2FoldChange: 0.6993446403<br />-log10(padj): 1.598990e-01<br />log10(baseMean): 0.9426515<br />gene: FBgn0037562","log2FoldChange: 0.1345610768<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 2.6748549<br />gene: FBgn0037566","log2FoldChange: 0.1710162644<br />-log10(padj): 1.144468e-01<br />log10(baseMean): 2.1856224<br />gene: FBgn0037567","log2FoldChange: 0.1403506059<br />-log10(padj): 8.973024e-02<br />log10(baseMean): 2.4728194<br />gene: FBgn0037569","log2FoldChange: 0.0139852001<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.9818822<br />gene: FBgn0037573","log2FoldChange: -0.3215788295<br />-log10(padj): 3.779447e-01<br />log10(baseMean): 2.0646467<br />gene: FBgn0037574","log2FoldChange: -0.0465353882<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.3360614<br />gene: FBgn0037578","log2FoldChange: 0.2622220588<br />-log10(padj): 7.660673e-01<br />log10(baseMean): 3.3423259<br />gene: FBgn0037580","log2FoldChange: 0.3279074418<br />-log10(padj): 3.259289e-01<br />log10(baseMean): 1.8520193<br />gene: FBgn0037581","log2FoldChange: 0.2981097812<br />-log10(padj): 2.545470e-01<br />log10(baseMean): 2.6336592<br />gene: FBgn0037583","log2FoldChange: 0.5676169508<br />-log10(padj): 2.158958e-01<br />log10(baseMean): 1.1858520<br />gene: FBgn0037584","log2FoldChange: -0.3317060065<br />-log10(padj): 1.082628e+00<br />log10(baseMean): 2.5918014<br />gene: FBgn0037592","log2FoldChange: 0.1488056406<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 1.7337525<br />gene: FBgn0037602","log2FoldChange: -0.0583938169<br />-log10(padj): 3.395903e-02<br />log10(baseMean): 2.2853318<br />gene: FBgn0037603","log2FoldChange: -0.3166599372<br />-log10(padj): 8.619033e-01<br />log10(baseMean): 2.4387618<br />gene: FBgn0037606","log2FoldChange: 0.5511509192<br />-log10(padj): 2.608705e+00<br />log10(baseMean): 3.7309798<br />gene: FBgn0037607","log2FoldChange: -0.0071160260<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.5347383<br />gene: FBgn0037608","log2FoldChange: -0.0184875791<br />-log10(padj): 1.149051e-02<br />log10(baseMean): 2.8157792<br />gene: FBgn0037609","log2FoldChange: 0.1049901786<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 2.3392458<br />gene: FBgn0037610","log2FoldChange: -0.0467372201<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 1.7717743<br />gene: FBgn0037611","log2FoldChange: -0.2118517732<br />-log10(padj): 2.672544e-01<br />log10(baseMean): 3.0539431<br />gene: FBgn0037612","log2FoldChange: 0.0972593764<br />-log10(padj): 5.508672e-02<br />log10(baseMean): 2.1084112<br />gene: FBgn0037613","log2FoldChange: -0.1262237649<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 2.5205589<br />gene: FBgn0037614","log2FoldChange: 0.1646763398<br />-log10(padj): 3.684901e-02<br />log10(baseMean): 1.2877901<br />gene: FBgn0037616","log2FoldChange: -0.0164651347<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.3392504<br />gene: FBgn0037617","log2FoldChange: 0.0947905519<br />-log10(padj): 4.853923e-02<br />log10(baseMean): 2.0773979<br />gene: FBgn0037619","log2FoldChange: 0.0577773433<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.4178080<br />gene: FBgn0037620","log2FoldChange: -0.1371577389<br />-log10(padj): 1.139380e-01<br />log10(baseMean): 2.8525088<br />gene: FBgn0037621","log2FoldChange: 0.0796740685<br />-log10(padj): 6.938076e-02<br />log10(baseMean): 2.8982418<br />gene: FBgn0037622","log2FoldChange: -0.4847134610<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 1.2260232<br />gene: FBgn0037623","log2FoldChange: 0.2102303514<br />-log10(padj): 8.315530e-01<br />log10(baseMean): 3.2113863<br />gene: FBgn0037624","log2FoldChange: 0.5073808015<br />-log10(padj): 3.135380e-01<br />log10(baseMean): 1.4378977<br />gene: FBgn0037630","log2FoldChange: 0.2822723169<br />-log10(padj): 6.972166e-01<br />log10(baseMean): 3.8533038<br />gene: FBgn0037632","log2FoldChange: -0.2107745879<br />-log10(padj): 5.199177e-01<br />log10(baseMean): 3.0632781<br />gene: FBgn0037633","log2FoldChange: -0.1047732022<br />-log10(padj): 4.687913e-02<br />log10(baseMean): 2.2057189<br />gene: FBgn0037634","log2FoldChange: -1.0818668144<br />-log10(padj): 7.977016e+00<br />log10(baseMean): 2.2240641<br />gene: FBgn0037635","log2FoldChange: -0.1455424198<br />-log10(padj): 3.075722e-02<br />log10(baseMean): 4.1937867<br />gene: FBgn0037636","log2FoldChange: 0.1340191951<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.8778051<br />gene: FBgn0037637","log2FoldChange: -0.1705545807<br />-log10(padj): 2.962915e-01<br />log10(baseMean): 3.0135413<br />gene: FBgn0037638","log2FoldChange: 0.7627630044<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 0.8250858<br />gene: FBgn0037640","log2FoldChange: -0.0392235374<br />-log10(padj): 2.703040e-02<br />log10(baseMean): 3.3638524<br />gene: FBgn0037643","log2FoldChange: 0.1037163936<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.9370743<br />gene: FBgn0037644","log2FoldChange: -2.8071764244<br />-log10(padj): 1.020948e+01<br />log10(baseMean): 1.5978878<br />gene: FBgn0037646","log2FoldChange: 0.1901133784<br />-log10(padj): 2.244703e-01<br />log10(baseMean): 2.5970339<br />gene: FBgn0037647","log2FoldChange: -0.0484078924<br />-log10(padj): 3.647743e-02<br />log10(baseMean): 2.6074881<br />gene: FBgn0037648","log2FoldChange: -0.0393473262<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 2.6414219<br />gene: FBgn0037652","log2FoldChange: 0.1130052533<br />-log10(padj): 1.234181e-01<br />log10(baseMean): 2.8271759<br />gene: FBgn0037653","log2FoldChange: -0.1776455959<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.0858979<br />gene: FBgn0037654","log2FoldChange: -0.1526890246<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.2946045<br />gene: FBgn0037655","log2FoldChange: 0.1264906604<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.1656292<br />gene: FBgn0037656","log2FoldChange: -0.0697794363<br />-log10(padj): 8.337751e-02<br />log10(baseMean): 3.0174366<br />gene: FBgn0037657","log2FoldChange: -0.2179161175<br />-log10(padj): 6.899592e-01<br />log10(baseMean): 3.3304133<br />gene: FBgn0037659","log2FoldChange: 0.0428166039<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.6282536<br />gene: FBgn0037660","log2FoldChange: 0.0091118806<br />-log10(padj): 3.666988e-03<br />log10(baseMean): 2.2177271<br />gene: FBgn0037661","log2FoldChange: 0.2102263792<br />-log10(padj): 2.116949e-02<br />log10(baseMean): 1.1105463<br />gene: FBgn0037662","log2FoldChange: 0.2523957992<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.0792417<br />gene: FBgn0037667","log2FoldChange: -0.1619003797<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 2.1627289<br />gene: FBgn0037669","log2FoldChange: 0.0230827742<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 2.0279195<br />gene: FBgn0037670","log2FoldChange: 0.1362188359<br />-log10(padj): 1.926605e-01<br />log10(baseMean): 3.5871291<br />gene: FBgn0037671","log2FoldChange: 0.7643083950<br />-log10(padj): 1.748774e+00<br />log10(baseMean): 1.9539214<br />gene: FBgn0037672","log2FoldChange: 1.7376789610<br />-log10(padj): 4.450481e+00<br />log10(baseMean): 1.5481693<br />gene: FBgn0037678","log2FoldChange: 0.2428858655<br />-log10(padj): 2.727418e-01<br />log10(baseMean): 2.2190999<br />gene: FBgn0037679","log2FoldChange: 0.7701252708<br />-log10(padj): 6.265868e+00<br />log10(baseMean): 2.6199647<br />gene: FBgn0037680","log2FoldChange: -0.8499489378<br />-log10(padj): 6.615072e+00<br />log10(baseMean): 3.1566539<br />gene: FBgn0037683","log2FoldChange: -0.5232293715<br />-log10(padj): 2.072623e-01<br />log10(baseMean): 1.4796299<br />gene: FBgn0037684","log2FoldChange: -0.1801088518<br />-log10(padj): 2.278202e-01<br />log10(baseMean): 4.2026126<br />gene: FBgn0037686","log2FoldChange: 0.0228285586<br />-log10(padj): 9.732012e-03<br />log10(baseMean): 2.4128008<br />gene: FBgn0037687","log2FoldChange: 0.0677822488<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.3419883<br />gene: FBgn0037688","log2FoldChange: 0.1247055756<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.0678265<br />gene: FBgn0037696","log2FoldChange: 0.2270330324<br />-log10(padj): 2.570293e-02<br />log10(baseMean): 0.9135379<br />gene: FBgn0037698","log2FoldChange: 0.2419429822<br />-log10(padj): 4.600252e-01<br />log10(baseMean): 2.8848198<br />gene: FBgn0037700","log2FoldChange: -0.1958250487<br />-log10(padj): 6.634256e-01<br />log10(baseMean): 3.6201389<br />gene: FBgn0037702","log2FoldChange: -0.2229543946<br />-log10(padj): 5.431711e-01<br />log10(baseMean): 2.6569245<br />gene: FBgn0037703","log2FoldChange: -0.6418757748<br />-log10(padj): 2.355266e+00<br />log10(baseMean): 2.5609298<br />gene: FBgn0037705","log2FoldChange: -0.0406417296<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.8163608<br />gene: FBgn0037707","log2FoldChange: 0.0398976934<br />-log10(padj): 1.083072e-02<br />log10(baseMean): 1.9514599<br />gene: FBgn0037708","log2FoldChange: -0.1679682093<br />-log10(padj): 1.176799e-01<br />log10(baseMean): 2.4435245<br />gene: FBgn0037709","log2FoldChange: -0.0101132982<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 2.6889506<br />gene: FBgn0037710","log2FoldChange: -0.3096043470<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 1.8178568<br />gene: FBgn0037712","log2FoldChange: -0.1733466188<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.7795164<br />gene: FBgn0037713","log2FoldChange: -0.1235092276<br />-log10(padj): 3.738336e-02<br />log10(baseMean): 1.6164944<br />gene: FBgn0037714","log2FoldChange: -0.2974751945<br />-log10(padj): 5.064448e-01<br />log10(baseMean): 2.3591350<br />gene: FBgn0037715","log2FoldChange: -0.1077489824<br />-log10(padj): 7.154737e-02<br />log10(baseMean): 2.5636625<br />gene: FBgn0037716","log2FoldChange: 0.4579372711<br />-log10(padj): 2.569035e+00<br />log10(baseMean): 2.7099663<br />gene: FBgn0037717","log2FoldChange: -0.3382037722<br />-log10(padj): 2.212696e+00<br />log10(baseMean): 3.4187968<br />gene: FBgn0037718","log2FoldChange: -0.2475761868<br />-log10(padj): 5.044849e-01<br />log10(baseMean): 2.6755188<br />gene: FBgn0037719","log2FoldChange: -0.0246719549<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.1166068<br />gene: FBgn0037720","log2FoldChange: 0.3445560806<br />-log10(padj): 5.992028e-02<br />log10(baseMean): 0.8704860<br />gene: FBgn0037721","log2FoldChange: -0.1659148068<br />-log10(padj): 1.696917e-01<br />log10(baseMean): 2.4462495<br />gene: FBgn0037722","log2FoldChange: 0.3157179811<br />-log10(padj): 5.051364e-01<br />log10(baseMean): 2.3668538<br />gene: FBgn0037723","log2FoldChange: 0.1363900057<br />-log10(padj): 1.891881e-02<br />log10(baseMean): 1.0000186<br />gene: FBgn0037726","log2FoldChange: 0.2472857621<br />-log10(padj): 5.294341e-01<br />log10(baseMean): 3.5281229<br />gene: FBgn0037728","log2FoldChange: 0.1604603163<br />-log10(padj): 2.538057e-02<br />log10(baseMean): 1.0604674<br />gene: FBgn0037730","log2FoldChange: -0.1411041145<br />-log10(padj): 1.240841e-01<br />log10(baseMean): 2.9488211<br />gene: FBgn0037734","log2FoldChange: -0.0187786843<br />-log10(padj): 5.614118e-03<br />log10(baseMean): 2.3985604<br />gene: FBgn0037737","log2FoldChange: 0.7639087494<br />-log10(padj): 5.603892e+00<br />log10(baseMean): 2.5532540<br />gene: FBgn0037739","log2FoldChange: 0.0494390061<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 2.5245636<br />gene: FBgn0037741","log2FoldChange: 0.3162714460<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 1.9797777<br />gene: FBgn0037742","log2FoldChange: 0.1135738876<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.7128350<br />gene: FBgn0037743","log2FoldChange: 0.2806311204<br />-log10(padj): 8.024285e-01<br />log10(baseMean): 2.7001160<br />gene: FBgn0037744","log2FoldChange: 0.1390374003<br />-log10(padj): 1.998419e-01<br />log10(baseMean): 2.7705405<br />gene: FBgn0037746","log2FoldChange: 0.2794622105<br />-log10(padj): 3.680154e-01<br />log10(baseMean): 2.4994144<br />gene: FBgn0037747","log2FoldChange: -0.0501345586<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.5190715<br />gene: FBgn0037749","log2FoldChange: -0.0404918338<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.5023857<br />gene: FBgn0037750","log2FoldChange: -0.0101705687<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.2571727<br />gene: FBgn0037751","log2FoldChange: -0.2869991385<br />-log10(padj): 2.036902e-01<br />log10(baseMean): 4.3316172<br />gene: FBgn0037752","log2FoldChange: 0.0919673267<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.1183509<br />gene: FBgn0037753","log2FoldChange: -2.3418551868<br />-log10(padj): 2.053927e+01<br />log10(baseMean): 2.4309570<br />gene: FBgn0037754","log2FoldChange: -0.2247096747<br />-log10(padj): 2.072149e-01<br />log10(baseMean): 2.7145381<br />gene: FBgn0037755","log2FoldChange: 0.0174219359<br />-log10(padj): 8.800874e-03<br />log10(baseMean): 3.4701730<br />gene: FBgn0037756","log2FoldChange: 0.3610451908<br />-log10(padj): 1.181582e+00<br />log10(baseMean): 3.0702206<br />gene: FBgn0037757","log2FoldChange: -0.1256953084<br />-log10(padj): 1.798294e-01<br />log10(baseMean): 3.1450012<br />gene: FBgn0037758","log2FoldChange: 0.2879933832<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.6088525<br />gene: FBgn0037759","log2FoldChange: -0.3529545178<br />-log10(padj): 1.727123e+00<br />log10(baseMean): 3.2550867<br />gene: FBgn0037760","log2FoldChange: -0.1205447473<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 2.9167621<br />gene: FBgn0037770","log2FoldChange: -0.1071105215<br />-log10(padj): 3.537453e-02<br />log10(baseMean): 1.9403161<br />gene: FBgn0037773","log2FoldChange: -0.1137379296<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.6610142<br />gene: FBgn0037777","log2FoldChange: 0.2628502983<br />-log10(padj): 2.435337e-01<br />log10(baseMean): 2.1435486<br />gene: FBgn0037778","log2FoldChange: -0.2830084880<br />-log10(padj): 5.931154e-01<br />log10(baseMean): 2.5123565<br />gene: FBgn0037779","log2FoldChange: 0.0643256482<br />-log10(padj): 5.004439e-02<br />log10(baseMean): 2.5795092<br />gene: FBgn0037780","log2FoldChange: -0.1602450991<br />-log10(padj): 6.702276e-02<br />log10(baseMean): 1.8377842<br />gene: FBgn0037781","log2FoldChange: 0.0177391377<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.1005489<br />gene: FBgn0037788","log2FoldChange: 0.0209717699<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 2.7998169<br />gene: FBgn0037792","log2FoldChange: 0.1138101252<br />-log10(padj): 5.444957e-02<br />log10(baseMean): 2.3682785<br />gene: FBgn0037794","log2FoldChange: -0.0652960176<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 0.8733610<br />gene: FBgn0037801","log2FoldChange: -0.2132040432<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 1.8993424<br />gene: FBgn0037802","log2FoldChange: 0.0560076133<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.9812488<br />gene: FBgn0037804","log2FoldChange: -0.1993839150<br />-log10(padj): 3.778552e-02<br />log10(baseMean): 1.1492126<br />gene: FBgn0037806","log2FoldChange: 0.0304371231<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.7132688<br />gene: FBgn0037807","log2FoldChange: -0.1747375815<br />-log10(padj): 3.353524e-01<br />log10(baseMean): 3.9239320<br />gene: FBgn0037808","log2FoldChange: -0.0603199954<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 2.2044602<br />gene: FBgn0037809","log2FoldChange: 0.1303955019<br />-log10(padj): 1.358389e-01<br />log10(baseMean): 3.6065976<br />gene: FBgn0037810","log2FoldChange: 0.0647599981<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.2747737<br />gene: FBgn0037811","log2FoldChange: 1.2609958159<br />-log10(padj): 1.102173e+00<br />log10(baseMean): 1.1476677<br />gene: FBgn0037812","log2FoldChange: 0.2026914756<br />-log10(padj): 3.648503e-01<br />log10(baseMean): 2.7386210<br />gene: FBgn0037814","log2FoldChange: 0.0208326326<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.2276575<br />gene: FBgn0037815","log2FoldChange: -0.1171462375<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.3913397<br />gene: FBgn0037816","log2FoldChange: 0.7576810362<br />-log10(padj): 7.184932e-01<br />log10(baseMean): 1.4727521<br />gene: FBgn0037817","log2FoldChange: -0.0126824196<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.9291663<br />gene: FBgn0037822","log2FoldChange: 0.1303264548<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 0.9881716<br />gene: FBgn0037824","log2FoldChange: -0.3122630548<br />-log10(padj): 4.603572e-01<br />log10(baseMean): 2.6616603<br />gene: FBgn0037831","log2FoldChange: 0.0612539257<br />-log10(padj): 5.156995e-02<br />log10(baseMean): 3.1957490<br />gene: FBgn0037834","log2FoldChange: 0.9106163594<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.0378144<br />gene: FBgn0037837","log2FoldChange: 0.2238796498<br />-log10(padj): 1.734551e-01<br />log10(baseMean): 1.9884791<br />gene: FBgn0037838","log2FoldChange: -0.0309666735<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.4083232<br />gene: FBgn0037842","log2FoldChange: 0.0269277121<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.1373085<br />gene: FBgn0037843","log2FoldChange: 0.7018872313<br />-log10(padj): 4.446617e+00<br />log10(baseMean): 2.3955005<br />gene: FBgn0037844","log2FoldChange: 0.1954747607<br />-log10(padj): 1.040791e-01<br />log10(baseMean): 1.8917084<br />gene: FBgn0037846","log2FoldChange: -0.3414611208<br />-log10(padj): 8.969608e-01<br />log10(baseMean): 2.4072955<br />gene: FBgn0037847","log2FoldChange: 0.3481635795<br />-log10(padj): 1.232503e+00<br />log10(baseMean): 2.8168572<br />gene: FBgn0037848","log2FoldChange: 0.6404032997<br />-log10(padj): 4.373437e+00<br />log10(baseMean): 2.5312183<br />gene: FBgn0037850","log2FoldChange: 0.3944961956<br />-log10(padj): 1.909538e+00<br />log10(baseMean): 2.8659301<br />gene: FBgn0037852","log2FoldChange: 0.1910864090<br />-log10(padj): 2.696819e-01<br />log10(baseMean): 3.1302009<br />gene: FBgn0037853","log2FoldChange: 0.0837474370<br />-log10(padj): 7.847011e-02<br />log10(baseMean): 2.7536294<br />gene: FBgn0037855","log2FoldChange: 0.2593247342<br />-log10(padj): 7.234091e-01<br />log10(baseMean): 2.6729415<br />gene: FBgn0037856","log2FoldChange: -0.8342949580<br />-log10(padj): 7.193492e+00<br />log10(baseMean): 2.5988112<br />gene: FBgn0037872","log2FoldChange: 0.2913789031<br />-log10(padj): 5.342235e-01<br />log10(baseMean): 2.9408357<br />gene: FBgn0037873","log2FoldChange: -0.1374094443<br />-log10(padj): 2.579744e-01<br />log10(baseMean): 4.2118004<br />gene: FBgn0037874","log2FoldChange: 0.1201918946<br />-log10(padj): 7.778007e-02<br />log10(baseMean): 2.9074766<br />gene: FBgn0037875","log2FoldChange: 0.2931164102<br />-log10(padj): 3.027248e-01<br />log10(baseMean): 2.0685124<br />gene: FBgn0037876","log2FoldChange: 0.1093198992<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 2.5766474<br />gene: FBgn0037877","log2FoldChange: -0.0017268325<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 3.0245377<br />gene: FBgn0037878","log2FoldChange: 0.3144504594<br />-log10(padj): 4.203212e-01<br />log10(baseMean): 2.2176546<br />gene: FBgn0037880","log2FoldChange: -0.0319734481<br />-log10(padj): 1.789957e-02<br />log10(baseMean): 2.9588785<br />gene: FBgn0037881","log2FoldChange: 0.2314441227<br />-log10(padj): 2.805785e-01<br />log10(baseMean): 2.3355254<br />gene: FBgn0037882","log2FoldChange: -0.2005443762<br />-log10(padj): 9.787838e-02<br />log10(baseMean): 1.8617066<br />gene: FBgn0037883","log2FoldChange: -0.4265557045<br />-log10(padj): 1.551315e+00<br />log10(baseMean): 2.9446487<br />gene: FBgn0037884","log2FoldChange: 0.2710423175<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 2.1258464<br />gene: FBgn0037885","log2FoldChange: 0.0221087369<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 2.7384350<br />gene: FBgn0037890","log2FoldChange: -0.0435146645<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.2046290<br />gene: FBgn0037891","log2FoldChange: 0.1132077974<br />-log10(padj): 7.625340e-02<br />log10(baseMean): 2.3242562<br />gene: FBgn0037892","log2FoldChange: 0.0979145121<br />-log10(padj): 8.484881e-02<br />log10(baseMean): 2.9649913<br />gene: FBgn0037893","log2FoldChange: 0.0482853276<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 3.2212308<br />gene: FBgn0037894","log2FoldChange: 0.9068362690<br />-log10(padj): 3.073435e+00<br />log10(baseMean): 1.8640418<br />gene: FBgn0037896","log2FoldChange: -0.1702609695<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.0264681<br />gene: FBgn0037897","log2FoldChange: 0.8303303328<br />-log10(padj): 5.092879e-01<br />log10(baseMean): 1.2240757<br />gene: FBgn0037898","log2FoldChange: -0.2762629556<br />-log10(padj): 4.891661e-01<br />log10(baseMean): 3.4404242<br />gene: FBgn0037899","log2FoldChange: -0.2179231878<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.4863955<br />gene: FBgn0037900","log2FoldChange: 0.4075480512<br />-log10(padj): 2.269907e+00<br />log10(baseMean): 2.7429727<br />gene: FBgn0037901","log2FoldChange: 0.3471637398<br />-log10(padj): 3.281711e-01<br />log10(baseMean): 1.9192097<br />gene: FBgn0037902","log2FoldChange: 0.2271215459<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 1.6455116<br />gene: FBgn0037906","log2FoldChange: 0.0759471464<br />-log10(padj): 4.491832e-02<br />log10(baseMean): 2.2970074<br />gene: FBgn0037911","log2FoldChange: -1.1869052652<br />-log10(padj): 3.319132e-01<br />log10(baseMean): 1.1896801<br />gene: FBgn0037912","log2FoldChange: -0.0313693905<br />-log10(padj): 3.581171e-03<br />log10(baseMean): 2.6908228<br />gene: FBgn0037913","log2FoldChange: 0.1191393234<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 1.8888433<br />gene: FBgn0037915","log2FoldChange: -0.3080045105<br />-log10(padj): 1.638757e-01<br />log10(baseMean): 1.6537014<br />gene: FBgn0037916","log2FoldChange: -0.4160321995<br />-log10(padj): 1.702905e+00<br />log10(baseMean): 2.8278937<br />gene: FBgn0037917","log2FoldChange: -0.0099914791<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 3.0746333<br />gene: FBgn0037918","log2FoldChange: 0.0763590433<br />-log10(padj): 2.959986e-02<br />log10(baseMean): 2.0409040<br />gene: FBgn0037920","log2FoldChange: -0.1175894911<br />-log10(padj): 3.504321e-02<br />log10(baseMean): 1.5052100<br />gene: FBgn0037921","log2FoldChange: 0.4542676335<br />-log10(padj): 2.504376e-01<br />log10(baseMean): 1.5119036<br />gene: FBgn0037923","log2FoldChange: -0.1363827204<br />-log10(padj): 1.672272e-01<br />log10(baseMean): 2.9811503<br />gene: FBgn0037924","log2FoldChange: 0.1351758050<br />-log10(padj): 1.845723e-01<br />log10(baseMean): 3.2843630<br />gene: FBgn0037926","log2FoldChange: 0.1460390674<br />-log10(padj): 2.227498e-01<br />log10(baseMean): 3.3556038<br />gene: FBgn0037930","log2FoldChange: 0.0543750230<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.5580065<br />gene: FBgn0037931","log2FoldChange: -0.1096080325<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.3042894<br />gene: FBgn0037933","log2FoldChange: -0.1944431390<br />-log10(padj): 8.549806e-02<br />log10(baseMean): 1.8248263<br />gene: FBgn0037934","log2FoldChange: -1.0676180470<br />-log10(padj): 8.195797e-01<br />log10(baseMean): 1.3654511<br />gene: FBgn0037935","log2FoldChange: -0.5433304061<br />-log10(padj): 9.019567e-02<br />log10(baseMean): 0.8178711<br />gene: FBgn0037937","log2FoldChange: -0.2098805912<br />-log10(padj): 3.647743e-02<br />log10(baseMean): 1.0731632<br />gene: FBgn0037938","log2FoldChange: 0.2090209181<br />-log10(padj): 1.475558e-01<br />log10(baseMean): 2.1997052<br />gene: FBgn0037942","log2FoldChange: -0.1327256855<br />-log10(padj): 1.788166e-01<br />log10(baseMean): 2.7361353<br />gene: FBgn0037943","log2FoldChange: -0.0509071728<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 2.9509099<br />gene: FBgn0037944","log2FoldChange: -0.2753910817<br />-log10(padj): 4.090614e-01<br />log10(baseMean): 2.5176660<br />gene: FBgn0037949","log2FoldChange: 0.0647210975<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 1.1197522<br />gene: FBgn0037950","log2FoldChange: 0.0066215366<br />-log10(padj): 4.196143e-03<br />log10(baseMean): 2.8427429<br />gene: FBgn0037955","log2FoldChange: -1.0433949367<br />-log10(padj): 3.630513e-01<br />log10(baseMean): 0.9255761<br />gene: FBgn0037956","log2FoldChange: 0.0147128925<br />-log10(padj): 7.066096e-03<br />log10(baseMean): 2.6466973<br />gene: FBgn0037958","log2FoldChange: 0.1719796942<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 2.4565947<br />gene: FBgn0037960","log2FoldChange: 0.2223473757<br />-log10(padj): 5.692288e-01<br />log10(baseMean): 3.0351068<br />gene: FBgn0037963","log2FoldChange: -0.1280046108<br />-log10(padj): 1.099102e-01<br />log10(baseMean): 2.4314218<br />gene: FBgn0037973","log2FoldChange: -0.3065703534<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 2.3437575<br />gene: FBgn0037974","log2FoldChange: 0.2480440320<br />-log10(padj): 2.158135e-01<br />log10(baseMean): 2.5050029<br />gene: FBgn0037975","log2FoldChange: 0.0446479143<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 2.5312904<br />gene: FBgn0037978","log2FoldChange: -0.1624690028<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 2.8883041<br />gene: FBgn0037979","log2FoldChange: -0.1258144249<br />-log10(padj): 8.267977e-02<br />log10(baseMean): 2.2453793<br />gene: FBgn0037980","log2FoldChange: -0.1543967428<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 2.1888493<br />gene: FBgn0037981","log2FoldChange: -0.0660426160<br />-log10(padj): 6.900827e-02<br />log10(baseMean): 3.9916741<br />gene: FBgn0037989","log2FoldChange: 0.1923335505<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 2.8728064<br />gene: FBgn0037998","log2FoldChange: 0.2207251118<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.9615920<br />gene: FBgn0037999","log2FoldChange: 0.1235698958<br />-log10(padj): 1.117342e-01<br />log10(baseMean): 2.5240889<br />gene: FBgn0038013","log2FoldChange: 0.1058851647<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 1.4165520<br />gene: FBgn0038014","log2FoldChange: -0.0968214932<br />-log10(padj): 1.325599e-01<br />log10(baseMean): 3.2850067<br />gene: FBgn0038016","log2FoldChange: 0.3969567125<br />-log10(padj): 8.280480e-01<br />log10(baseMean): 2.9921229<br />gene: FBgn0038020","log2FoldChange: 0.0284107403<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.6168662<br />gene: FBgn0038034","log2FoldChange: 0.0992853245<br />-log10(padj): 7.211131e-02<br />log10(baseMean): 2.6717823<br />gene: FBgn0038035","log2FoldChange: -0.0418091555<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.7527606<br />gene: FBgn0038037","log2FoldChange: -0.0962572707<br />-log10(padj): 8.188638e-02<br />log10(baseMean): 2.8904192<br />gene: FBgn0038038","log2FoldChange: -0.1096009085<br />-log10(padj): 7.713327e-02<br />log10(baseMean): 2.9935043<br />gene: FBgn0038039","log2FoldChange: 0.4693816536<br />-log10(padj): 1.751247e-01<br />log10(baseMean): 1.2367045<br />gene: FBgn0038042","log2FoldChange: 0.5387808437<br />-log10(padj): 1.702834e+00<br />log10(baseMean): 2.5699683<br />gene: FBgn0038043","log2FoldChange: 0.2703350676<br />-log10(padj): 7.597214e-01<br />log10(baseMean): 2.6986832<br />gene: FBgn0038046","log2FoldChange: -0.1015719527<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 1.8658504<br />gene: FBgn0038047","log2FoldChange: 0.5141547415<br />-log10(padj): 1.920995e+00<br />log10(baseMean): 2.8610500<br />gene: FBgn0038049","log2FoldChange: -0.0041547491<br />-log10(padj): 2.725201e-03<br />log10(baseMean): 2.8408387<br />gene: FBgn0038053","log2FoldChange: 0.1807071723<br />-log10(padj): 1.942147e-01<br />log10(baseMean): 2.4567846<br />gene: FBgn0038055","log2FoldChange: -0.0305600248<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.5487538<br />gene: FBgn0038056","log2FoldChange: -0.0067051409<br />-log10(padj): 3.005491e-03<br />log10(baseMean): 2.1356461<br />gene: FBgn0038057","log2FoldChange: -0.1084990636<br />-log10(padj): 6.971958e-02<br />log10(baseMean): 2.4102935<br />gene: FBgn0038058","log2FoldChange: -0.1764013832<br />-log10(padj): 3.592539e-01<br />log10(baseMean): 3.3545210<br />gene: FBgn0038065","log2FoldChange: 0.0740478189<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.1021439<br />gene: FBgn0038067","log2FoldChange: 0.5731902435<br />-log10(padj): 2.074624e-01<br />log10(baseMean): 1.2205578<br />gene: FBgn0038074","log2FoldChange: 0.0168201516<br />-log10(padj): 4.621579e-03<br />log10(baseMean): 2.3741335<br />gene: FBgn0038080","log2FoldChange: -0.8634282198<br />-log10(padj): 2.033815e-01<br />log10(baseMean): 0.7979233<br />gene: FBgn0038083","log2FoldChange: -0.5676145962<br />-log10(padj): 2.418834e+00<br />log10(baseMean): 2.3462834<br />gene: FBgn0038088","log2FoldChange: 0.6635631043<br />-log10(padj): 3.101635e-01<br />log10(baseMean): 1.1875718<br />gene: FBgn0038092","log2FoldChange: -0.5863940565<br />-log10(padj): 1.604900e-01<br />log10(baseMean): 1.0319223<br />gene: FBgn0038095","log2FoldChange: -0.1680675386<br />-log10(padj): 1.230588e-01<br />log10(baseMean): 2.7063839<br />gene: FBgn0038098","log2FoldChange: -0.1528091008<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.8190827<br />gene: FBgn0038099","log2FoldChange: -0.3124984409<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.8816224<br />gene: FBgn0038100","log2FoldChange: -0.2074794743<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.6036072<br />gene: FBgn0038102","log2FoldChange: -0.0951886061<br />-log10(padj): 1.009422e-01<br />log10(baseMean): 2.7764400<br />gene: FBgn0038105","log2FoldChange: 0.0195371429<br />-log10(padj): 1.305418e-02<br />log10(baseMean): 2.7905299<br />gene: FBgn0038106","log2FoldChange: 0.4363515622<br />-log10(padj): 1.815103e+00<br />log10(baseMean): 3.1074042<br />gene: FBgn0038107","log2FoldChange: 0.0181704154<br />-log10(padj): 9.466923e-03<br />log10(baseMean): 3.7587483<br />gene: FBgn0038108","log2FoldChange: 0.0458386518<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 1.5878003<br />gene: FBgn0038109","log2FoldChange: 0.1177722082<br />-log10(padj): 1.022245e-01<br />log10(baseMean): 2.6949946<br />gene: FBgn0038110","log2FoldChange: 0.0207252968<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.1547258<br />gene: FBgn0038111","log2FoldChange: -0.4075023364<br />-log10(padj): 1.920995e+00<br />log10(baseMean): 2.7014892<br />gene: FBgn0038118","log2FoldChange: -0.0411939755<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.0886591<br />gene: FBgn0038128","log2FoldChange: -0.2355489034<br />-log10(padj): 3.121768e-01<br />log10(baseMean): 2.4775818<br />gene: FBgn0038129","log2FoldChange: 0.9584535424<br />-log10(padj): 1.332998e+00<br />log10(baseMean): 1.4522833<br />gene: FBgn0038143","log2FoldChange: 1.7094727534<br />-log10(padj): 8.389397e-01<br />log10(baseMean): 0.8486794<br />gene: FBgn0038144","log2FoldChange: -0.2883561835<br />-log10(padj): 1.256636e+00<br />log10(baseMean): 3.7949096<br />gene: FBgn0038145","log2FoldChange: -0.2535949752<br />-log10(padj): 6.468697e-01<br />log10(baseMean): 2.9981306<br />gene: FBgn0038146","log2FoldChange: 1.0499492222<br />-log10(padj): 1.392437e+01<br />log10(baseMean): 2.9971583<br />gene: FBgn0038149","log2FoldChange: 0.1856772285<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 1.8758596<br />gene: FBgn0038156","log2FoldChange: 0.0345314762<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 2.5758683<br />gene: FBgn0038166","log2FoldChange: -0.1114329934<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 2.8698578<br />gene: FBgn0038167","log2FoldChange: 0.1180277845<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 2.8618729<br />gene: FBgn0038168","log2FoldChange: -0.5397694384<br />-log10(padj): 9.412115e-02<br />log10(baseMean): 0.8511681<br />gene: FBgn0038170","log2FoldChange: -0.0620056826<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 2.3821232<br />gene: FBgn0038183","log2FoldChange: 0.2978916570<br />-log10(padj): 2.202451e-01<br />log10(baseMean): 1.8284481<br />gene: FBgn0038194","log2FoldChange: -0.0618274108<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 3.2388015<br />gene: FBgn0038195","log2FoldChange: -0.0456876305<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.6672331<br />gene: FBgn0038196","log2FoldChange: -0.1181939703<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 2.6949333<br />gene: FBgn0038197","log2FoldChange: 2.3942229360<br />-log10(padj): 1.239540e+01<br />log10(baseMean): 1.7944347<br />gene: FBgn0038198","log2FoldChange: 0.0701416259<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 2.7759290<br />gene: FBgn0038206","log2FoldChange: 0.0593127284<br />-log10(padj): 5.674078e-02<br />log10(baseMean): 2.7640060<br />gene: FBgn0038220","log2FoldChange: 0.7291934694<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 0.9301153<br />gene: FBgn0038221","log2FoldChange: -0.1344979745<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 2.6859964<br />gene: FBgn0038223","log2FoldChange: 0.1953935478<br />-log10(padj): 3.662788e-01<br />log10(baseMean): 3.0365417<br />gene: FBgn0038224","log2FoldChange: -0.0812800277<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.1234301<br />gene: FBgn0038233","log2FoldChange: 0.0811106869<br />-log10(padj): 3.492174e-02<br />log10(baseMean): 2.2243543<br />gene: FBgn0038234","log2FoldChange: -0.1007530434<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 1.8623392<br />gene: FBgn0038235","log2FoldChange: -2.8734421734<br />-log10(padj): 4.977094e+00<br />log10(baseMean): 1.3174725<br />gene: FBgn0038237","log2FoldChange: 0.0252368274<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 2.8772435<br />gene: FBgn0038243","log2FoldChange: 0.0101177094<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 3.3422913<br />gene: FBgn0038244","log2FoldChange: -0.4532514788<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 1.0878743<br />gene: FBgn0038248","log2FoldChange: -0.3613897474<br />-log10(padj): 3.284321e-01<br />log10(baseMean): 2.4301766<br />gene: FBgn0038250","log2FoldChange: 0.1182027697<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 2.4779459<br />gene: FBgn0038251","log2FoldChange: -0.6162364575<br />-log10(padj): 2.557526e+00<br />log10(baseMean): 3.0317888<br />gene: FBgn0038256","log2FoldChange: 0.4230956661<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 1.0286112<br />gene: FBgn0038257","log2FoldChange: 0.1258459260<br />-log10(padj): 2.830218e-02<br />log10(baseMean): 1.5436862<br />gene: FBgn0038258","log2FoldChange: -0.6509339151<br />-log10(padj): 1.051583e+00<br />log10(baseMean): 1.7695505<br />gene: FBgn0038260","log2FoldChange: 1.5066439584<br />-log10(padj): 2.784527e+00<br />log10(baseMean): 1.4644044<br />gene: FBgn0038261","log2FoldChange: 0.1487459752<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.2916274<br />gene: FBgn0038268","log2FoldChange: -0.1729801451<br />-log10(padj): 3.240448e-01<br />log10(baseMean): 2.8446289<br />gene: FBgn0038269","log2FoldChange: -0.0784011565<br />-log10(padj): 4.442726e-02<br />log10(baseMean): 3.7050467<br />gene: FBgn0038271","log2FoldChange: 0.0591218226<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.7651513<br />gene: FBgn0038272","log2FoldChange: 0.0088156803<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.8742081<br />gene: FBgn0038274","log2FoldChange: -0.2438942075<br />-log10(padj): 3.144011e-01<br />log10(baseMean): 2.4466589<br />gene: FBgn0038275","log2FoldChange: 0.3873584015<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 1.0909782<br />gene: FBgn0038277","log2FoldChange: -0.1452336398<br />-log10(padj): 1.870176e-01<br />log10(baseMean): 3.2050251<br />gene: FBgn0038286","log2FoldChange: 0.3049667940<br />-log10(padj): 5.568602e-01<br />log10(baseMean): 2.5192571<br />gene: FBgn0038289","log2FoldChange: -0.4052831588<br />-log10(padj): 2.078772e+00<br />log10(baseMean): 2.9902360<br />gene: FBgn0038290","log2FoldChange: -0.2946280541<br />-log10(padj): 5.671078e-01<br />log10(baseMean): 2.4333579<br />gene: FBgn0038291","log2FoldChange: 0.0092395469<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 0.8463234<br />gene: FBgn0038292","log2FoldChange: 0.6125690399<br />-log10(padj): 6.265868e+00<br />log10(baseMean): 3.5186246<br />gene: FBgn0038293","log2FoldChange: 0.1138190882<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.3666048<br />gene: FBgn0038296","log2FoldChange: -0.2651224084<br />-log10(padj): 6.144944e-02<br />log10(baseMean): 1.1257262<br />gene: FBgn0038299","log2FoldChange: 0.0176490017<br />-log10(padj): 8.451393e-03<br />log10(baseMean): 2.7121258<br />gene: FBgn0038300","log2FoldChange: 0.1171458582<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.3917001<br />gene: FBgn0038301","log2FoldChange: 0.1134010067<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.4941890<br />gene: FBgn0038302","log2FoldChange: -0.0468702136<br />-log10(padj): 3.197684e-02<br />log10(baseMean): 2.8156774<br />gene: FBgn0038303","log2FoldChange: 0.2474516374<br />-log10(padj): 7.812115e-01<br />log10(baseMean): 2.7638348<br />gene: FBgn0038304","log2FoldChange: -0.0949681430<br />-log10(padj): 8.164038e-02<br />log10(baseMean): 2.6966908<br />gene: FBgn0038306","log2FoldChange: 0.5697127831<br />-log10(padj): 8.721040e-01<br />log10(baseMean): 1.7954167<br />gene: FBgn0038309","log2FoldChange: -0.0108214211<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.7184888<br />gene: FBgn0038311","log2FoldChange: -0.0758953229<br />-log10(padj): 3.995181e-02<br />log10(baseMean): 2.2080651<br />gene: FBgn0038312","log2FoldChange: 0.1628268635<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 2.4323104<br />gene: FBgn0038313","log2FoldChange: -0.0347813996<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.5230101<br />gene: FBgn0038316","log2FoldChange: 0.0028584637<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 3.2902205<br />gene: FBgn0038318","log2FoldChange: 0.0083375312<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.4757610<br />gene: FBgn0038319","log2FoldChange: -0.0896215270<br />-log10(padj): 1.207488e-01<br />log10(baseMean): 3.6958511<br />gene: FBgn0038320","log2FoldChange: 0.1349902187<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.2018678<br />gene: FBgn0038321","log2FoldChange: -0.0315844700<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 1.8320677<br />gene: FBgn0038323","log2FoldChange: 0.3616937595<br />-log10(padj): 6.393582e-01<br />log10(baseMean): 2.3503134<br />gene: FBgn0038324","log2FoldChange: -0.0996144259<br />-log10(padj): 9.723018e-02<br />log10(baseMean): 2.5183535<br />gene: FBgn0038325","log2FoldChange: -0.3122190925<br />-log10(padj): 7.082080e-01<br />log10(baseMean): 2.7592245<br />gene: FBgn0038326","log2FoldChange: 0.2132209411<br />-log10(padj): 2.704925e-01<br />log10(baseMean): 2.5040765<br />gene: FBgn0038327","log2FoldChange: 0.1545596202<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 2.2776870<br />gene: FBgn0038330","log2FoldChange: 0.0232494824<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.7910659<br />gene: FBgn0038331","log2FoldChange: 0.0045655339<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.5182593<br />gene: FBgn0038332","log2FoldChange: 0.8918536168<br />-log10(padj): 6.794176e+00<br />log10(baseMean): 2.6311959<br />gene: FBgn0038341","log2FoldChange: 0.1492168023<br />-log10(padj): 2.953616e-01<br />log10(baseMean): 3.3399704<br />gene: FBgn0038344","log2FoldChange: -0.1409978151<br />-log10(padj): 2.231112e-01<br />log10(baseMean): 2.8554190<br />gene: FBgn0038346","log2FoldChange: 0.3223229231<br />-log10(padj): 1.443637e+00<br />log10(baseMean): 3.4831095<br />gene: FBgn0038347","log2FoldChange: 0.2573823753<br />-log10(padj): 2.635552e-01<br />log10(baseMean): 2.0559427<br />gene: FBgn0038348","log2FoldChange: -0.3239516235<br />-log10(padj): 1.115528e+00<br />log10(baseMean): 3.3619581<br />gene: FBgn0038349","log2FoldChange: -1.6327533578<br />-log10(padj): 8.072317e+00<br />log10(baseMean): 1.9884621<br />gene: FBgn0038351","log2FoldChange: -0.2749024170<br />-log10(padj): 1.826383e-01<br />log10(baseMean): 2.2705236<br />gene: FBgn0038353","log2FoldChange: -0.2171178965<br />-log10(padj): 3.932959e-01<br />log10(baseMean): 2.6696855<br />gene: FBgn0038360","log2FoldChange: -0.0988271402<br />-log10(padj): 1.315175e-02<br />log10(baseMean): 0.9150617<br />gene: FBgn0038361","log2FoldChange: 0.4934133920<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 2.0578596<br />gene: FBgn0038363","log2FoldChange: -0.2230414319<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 3.5727914<br />gene: FBgn0038364","log2FoldChange: 0.2087890897<br />-log10(padj): 8.267977e-02<br />log10(baseMean): 1.6903240<br />gene: FBgn0038365","log2FoldChange: 0.2650811995<br />-log10(padj): 7.577305e-02<br />log10(baseMean): 1.3530838<br />gene: FBgn0038366","log2FoldChange: 0.0355820333<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.9478885<br />gene: FBgn0038369","log2FoldChange: -0.0092107658<br />-log10(padj): 3.581171e-03<br />log10(baseMean): 1.9305649<br />gene: FBgn0038371","log2FoldChange: 0.0412595577<br />-log10(padj): 9.688812e-03<br />log10(baseMean): 1.5481017<br />gene: FBgn0038373","log2FoldChange: -0.0305291343<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 3.3424479<br />gene: FBgn0038376","log2FoldChange: -1.1215227526<br />-log10(padj): 6.678690e+00<br />log10(baseMean): 2.1992938<br />gene: FBgn0038381","log2FoldChange: -0.0061690724<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.5085901<br />gene: FBgn0038387","log2FoldChange: -0.1385035918<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.6722819<br />gene: FBgn0038388","log2FoldChange: -0.0367151161<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.3109147<br />gene: FBgn0038389","log2FoldChange: -0.0536080611<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.2500218<br />gene: FBgn0038390","log2FoldChange: -0.4794022144<br />-log10(padj): 1.152376e+00<br />log10(baseMean): 2.6358467<br />gene: FBgn0038391","log2FoldChange: 0.5722113314<br />-log10(padj): 9.131138e-02<br />log10(baseMean): 0.8204911<br />gene: FBgn0038392","log2FoldChange: 0.7092291868<br />-log10(padj): 1.545900e-01<br />log10(baseMean): 0.8352955<br />gene: FBgn0038394","log2FoldChange: -0.2377777400<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.9194262<br />gene: FBgn0038395","log2FoldChange: 0.0574565262<br />-log10(padj): 3.231150e-02<br />log10(baseMean): 2.4054425<br />gene: FBgn0038396","log2FoldChange: -0.2287005637<br />-log10(padj): 9.620880e-02<br />log10(baseMean): 1.6416061<br />gene: FBgn0038397","log2FoldChange: 0.0594292182<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.2616762<br />gene: FBgn0038400","log2FoldChange: -0.0872792347<br />-log10(padj): 6.482097e-02<br />log10(baseMean): 2.4886884<br />gene: FBgn0038401","log2FoldChange: -0.0912182383<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 0.8120456<br />gene: FBgn0038402","log2FoldChange: 0.5794017149<br />-log10(padj): 1.024462e-01<br />log10(baseMean): 0.8153000<br />gene: FBgn0038404","log2FoldChange: -0.0618946389<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.8317440<br />gene: FBgn0038407","log2FoldChange: 0.7408499037<br />-log10(padj): 7.337993e-01<br />log10(baseMean): 1.6700454<br />gene: FBgn0038414","log2FoldChange: 0.6764784864<br />-log10(padj): 4.287366e-01<br />log10(baseMean): 1.5319946<br />gene: FBgn0038415","log2FoldChange: 0.8664271448<br />-log10(padj): 6.959054e-01<br />log10(baseMean): 1.6216372<br />gene: FBgn0038416","log2FoldChange: -0.1139947823<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 2.3089532<br />gene: FBgn0038418","log2FoldChange: -0.5105757475<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 0.8806983<br />gene: FBgn0038419","log2FoldChange: 0.3288905380<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.0068764<br />gene: FBgn0038420","log2FoldChange: 0.0525032217<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 1.6305682<br />gene: FBgn0038421","log2FoldChange: -0.1324299450<br />-log10(padj): 1.163594e-01<br />log10(baseMean): 2.7787414<br />gene: FBgn0038424","log2FoldChange: 0.2570410380<br />-log10(padj): 2.169046e-01<br />log10(baseMean): 2.1111839<br />gene: FBgn0038425","log2FoldChange: 0.0823975953<br />-log10(padj): 4.369314e-02<br />log10(baseMean): 2.2596303<br />gene: FBgn0038426","log2FoldChange: -0.0236645155<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.9664939<br />gene: FBgn0038427","log2FoldChange: 0.2674045196<br />-log10(padj): 5.773835e-01<br />log10(baseMean): 2.6758950<br />gene: FBgn0038428","log2FoldChange: 0.1184736483<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 2.0355456<br />gene: FBgn0038429","log2FoldChange: -0.1146231454<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.6941601<br />gene: FBgn0038431","log2FoldChange: 0.2099015857<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 2.4317585<br />gene: FBgn0038432","log2FoldChange: 0.3414910082<br />-log10(padj): 1.940753e-01<br />log10(baseMean): 1.6338124<br />gene: FBgn0038435","log2FoldChange: 0.6754035317<br />-log10(padj): 5.414132e+00<br />log10(baseMean): 2.7943840<br />gene: FBgn0038436","log2FoldChange: 0.0594463899<br />-log10(padj): 1.897739e-02<br />log10(baseMean): 1.7728825<br />gene: FBgn0038437","log2FoldChange: -0.1123774011<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 2.9671233<br />gene: FBgn0038438","log2FoldChange: 0.0218275714<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.0555261<br />gene: FBgn0038446","log2FoldChange: 0.2923512342<br />-log10(padj): 5.630449e-01<br />log10(baseMean): 2.9024360<br />gene: FBgn0038453","log2FoldChange: -0.0829251395<br />-log10(padj): 4.281389e-02<br />log10(baseMean): 2.3693987<br />gene: FBgn0038454","log2FoldChange: -1.4362892015<br />-log10(padj): 8.561056e-01<br />log10(baseMean): 0.9878985<br />gene: FBgn0038460","log2FoldChange: 0.1884503059<br />-log10(padj): 1.070810e-01<br />log10(baseMean): 2.4361199<br />gene: FBgn0038461","log2FoldChange: 0.4612242419<br />-log10(padj): 4.194621e-01<br />log10(baseMean): 2.5459666<br />gene: FBgn0038462","log2FoldChange: 0.0699066748<br />-log10(padj): 6.267069e-02<br />log10(baseMean): 2.6383693<br />gene: FBgn0038463","log2FoldChange: -0.0531233346<br />-log10(padj): 4.675296e-02<br />log10(baseMean): 3.0300413<br />gene: FBgn0038464","log2FoldChange: -0.3624096779<br />-log10(padj): 1.665850e+00<br />log10(baseMean): 3.7845353<br />gene: FBgn0038465","log2FoldChange: -0.0248201952<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 1.0403281<br />gene: FBgn0038466","log2FoldChange: 0.1268413185<br />-log10(padj): 9.780621e-02<br />log10(baseMean): 2.6680155<br />gene: FBgn0038467","log2FoldChange: -0.2311204519<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 0.9978386<br />gene: FBgn0038469","log2FoldChange: 0.2233978121<br />-log10(padj): 5.732811e-02<br />log10(baseMean): 2.6941370<br />gene: FBgn0038470","log2FoldChange: -0.0800898009<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 2.3943220<br />gene: FBgn0038471","log2FoldChange: 0.0140839411<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.3567678<br />gene: FBgn0038472","log2FoldChange: -0.0727456969<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.0145164<br />gene: FBgn0038473","log2FoldChange: 0.1165248680<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.3933616<br />gene: FBgn0038474","log2FoldChange: 0.0884810152<br />-log10(padj): 8.585861e-02<br />log10(baseMean): 2.6077452<br />gene: FBgn0038475","log2FoldChange: 0.6755480150<br />-log10(padj): 7.787263e+00<br />log10(baseMean): 3.1359045<br />gene: FBgn0038476","log2FoldChange: 0.1297577445<br />-log10(padj): 1.337431e-01<br />log10(baseMean): 2.6255813<br />gene: FBgn0038478","log2FoldChange: -1.3812010977<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 1.4368652<br />gene: FBgn0038484","log2FoldChange: -0.0116955122<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.7088088<br />gene: FBgn0038488","log2FoldChange: -0.0675675572<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 1.9460030<br />gene: FBgn0038490","log2FoldChange: 0.0929072321<br />-log10(padj): 3.829192e-02<br />log10(baseMean): 2.6242067<br />gene: FBgn0038491","log2FoldChange: -0.1404660159<br />-log10(padj): 2.343861e-01<br />log10(baseMean): 2.9217725<br />gene: FBgn0038499","log2FoldChange: -0.0084589505<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.8521035<br />gene: FBgn0038501","log2FoldChange: -0.0719709167<br />-log10(padj): 9.410274e-02<br />log10(baseMean): 3.7270716<br />gene: FBgn0038504","log2FoldChange: -0.3149421019<br />-log10(padj): 7.724669e-01<br />log10(baseMean): 2.7070895<br />gene: FBgn0038515","log2FoldChange: -0.0770699711<br />-log10(padj): 4.021223e-02<br />log10(baseMean): 2.3018480<br />gene: FBgn0038516","log2FoldChange: 0.0623898021<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.8094793<br />gene: FBgn0038519","log2FoldChange: 0.0329318212<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.5991052<br />gene: FBgn0038524","log2FoldChange: 0.7138157333<br />-log10(padj): 5.355160e-01<br />log10(baseMean): 1.3590736<br />gene: FBgn0038527","log2FoldChange: 1.3954888618<br />-log10(padj): 1.158817e+01<br />log10(baseMean): 2.1835671<br />gene: FBgn0038528","log2FoldChange: 0.8994847832<br />-log10(padj): 1.003309e+00<br />log10(baseMean): 1.4919706<br />gene: FBgn0038530","log2FoldChange: 0.0253662639<br />-log10(padj): 8.177369e-03<br />log10(baseMean): 2.6473157<br />gene: FBgn0038532","log2FoldChange: -0.2074833379<br />-log10(padj): 2.750538e-01<br />log10(baseMean): 3.0620767<br />gene: FBgn0038533","log2FoldChange: -0.0154451409<br />-log10(padj): 1.025454e-02<br />log10(baseMean): 3.7152387<br />gene: FBgn0038535","log2FoldChange: 0.0700138293<br />-log10(padj): 4.633512e-02<br />log10(baseMean): 2.7929278<br />gene: FBgn0038536","log2FoldChange: -0.6965239310<br />-log10(padj): 1.353758e-01<br />log10(baseMean): 0.8620729<br />gene: FBgn0038544","log2FoldChange: 1.0276690893<br />-log10(padj): 2.595733e+00<br />log10(baseMean): 1.9137163<br />gene: FBgn0038545","log2FoldChange: -0.1592497583<br />-log10(padj): 1.467644e-01<br />log10(baseMean): 2.7689713<br />gene: FBgn0038546","log2FoldChange: -0.0400767470<br />-log10(padj): 7.820026e-03<br />log10(baseMean): 1.5199123<br />gene: FBgn0038547","log2FoldChange: -0.0074493584<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.0636123<br />gene: FBgn0038548","log2FoldChange: 0.0355201679<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.1052692<br />gene: FBgn0038549","log2FoldChange: -0.2225623633<br />-log10(padj): 5.580254e-01<br />log10(baseMean): 2.6710438<br />gene: FBgn0038551","log2FoldChange: -0.2389445235<br />-log10(padj): 3.621771e-01<br />log10(baseMean): 3.3246720<br />gene: FBgn0038552","log2FoldChange: 0.7701133617<br />-log10(padj): 8.798850e-01<br />log10(baseMean): 1.6126215<br />gene: FBgn0038554","log2FoldChange: 0.0627748945<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 1.9162923<br />gene: FBgn0038564","log2FoldChange: 0.0461634014<br />-log10(padj): 2.945997e-02<br />log10(baseMean): 2.7415533<br />gene: FBgn0038569","log2FoldChange: -0.4686386872<br />-log10(padj): 9.608974e-02<br />log10(baseMean): 0.9925765<br />gene: FBgn0038575","log2FoldChange: 0.1143319257<br />-log10(padj): 1.385050e-01<br />log10(baseMean): 2.6862302<br />gene: FBgn0038576","log2FoldChange: 0.2197034137<br />-log10(padj): 3.784250e-01<br />log10(baseMean): 2.3995328<br />gene: FBgn0038577","log2FoldChange: 0.0403193549<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.7167449<br />gene: FBgn0038578","log2FoldChange: -0.2592775114<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 0.9126837<br />gene: FBgn0038581","log2FoldChange: 0.3170694507<br />-log10(padj): 2.006360e-01<br />log10(baseMean): 1.9188691<br />gene: FBgn0038582","log2FoldChange: 0.0168227475<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.2545016<br />gene: FBgn0038583","log2FoldChange: 0.0519646033<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 2.5976887<br />gene: FBgn0038584","log2FoldChange: -0.1441296373<br />-log10(padj): 1.811904e-01<br />log10(baseMean): 2.6882138<br />gene: FBgn0038585","log2FoldChange: 0.0789407086<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 2.3485250<br />gene: FBgn0038586","log2FoldChange: -0.1488833593<br />-log10(padj): 1.983976e-01<br />log10(baseMean): 3.3665023<br />gene: FBgn0038587","log2FoldChange: 0.3487943284<br />-log10(padj): 6.923129e-01<br />log10(baseMean): 2.1678404<br />gene: FBgn0038588","log2FoldChange: -0.5097158187<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.1192639<br />gene: FBgn0038589","log2FoldChange: 0.1075936130<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.9729222<br />gene: FBgn0038590","log2FoldChange: 0.0556026007<br />-log10(padj): 4.949434e-02<br />log10(baseMean): 2.7438843<br />gene: FBgn0038593","log2FoldChange: -0.6999810801<br />-log10(padj): 1.758016e+00<br />log10(baseMean): 2.1710837<br />gene: FBgn0038595","log2FoldChange: -0.0517488525<br />-log10(padj): 3.647743e-02<br />log10(baseMean): 2.8951864<br />gene: FBgn0038597","log2FoldChange: -0.2285958098<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.2801836<br />gene: FBgn0038598","log2FoldChange: -0.1324207667<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 2.5403404<br />gene: FBgn0038601","log2FoldChange: -1.2603597637<br />-log10(padj): 1.471315e+00<br />log10(baseMean): 1.3026704<br />gene: FBgn0038603","log2FoldChange: -0.5438826967<br />-log10(padj): 1.507634e+00<br />log10(baseMean): 2.5983699<br />gene: FBgn0038608","log2FoldChange: -0.0657296482<br />-log10(padj): 3.537453e-02<br />log10(baseMean): 2.5970074<br />gene: FBgn0038609","log2FoldChange: -0.0382494013<br />-log10(padj): 1.056523e-02<br />log10(baseMean): 2.0340010<br />gene: FBgn0038612","log2FoldChange: -0.2675699493<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 2.4253372<br />gene: FBgn0038617","log2FoldChange: -0.1186938486<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.1679530<br />gene: FBgn0038619","log2FoldChange: -0.4021045232<br />-log10(padj): 1.346571e-01<br />log10(baseMean): 1.2664513<br />gene: FBgn0038621","log2FoldChange: 1.3186461456<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 0.8613108<br />gene: FBgn0038629","log2FoldChange: 0.1321245411<br />-log10(padj): 2.941089e-02<br />log10(baseMean): 1.3366450<br />gene: FBgn0038633","log2FoldChange: -0.2612048775<br />-log10(padj): 9.518441e-01<br />log10(baseMean): 2.9072158<br />gene: FBgn0038638","log2FoldChange: 0.0388305399<br />-log10(padj): 2.092265e-02<br />log10(baseMean): 2.4751648<br />gene: FBgn0038639","log2FoldChange: 0.0555912083<br />-log10(padj): 3.267959e-02<br />log10(baseMean): 2.3279539<br />gene: FBgn0038640","log2FoldChange: -0.2534461101<br />-log10(padj): 3.535967e-01<br />log10(baseMean): 2.5121830<br />gene: FBgn0038649","log2FoldChange: -0.2875263346<br />-log10(padj): 1.181582e+00<br />log10(baseMean): 3.3928801<br />gene: FBgn0038651","log2FoldChange: 0.0403491340<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.3799645<br />gene: FBgn0038652","log2FoldChange: -0.1235286229<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 0.8175456<br />gene: FBgn0038653","log2FoldChange: -0.7828079888<br />-log10(padj): 1.666118e-01<br />log10(baseMean): 0.8573411<br />gene: FBgn0038658","log2FoldChange: -0.2336266206<br />-log10(padj): 5.693002e-01<br />log10(baseMean): 2.9412335<br />gene: FBgn0038659","log2FoldChange: -0.4819263953<br />-log10(padj): 2.992740e+00<br />log10(baseMean): 3.2161139<br />gene: FBgn0038660","log2FoldChange: -0.1485050446<br />-log10(padj): 1.351620e-01<br />log10(baseMean): 2.4797364<br />gene: FBgn0038662","log2FoldChange: -0.1914104195<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.5412444<br />gene: FBgn0038666","log2FoldChange: -0.2442729497<br />-log10(padj): 4.101715e-01<br />log10(baseMean): 2.5536017<br />gene: FBgn0038667","log2FoldChange: -0.0328434034<br />-log10(padj): 1.356040e-02<br />log10(baseMean): 2.1357664<br />gene: FBgn0038672","log2FoldChange: 0.0636944256<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 2.1284866<br />gene: FBgn0038673","log2FoldChange: -0.7195950672<br />-log10(padj): 1.827227e-01<br />log10(baseMean): 0.8930827<br />gene: FBgn0038674","log2FoldChange: 0.0957831284<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.3997903<br />gene: FBgn0038675","log2FoldChange: -0.2058492638<br />-log10(padj): 1.487293e-01<br />log10(baseMean): 2.2279652<br />gene: FBgn0038678","log2FoldChange: 0.2674125671<br />-log10(padj): 1.942147e-01<br />log10(baseMean): 2.0177821<br />gene: FBgn0038679","log2FoldChange: -0.1122636935<br />-log10(padj): 6.900707e-02<br />log10(baseMean): 2.9179373<br />gene: FBgn0038680","log2FoldChange: -0.3524966824<br />-log10(padj): 4.866456e-01<br />log10(baseMean): 2.3448384<br />gene: FBgn0038681","log2FoldChange: -0.3420014091<br />-log10(padj): 1.435432e-01<br />log10(baseMean): 1.5387977<br />gene: FBgn0038682","log2FoldChange: -0.0626883286<br />-log10(padj): 5.645306e-02<br />log10(baseMean): 2.8048971<br />gene: FBgn0038683","log2FoldChange: -0.0197317644<br />-log10(padj): 1.079808e-02<br />log10(baseMean): 2.6154615<br />gene: FBgn0038686","log2FoldChange: -0.3397090382<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.3915850<br />gene: FBgn0038690","log2FoldChange: -0.4023185094<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 0.7979958<br />gene: FBgn0038691","log2FoldChange: 0.1100212408<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.3201728<br />gene: FBgn0038692","log2FoldChange: -0.2716323203<br />-log10(padj): 1.919609e-01<br />log10(baseMean): 1.7960819<br />gene: FBgn0038693","log2FoldChange: 0.0160253621<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.7323821<br />gene: FBgn0038704","log2FoldChange: 0.4371998840<br />-log10(padj): 2.296369e-01<br />log10(baseMean): 1.6129691<br />gene: FBgn0038705","log2FoldChange: 0.1799788406<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 1.2207680<br />gene: FBgn0038717","log2FoldChange: 0.6810915453<br />-log10(padj): 4.411565e+00<br />log10(baseMean): 2.4079492<br />gene: FBgn0038720","log2FoldChange: -0.3314611565<br />-log10(padj): 6.314528e-01<br />log10(baseMean): 2.9067411<br />gene: FBgn0038721","log2FoldChange: -0.0029297867<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 2.7222870<br />gene: FBgn0038722","log2FoldChange: 0.2204331208<br />-log10(padj): 3.825185e-01<br />log10(baseMean): 2.8183361<br />gene: FBgn0038723","log2FoldChange: -0.1412092871<br />-log10(padj): 3.718272e-02<br />log10(baseMean): 1.4711832<br />gene: FBgn0038725","log2FoldChange: 0.0132216684<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 2.6023165<br />gene: FBgn0038735","log2FoldChange: -0.1084920433<br />-log10(padj): 1.942147e-01<br />log10(baseMean): 3.2857616<br />gene: FBgn0038736","log2FoldChange: 0.1432646557<br />-log10(padj): 9.131138e-02<br />log10(baseMean): 2.1882244<br />gene: FBgn0038737","log2FoldChange: -0.0300762299<br />-log10(padj): 2.599341e-02<br />log10(baseMean): 3.4180046<br />gene: FBgn0038738","log2FoldChange: 0.1107713873<br />-log10(padj): 5.183965e-02<br />log10(baseMean): 2.5784548<br />gene: FBgn0038739","log2FoldChange: -0.0357107985<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.2013841<br />gene: FBgn0038741","log2FoldChange: 0.0698704338<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.9766053<br />gene: FBgn0038742","log2FoldChange: -0.6135337192<br />-log10(padj): 7.105856e-01<br />log10(baseMean): 1.6085378<br />gene: FBgn0038744","log2FoldChange: -0.3512641049<br />-log10(padj): 2.212696e+00<br />log10(baseMean): 3.1694652<br />gene: FBgn0038745","log2FoldChange: -0.2287766932<br />-log10(padj): 3.239818e-01<br />log10(baseMean): 2.8213523<br />gene: FBgn0038746","log2FoldChange: -0.0666593885<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 3.3415922<br />gene: FBgn0038747","log2FoldChange: 0.1865166550<br />-log10(padj): 1.938633e-01<br />log10(baseMean): 2.3356214<br />gene: FBgn0038755","log2FoldChange: -0.0947964832<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 2.8223576<br />gene: FBgn0038760","log2FoldChange: -0.0414435784<br />-log10(padj): 3.275836e-02<br />log10(baseMean): 2.9024618<br />gene: FBgn0038763","log2FoldChange: 0.3015615842<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 1.9196714<br />gene: FBgn0038765","log2FoldChange: 0.2107778347<br />-log10(padj): 1.106085e-01<br />log10(baseMean): 1.8386118<br />gene: FBgn0038766","log2FoldChange: 0.1642683368<br />-log10(padj): 1.478490e-01<br />log10(baseMean): 2.4318536<br />gene: FBgn0038767","log2FoldChange: -0.0948410906<br />-log10(padj): 7.995419e-02<br />log10(baseMean): 2.9406031<br />gene: FBgn0038768","log2FoldChange: -0.0962519316<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.4617602<br />gene: FBgn0038769","log2FoldChange: -0.0543371275<br />-log10(padj): 4.555352e-02<br />log10(baseMean): 3.2358872<br />gene: FBgn0038771","log2FoldChange: -0.1025892761<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.5111018<br />gene: FBgn0038772","log2FoldChange: -0.4368663693<br />-log10(padj): 6.621192e-01<br />log10(baseMean): 3.2923767<br />gene: FBgn0038780","log2FoldChange: 0.3674478657<br />-log10(padj): 6.428300e-02<br />log10(baseMean): 0.8480237<br />gene: FBgn0038784","log2FoldChange: 0.0298351487<br />-log10(padj): 2.344795e-02<br />log10(baseMean): 3.6783184<br />gene: FBgn0038787","log2FoldChange: -0.1769769104<br />-log10(padj): 2.507520e-01<br />log10(baseMean): 2.7153286<br />gene: FBgn0038788","log2FoldChange: 0.1958674350<br />-log10(padj): 3.024818e-01<br />log10(baseMean): 2.6753853<br />gene: FBgn0038803","log2FoldChange: 0.7934575893<br />-log10(padj): 3.144982e+00<br />log10(baseMean): 2.0430920<br />gene: FBgn0038804","log2FoldChange: 0.7539351833<br />-log10(padj): 9.205394e+00<br />log10(baseMean): 2.8746576<br />gene: FBgn0038805","log2FoldChange: -0.0241148295<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.4213813<br />gene: FBgn0038806","log2FoldChange: 0.0873633321<br />-log10(padj): 4.853923e-02<br />log10(baseMean): 2.3963568<br />gene: FBgn0038808","log2FoldChange: -0.1184615867<br />-log10(padj): 1.707397e-01<br />log10(baseMean): 3.3167464<br />gene: FBgn0038809","log2FoldChange: 0.0689879996<br />-log10(padj): 6.642066e-02<br />log10(baseMean): 2.9398045<br />gene: FBgn0038810","log2FoldChange: -0.1274499092<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.5261657<br />gene: FBgn0038811","log2FoldChange: 0.7048396582<br />-log10(padj): 3.352911e+00<br />log10(baseMean): 2.2424801<br />gene: FBgn0038815","log2FoldChange: -0.2269962858<br />-log10(padj): 3.247082e-01<br />log10(baseMean): 2.6134622<br />gene: FBgn0038816","log2FoldChange: 0.4709509936<br />-log10(padj): 2.845705e-01<br />log10(baseMean): 1.5071039<br />gene: FBgn0038821","log2FoldChange: -0.1404007833<br />-log10(padj): 6.554086e-02<br />log10(baseMean): 3.1252696<br />gene: FBgn0038826","log2FoldChange: 0.0316872738<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.6651203<br />gene: FBgn0038827","log2FoldChange: 0.9979650921<br />-log10(padj): 3.633488e-01<br />log10(baseMean): 0.9020834<br />gene: FBgn0038828","log2FoldChange: 0.2990304261<br />-log10(padj): 6.929979e-01<br />log10(baseMean): 2.8642027<br />gene: FBgn0038829","log2FoldChange: -0.0485523345<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.0037468<br />gene: FBgn0038830","log2FoldChange: -2.5410105398<br />-log10(padj): 3.271450e+01<br />log10(baseMean): 2.4798795<br />gene: FBgn0038832","log2FoldChange: -0.0378919762<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 4.2578420<br />gene: FBgn0038834","log2FoldChange: -0.2229828080<br />-log10(padj): 5.626611e-01<br />log10(baseMean): 3.1933916<br />gene: FBgn0038842","log2FoldChange: 0.0338201016<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.1071508<br />gene: FBgn0038846","log2FoldChange: 0.0212972005<br />-log10(padj): 4.015482e-03<br />log10(baseMean): 1.3196497<br />gene: FBgn0038851","log2FoldChange: 0.3212816117<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 0.9772738<br />gene: FBgn0038852","log2FoldChange: -0.1190742779<br />-log10(padj): 1.788166e-01<br />log10(baseMean): 3.3034612<br />gene: FBgn0038853","log2FoldChange: 0.0279265066<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 3.0053423<br />gene: FBgn0038854","log2FoldChange: -0.0338185349<br />-log10(padj): 2.327121e-02<br />log10(baseMean): 2.8311958<br />gene: FBgn0038855","log2FoldChange: 0.0520050489<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 2.6706265<br />gene: FBgn0038856","log2FoldChange: -0.0445245638<br />-log10(padj): 2.891818e-02<br />log10(baseMean): 2.5989121<br />gene: FBgn0038857","log2FoldChange: 0.4074027822<br />-log10(padj): 7.485074e-01<br />log10(baseMean): 2.1342807<br />gene: FBgn0038858","log2FoldChange: -0.1966364756<br />-log10(padj): 1.310012e-01<br />log10(baseMean): 2.2838188<br />gene: FBgn0038860","log2FoldChange: 0.2048803502<br />-log10(padj): 1.394616e-01<br />log10(baseMean): 2.0605176<br />gene: FBgn0038861","log2FoldChange: -0.0716038667<br />-log10(padj): 7.995419e-02<br />log10(baseMean): 2.8838904<br />gene: FBgn0038862","log2FoldChange: -1.0373015533<br />-log10(padj): 8.788483e+00<br />log10(baseMean): 2.4514698<br />gene: FBgn0038865","log2FoldChange: 0.2399241571<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 1.6071431<br />gene: FBgn0038866","log2FoldChange: 0.0534330967<br />-log10(padj): 3.792700e-02<br />log10(baseMean): 2.6942554<br />gene: FBgn0038868","log2FoldChange: -0.1748339585<br />-log10(padj): 1.766420e-01<br />log10(baseMean): 2.6819800<br />gene: FBgn0038869","log2FoldChange: 0.2449307521<br />-log10(padj): 7.399142e-01<br />log10(baseMean): 2.9121462<br />gene: FBgn0038870","log2FoldChange: 0.0325826835<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 2.1706873<br />gene: FBgn0038871","log2FoldChange: -0.1809105255<br />-log10(padj): 3.673593e-01<br />log10(baseMean): 3.2313833<br />gene: FBgn0038872","log2FoldChange: 0.0830790020<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 1.3287752<br />gene: FBgn0038873","log2FoldChange: -0.6830440280<br />-log10(padj): 1.013956e+00<br />log10(baseMean): 2.0161640<br />gene: FBgn0038874","log2FoldChange: 0.2369942886<br />-log10(padj): 2.674975e-01<br />log10(baseMean): 2.3926740<br />gene: FBgn0038876","log2FoldChange: -0.7001298604<br />-log10(padj): 5.254594e+00<br />log10(baseMean): 2.9926886<br />gene: FBgn0038877","log2FoldChange: -0.1254384341<br />-log10(padj): 1.938633e-01<br />log10(baseMean): 3.0653036<br />gene: FBgn0038881","log2FoldChange: -0.1267348121<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.5716692<br />gene: FBgn0038889","log2FoldChange: -0.3355822539<br />-log10(padj): 8.238489e-01<br />log10(baseMean): 3.3060473<br />gene: FBgn0038890","log2FoldChange: 0.1781383354<br />-log10(padj): 1.933346e-01<br />log10(baseMean): 2.7517073<br />gene: FBgn0038893","log2FoldChange: -0.8953944170<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 0.9017787<br />gene: FBgn0038894","log2FoldChange: -0.1142369417<br />-log10(padj): 5.107134e-02<br />log10(baseMean): 1.9549140<br />gene: FBgn0038903","log2FoldChange: -0.4118474621<br />-log10(padj): 7.096361e-01<br />log10(baseMean): 2.2297016<br />gene: FBgn0038909","log2FoldChange: -0.5303977810<br />-log10(padj): 3.132872e+00<br />log10(baseMean): 2.7563549<br />gene: FBgn0038912","log2FoldChange: 0.1841209152<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 0.8366916<br />gene: FBgn0038916","log2FoldChange: 0.3009058729<br />-log10(padj): 3.628058e-01<br />log10(baseMean): 2.1463795<br />gene: FBgn0038917","log2FoldChange: -0.0360185465<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.9063207<br />gene: FBgn0038922","log2FoldChange: 0.0348729496<br />-log10(padj): 1.557024e-02<br />log10(baseMean): 2.2087373<br />gene: FBgn0038923","log2FoldChange: 0.1070211340<br />-log10(padj): 8.129830e-02<br />log10(baseMean): 2.4668197<br />gene: FBgn0038924","log2FoldChange: 0.0151474307<br />-log10(padj): 7.932954e-03<br />log10(baseMean): 2.7878906<br />gene: FBgn0038925","log2FoldChange: 0.4462663195<br />-log10(padj): 1.605835e-01<br />log10(baseMean): 1.3038936<br />gene: FBgn0038926","log2FoldChange: -0.1884807970<br />-log10(padj): 4.253737e-01<br />log10(baseMean): 2.9449654<br />gene: FBgn0038927","log2FoldChange: -0.0849579288<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 2.2871906<br />gene: FBgn0038928","log2FoldChange: -0.0262369989<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.2779163<br />gene: FBgn0038947","log2FoldChange: 0.1384212928<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.8769682<br />gene: FBgn0038948","log2FoldChange: -0.1106518745<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.7278041<br />gene: FBgn0038950","log2FoldChange: 0.4312045071<br />-log10(padj): 6.722414e-01<br />log10(baseMean): 2.0746696<br />gene: FBgn0038951","log2FoldChange: 0.5349931584<br />-log10(padj): 5.845809e-01<br />log10(baseMean): 2.2917230<br />gene: FBgn0038952","log2FoldChange: 0.2997047431<br />-log10(padj): 5.710417e-01<br />log10(baseMean): 2.9057869<br />gene: FBgn0038953","log2FoldChange: -0.5569538586<br />-log10(padj): 5.580254e-01<br />log10(baseMean): 1.6517444<br />gene: FBgn0038957","log2FoldChange: -0.0752038331<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.4686313<br />gene: FBgn0038961","log2FoldChange: 0.0305394580<br />-log10(padj): 2.023076e-02<br />log10(baseMean): 3.2580929<br />gene: FBgn0038964","log2FoldChange: -0.0336510573<br />-log10(padj): 1.671862e-02<br />log10(baseMean): 2.4472957<br />gene: FBgn0038965","log2FoldChange: -0.3497629580<br />-log10(padj): 1.049229e+00<br />log10(baseMean): 2.5303876<br />gene: FBgn0038966","log2FoldChange: -0.1734646548<br />-log10(padj): 1.770380e-01<br />log10(baseMean): 3.0547545<br />gene: FBgn0038968","log2FoldChange: 0.3324131645<br />-log10(padj): 1.020122e+00<br />log10(baseMean): 2.5855832<br />gene: FBgn0038972","log2FoldChange: -0.2449932815<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 1.3336516<br />gene: FBgn0038973","log2FoldChange: 0.0941369931<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.2160187<br />gene: FBgn0038974","log2FoldChange: -1.1598449002<br />-log10(padj): 6.769701e-01<br />log10(baseMean): 1.0642642<br />gene: FBgn0038975","log2FoldChange: 0.0119716822<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.7163162<br />gene: FBgn0038976","log2FoldChange: 0.2900189620<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 0.9492998<br />gene: FBgn0038977","log2FoldChange: 0.0334007085<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 3.0087375<br />gene: FBgn0038981","log2FoldChange: 0.0596840491<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 3.1745822<br />gene: FBgn0038984","log2FoldChange: -0.2558807102<br />-log10(padj): 6.577099e-01<br />log10(baseMean): 2.5998680<br />gene: FBgn0038989","log2FoldChange: -0.0561602998<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 0.9470578<br />gene: FBgn0038993","log2FoldChange: -0.0028522724<br />-log10(padj): 1.734822e-03<br />log10(baseMean): 2.7561228<br />gene: FBgn0038996","log2FoldChange: -0.6291966880<br />-log10(padj): 4.958183e+00<br />log10(baseMean): 2.6794467<br />gene: FBgn0039000","log2FoldChange: 0.1056343636<br />-log10(padj): 1.126337e-01<br />log10(baseMean): 2.8060044<br />gene: FBgn0039003","log2FoldChange: 0.0096837885<br />-log10(padj): 4.818680e-03<br />log10(baseMean): 2.8653798<br />gene: FBgn0039004","log2FoldChange: 0.7994135715<br />-log10(padj): 3.104994e-01<br />log10(baseMean): 1.0687776<br />gene: FBgn0039005","log2FoldChange: 0.3653103141<br />-log10(padj): 1.284522e+00<br />log10(baseMean): 2.7973143<br />gene: FBgn0039006","log2FoldChange: -0.0885168650<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 2.3001427<br />gene: FBgn0039013","log2FoldChange: 0.0679899385<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 3.0671746<br />gene: FBgn0039016","log2FoldChange: 0.0045998421<br />-log10(padj): 2.301060e-03<br />log10(baseMean): 2.1476718<br />gene: FBgn0039017","log2FoldChange: 0.0277532548<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.5162164<br />gene: FBgn0039018","log2FoldChange: 0.3372728184<br />-log10(padj): 6.929979e-01<br />log10(baseMean): 2.4215980<br />gene: FBgn0039019","log2FoldChange: -0.1239163020<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.9337558<br />gene: FBgn0039020","log2FoldChange: -0.3099595511<br />-log10(padj): 2.690434e-01<br />log10(baseMean): 1.9132487<br />gene: FBgn0039022","log2FoldChange: -0.3741701592<br />-log10(padj): 3.710694e-01<br />log10(baseMean): 2.0481174<br />gene: FBgn0039023","log2FoldChange: -0.2463015048<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.0820728<br />gene: FBgn0039024","log2FoldChange: -0.2077317414<br />-log10(padj): 2.836072e-01<br />log10(baseMean): 2.2890888<br />gene: FBgn0039025","log2FoldChange: -0.4010967157<br />-log10(padj): 2.034398e+00<br />log10(baseMean): 3.4537385<br />gene: FBgn0039026","log2FoldChange: -1.0208402455<br />-log10(padj): 2.532958e-01<br />log10(baseMean): 0.8178223<br />gene: FBgn0039043","log2FoldChange: 0.1989150900<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 2.4216039<br />gene: FBgn0039044","log2FoldChange: -0.4211586007<br />-log10(padj): 1.165532e+00<br />log10(baseMean): 3.0021673<br />gene: FBgn0039045","log2FoldChange: 0.0843252999<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.3843980<br />gene: FBgn0039049","log2FoldChange: 0.2556977684<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 1.4875270<br />gene: FBgn0039051","log2FoldChange: 1.2640475593<br />-log10(padj): 7.720311e-01<br />log10(baseMean): 1.0865543<br />gene: FBgn0039052","log2FoldChange: -1.1588750416<br />-log10(padj): 2.944630e-01<br />log10(baseMean): 0.9617983<br />gene: FBgn0039054","log2FoldChange: -0.1280501172<br />-log10(padj): 2.024417e-01<br />log10(baseMean): 3.0329153<br />gene: FBgn0039055","log2FoldChange: 0.0265105441<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 3.2251269<br />gene: FBgn0039056","log2FoldChange: -0.3425900391<br />-log10(padj): 1.270824e-01<br />log10(baseMean): 1.5432858<br />gene: FBgn0039058","log2FoldChange: 0.0228969376<br />-log10(padj): 4.547574e-03<br />log10(baseMean): 1.4753297<br />gene: FBgn0039059","log2FoldChange: -0.9711225530<br />-log10(padj): 3.172190e+00<br />log10(baseMean): 1.9422517<br />gene: FBgn0039060","log2FoldChange: -0.4232008967<br />-log10(padj): 1.636011e+00<br />log10(baseMean): 3.0235871<br />gene: FBgn0039061","log2FoldChange: -0.7522213394<br />-log10(padj): 2.903845e-01<br />log10(baseMean): 1.2446035<br />gene: FBgn0039064","log2FoldChange: 0.1059041486<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.3039794<br />gene: FBgn0039065","log2FoldChange: 0.1183008184<br />-log10(padj): 1.602979e-01<br />log10(baseMean): 2.9310712<br />gene: FBgn0039066","log2FoldChange: -0.0756947941<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 2.4911246<br />gene: FBgn0039067","log2FoldChange: 0.1720562636<br />-log10(padj): 6.861476e-02<br />log10(baseMean): 1.8458557<br />gene: FBgn0039068","log2FoldChange: -1.0658819061<br />-log10(padj): 1.630858e+00<br />log10(baseMean): 1.5281196<br />gene: FBgn0039069","log2FoldChange: 0.2554594803<br />-log10(padj): 4.501543e-02<br />log10(baseMean): 1.0180907<br />gene: FBgn0039071","log2FoldChange: 0.5296312202<br />-log10(padj): 1.169333e+00<br />log10(baseMean): 2.2378172<br />gene: FBgn0039073","log2FoldChange: -1.7376959983<br />-log10(padj): 2.049371e+00<br />log10(baseMean): 1.1938909<br />gene: FBgn0039094","log2FoldChange: -1.0198227806<br />-log10(padj): 6.872873e+00<br />log10(baseMean): 2.4892365<br />gene: FBgn0039098","log2FoldChange: -0.2692574137<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 1.6345231<br />gene: FBgn0039099","log2FoldChange: -0.9807804820<br />-log10(padj): 4.631146e+00<br />log10(baseMean): 2.4689586<br />gene: FBgn0039101","log2FoldChange: 0.0733773720<br />-log10(padj): 7.188587e-03<br />log10(baseMean): 0.8860869<br />gene: FBgn0039102","log2FoldChange: 0.5999402607<br />-log10(padj): 1.263777e+00<br />log10(baseMean): 2.2260559<br />gene: FBgn0039108","log2FoldChange: -1.3035462226<br />-log10(padj): 1.976616e+01<br />log10(baseMean): 2.6770627<br />gene: FBgn0039109","log2FoldChange: 0.1887883302<br />-log10(padj): 1.401391e-01<br />log10(baseMean): 2.9706449<br />gene: FBgn0039110","log2FoldChange: 0.0704409041<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.2985503<br />gene: FBgn0039111","log2FoldChange: 0.2676860914<br />-log10(padj): 5.266112e-01<br />log10(baseMean): 2.5246308<br />gene: FBgn0039112","log2FoldChange: 0.9480766300<br />-log10(padj): 9.612305e+00<br />log10(baseMean): 2.6760877<br />gene: FBgn0039113","log2FoldChange: -0.0556557290<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 2.0328897<br />gene: FBgn0039114","log2FoldChange: -0.0874460352<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.1787176<br />gene: FBgn0039115","log2FoldChange: -0.1689471611<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 2.1405509<br />gene: FBgn0039116","log2FoldChange: 0.1564522602<br />-log10(padj): 1.578071e-01<br />log10(baseMean): 2.6313611<br />gene: FBgn0039117","log2FoldChange: 0.0234082788<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 1.6615462<br />gene: FBgn0039118","log2FoldChange: -0.0291559415<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 3.5916574<br />gene: FBgn0039120","log2FoldChange: 0.3637391833<br />-log10(padj): 9.949048e-01<br />log10(baseMean): 3.0376643<br />gene: FBgn0039125","log2FoldChange: 0.1520248065<br />-log10(padj): 1.986652e-01<br />log10(baseMean): 2.5371715<br />gene: FBgn0039126","log2FoldChange: -0.0114308292<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.7081616<br />gene: FBgn0039127","log2FoldChange: -0.0556149862<br />-log10(padj): 1.891881e-02<br />log10(baseMean): 2.0477172<br />gene: FBgn0039128","log2FoldChange: 0.7158471588<br />-log10(padj): 5.405273e-01<br />log10(baseMean): 1.4970749<br />gene: FBgn0039129","log2FoldChange: -0.0038025080<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 3.2127573<br />gene: FBgn0039130","log2FoldChange: 0.0709220426<br />-log10(padj): 3.436600e-02<br />log10(baseMean): 2.6104586<br />gene: FBgn0039132","log2FoldChange: -0.2169797672<br />-log10(padj): 4.246612e-01<br />log10(baseMean): 2.8645406<br />gene: FBgn0039135","log2FoldChange: 0.0217339207<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.0337435<br />gene: FBgn0039136","log2FoldChange: -0.1985903095<br />-log10(padj): 2.666229e-01<br />log10(baseMean): 2.4746861<br />gene: FBgn0039137","log2FoldChange: 0.3367531001<br />-log10(padj): 9.221362e-01<br />log10(baseMean): 2.3767753<br />gene: FBgn0039139","log2FoldChange: -0.1211513865<br />-log10(padj): 2.136474e-01<br />log10(baseMean): 2.9112548<br />gene: FBgn0039140","log2FoldChange: 0.1114427324<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.8438110<br />gene: FBgn0039141","log2FoldChange: -0.2181168869<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 1.7068912<br />gene: FBgn0039145","log2FoldChange: 0.3428717427<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 1.7716862<br />gene: FBgn0039149","log2FoldChange: -0.0914904208<br />-log10(padj): 6.539875e-02<br />log10(baseMean): 2.6595089<br />gene: FBgn0039150","log2FoldChange: -0.5285955921<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 0.9057032<br />gene: FBgn0039152","log2FoldChange: 0.0760405018<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.1949970<br />gene: FBgn0039153","log2FoldChange: -0.2582761736<br />-log10(padj): 6.524351e-01<br />log10(baseMean): 2.6298621<br />gene: FBgn0039156","log2FoldChange: -0.2110141292<br />-log10(padj): 3.580114e-01<br />log10(baseMean): 2.6483139<br />gene: FBgn0039157","log2FoldChange: -0.0614673841<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.0077496<br />gene: FBgn0039158","log2FoldChange: 0.2669746379<br />-log10(padj): 1.333676e-01<br />log10(baseMean): 1.7152182<br />gene: FBgn0039159","log2FoldChange: -0.1054867286<br />-log10(padj): 1.208914e-01<br />log10(baseMean): 2.8035642<br />gene: FBgn0039160","log2FoldChange: 0.1398095613<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 2.6344971<br />gene: FBgn0039163","log2FoldChange: -0.2392546161<br />-log10(padj): 3.214162e-01<br />log10(baseMean): 2.7195442<br />gene: FBgn0039164","log2FoldChange: -0.1594798676<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.3046637<br />gene: FBgn0039165","log2FoldChange: -0.2301066131<br />-log10(padj): 2.265519e-01<br />log10(baseMean): 2.1383174<br />gene: FBgn0039167","log2FoldChange: -0.3369681980<br />-log10(padj): 1.218676e+00<br />log10(baseMean): 2.6209854<br />gene: FBgn0039169","log2FoldChange: 0.2256575561<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 1.2942290<br />gene: FBgn0039170","log2FoldChange: 0.1722834029<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 3.1800810<br />gene: FBgn0039172","log2FoldChange: 0.0967784503<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 2.8700549<br />gene: FBgn0039175","log2FoldChange: 2.3203729204<br />-log10(padj): 2.781733e+00<br />log10(baseMean): 1.1437916<br />gene: FBgn0039178","log2FoldChange: -0.0498495489<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 2.5765279<br />gene: FBgn0039179","log2FoldChange: 0.2235275582<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.0071177<br />gene: FBgn0039180","log2FoldChange: -0.1361949866<br />-log10(padj): 1.818729e-01<br />log10(baseMean): 3.0311994<br />gene: FBgn0039182","log2FoldChange: -0.0356339046<br />-log10(padj): 2.157692e-02<br />log10(baseMean): 2.9840235<br />gene: FBgn0039183","log2FoldChange: -0.1946453924<br />-log10(padj): 2.091906e-01<br />log10(baseMean): 3.1982761<br />gene: FBgn0039186","log2FoldChange: -0.0930858015<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 3.0336073<br />gene: FBgn0039187","log2FoldChange: -0.0559206903<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.5214480<br />gene: FBgn0039188","log2FoldChange: 0.0261876827<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.1445239<br />gene: FBgn0039189","log2FoldChange: 0.0377080966<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.3592970<br />gene: FBgn0039201","log2FoldChange: 0.1101670879<br />-log10(padj): 7.625340e-02<br />log10(baseMean): 2.7523772<br />gene: FBgn0039204","log2FoldChange: 0.2608295294<br />-log10(padj): 1.310195e-01<br />log10(baseMean): 1.7790418<br />gene: FBgn0039205","log2FoldChange: -0.0998313580<br />-log10(padj): 8.004060e-02<br />log10(baseMean): 2.8833902<br />gene: FBgn0039206","log2FoldChange: -0.0478870359<br />-log10(padj): 4.311384e-02<br />log10(baseMean): 3.3097545<br />gene: FBgn0039207","log2FoldChange: -0.3276119419<br />-log10(padj): 1.874347e+00<br />log10(baseMean): 3.6502791<br />gene: FBgn0039208","log2FoldChange: -0.5597457414<br />-log10(padj): 2.026002e+00<br />log10(baseMean): 3.2118565<br />gene: FBgn0039209","log2FoldChange: -0.0113963792<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.4611616<br />gene: FBgn0039210","log2FoldChange: -0.2212304481<br />-log10(padj): 3.628965e-01<br />log10(baseMean): 2.6409184<br />gene: FBgn0039212","log2FoldChange: 0.0685315366<br />-log10(padj): 8.933533e-02<br />log10(baseMean): 3.2343854<br />gene: FBgn0039213","log2FoldChange: 0.0034415836<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.5043100<br />gene: FBgn0039214","log2FoldChange: 0.0264208840<br />-log10(padj): 1.426905e-02<br />log10(baseMean): 3.0987272<br />gene: FBgn0039215","log2FoldChange: 0.1268272220<br />-log10(padj): 4.490020e-02<br />log10(baseMean): 2.2670142<br />gene: FBgn0039218","log2FoldChange: 0.3720191191<br />-log10(padj): 1.227248e+00<br />log10(baseMean): 2.6010535<br />gene: FBgn0039219","log2FoldChange: 0.1002309656<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 2.8456315<br />gene: FBgn0039223","log2FoldChange: 0.2283901832<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 1.7814392<br />gene: FBgn0039226","log2FoldChange: -0.0715655263<br />-log10(padj): 7.845574e-02<br />log10(baseMean): 3.1268844<br />gene: FBgn0039227","log2FoldChange: -0.0841254969<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 3.2076344<br />gene: FBgn0039229","log2FoldChange: -0.1700167061<br />-log10(padj): 1.571206e-01<br />log10(baseMean): 2.3700076<br />gene: FBgn0039233","log2FoldChange: -0.0861812423<br />-log10(padj): 9.728847e-02<br />log10(baseMean): 3.2851888<br />gene: FBgn0039234","log2FoldChange: -0.1186585731<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 3.0316672<br />gene: FBgn0039240","log2FoldChange: -0.1222175853<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.7326125<br />gene: FBgn0039241","log2FoldChange: -0.2141194474<br />-log10(padj): 2.588560e-01<br />log10(baseMean): 2.3101162<br />gene: FBgn0039249","log2FoldChange: -0.0425918576<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 3.0680235<br />gene: FBgn0039250","log2FoldChange: -0.2984477721<br />-log10(padj): 9.220293e-02<br />log10(baseMean): 1.4610454<br />gene: FBgn0039251","log2FoldChange: -0.0238188910<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.8339021<br />gene: FBgn0039252","log2FoldChange: 0.2781385149<br />-log10(padj): 3.774457e-01<br />log10(baseMean): 2.3254899<br />gene: FBgn0039254","log2FoldChange: -0.7426092186<br />-log10(padj): 8.815624e-01<br />log10(baseMean): 1.5577554<br />gene: FBgn0039255","log2FoldChange: -1.0525308153<br />-log10(padj): 1.520702e+01<br />log10(baseMean): 2.7176002<br />gene: FBgn0039257","log2FoldChange: 0.1914595201<br />-log10(padj): 2.249970e-01<br />log10(baseMean): 2.3991235<br />gene: FBgn0039258","log2FoldChange: 0.0948512815<br />-log10(padj): 4.328987e-02<br />log10(baseMean): 1.9737805<br />gene: FBgn0039259","log2FoldChange: -0.2592534224<br />-log10(padj): 1.000222e+00<br />log10(baseMean): 3.0367393<br />gene: FBgn0039260","log2FoldChange: 0.0114769116<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.9799922<br />gene: FBgn0039261","log2FoldChange: -0.0749894098<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 2.6977387<br />gene: FBgn0039265","log2FoldChange: 0.1816726787<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.9303247<br />gene: FBgn0039266","log2FoldChange: -0.1229954273<br />-log10(padj): 1.027193e-01<br />log10(baseMean): 2.4106494<br />gene: FBgn0039269","log2FoldChange: -0.2819366028<br />-log10(padj): 2.919246e-01<br />log10(baseMean): 2.0392989<br />gene: FBgn0039270","log2FoldChange: 0.0483422867<br />-log10(padj): 1.356040e-02<br />log10(baseMean): 1.9510068<br />gene: FBgn0039271","log2FoldChange: 0.4013050157<br />-log10(padj): 5.165763e-01<br />log10(baseMean): 2.3623685<br />gene: FBgn0039273","log2FoldChange: -0.2741648273<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 2.2965446<br />gene: FBgn0039274","log2FoldChange: -0.3865077298<br />-log10(padj): 7.520383e-01<br />log10(baseMean): 2.4676218<br />gene: FBgn0039290","log2FoldChange: -0.1429448911<br />-log10(padj): 4.176811e-02<br />log10(baseMean): 1.8980785<br />gene: FBgn0039291","log2FoldChange: 0.0863555289<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.7818295<br />gene: FBgn0039293","log2FoldChange: -0.4202453743<br />-log10(padj): 2.136153e-01<br />log10(baseMean): 1.5475552<br />gene: FBgn0039294","log2FoldChange: 0.3045288832<br />-log10(padj): 9.015875e-01<br />log10(baseMean): 2.7722188<br />gene: FBgn0039296","log2FoldChange: 0.1321419241<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 0.9438380<br />gene: FBgn0039297","log2FoldChange: -0.0786234362<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 4.0274031<br />gene: FBgn0039300","log2FoldChange: -0.0129386850<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 2.3865333<br />gene: FBgn0039301","log2FoldChange: -0.0513144646<br />-log10(padj): 3.945064e-02<br />log10(baseMean): 3.8100551<br />gene: FBgn0039302","log2FoldChange: -0.0994161882<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.2225013<br />gene: FBgn0039303","log2FoldChange: 0.1229332587<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 2.6703468<br />gene: FBgn0039304","log2FoldChange: 0.4476996556<br />-log10(padj): 6.458387e-01<br />log10(baseMean): 2.1817385<br />gene: FBgn0039305","log2FoldChange: -0.2897061504<br />-log10(padj): 7.932671e-01<br />log10(baseMean): 2.6273197<br />gene: FBgn0039306","log2FoldChange: -0.4768078518<br />-log10(padj): 1.115528e+00<br />log10(baseMean): 2.0715872<br />gene: FBgn0039321","log2FoldChange: 0.6230602766<br />-log10(padj): 7.109292e-01<br />log10(baseMean): 1.7592695<br />gene: FBgn0039323","log2FoldChange: -0.1828859684<br />-log10(padj): 1.047710e-01<br />log10(baseMean): 2.2156766<br />gene: FBgn0039328","log2FoldChange: 0.2278313980<br />-log10(padj): 1.526116e-01<br />log10(baseMean): 1.9258467<br />gene: FBgn0039329","log2FoldChange: -0.0529951392<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.3275170<br />gene: FBgn0039335","log2FoldChange: 0.1174074590<br />-log10(padj): 7.012969e-02<br />log10(baseMean): 2.0283442<br />gene: FBgn0039336","log2FoldChange: -0.0209347717<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 2.2076764<br />gene: FBgn0039337","log2FoldChange: -0.0481688912<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 3.4461967<br />gene: FBgn0039338","log2FoldChange: -0.0429310198<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.3263994<br />gene: FBgn0039339","log2FoldChange: 0.1706726955<br />-log10(padj): 2.978285e-01<br />log10(baseMean): 3.0973493<br />gene: FBgn0039341","log2FoldChange: 0.4482413662<br />-log10(padj): 1.391519e-01<br />log10(baseMean): 1.3281629<br />gene: FBgn0039347","log2FoldChange: 0.1070144053<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 2.9058431<br />gene: FBgn0039348","log2FoldChange: 0.0283341225<br />-log10(padj): 1.693673e-02<br />log10(baseMean): 2.5861617<br />gene: FBgn0039349","log2FoldChange: 0.1272403649<br />-log10(padj): 1.073550e-01<br />log10(baseMean): 2.2897667<br />gene: FBgn0039350","log2FoldChange: -0.3974724925<br />-log10(padj): 6.233936e-01<br />log10(baseMean): 2.6663607<br />gene: FBgn0039352","log2FoldChange: -0.6539851714<br />-log10(padj): 6.978278e-01<br />log10(baseMean): 1.6404003<br />gene: FBgn0039354","log2FoldChange: 0.2861435081<br />-log10(padj): 2.717944e-01<br />log10(baseMean): 1.9792998<br />gene: FBgn0039355","log2FoldChange: 0.5107236838<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 1.2773764<br />gene: FBgn0039356","log2FoldChange: 0.1999813909<br />-log10(padj): 9.877629e-02<br />log10(baseMean): 1.8533103<br />gene: FBgn0039357","log2FoldChange: -0.1627543255<br />-log10(padj): 2.944630e-01<br />log10(baseMean): 2.9947445<br />gene: FBgn0039358","log2FoldChange: -0.1345322804<br />-log10(padj): 1.340074e-01<br />log10(baseMean): 4.2881684<br />gene: FBgn0039359","log2FoldChange: 0.1337458640<br />-log10(padj): 1.106085e-01<br />log10(baseMean): 2.5446554<br />gene: FBgn0039360","log2FoldChange: -0.5877902973<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 1.0771024<br />gene: FBgn0039371","log2FoldChange: 0.2759504804<br />-log10(padj): 1.040319e+00<br />log10(baseMean): 2.8545402<br />gene: FBgn0039379","log2FoldChange: 0.6043667625<br />-log10(padj): 3.143762e+00<br />log10(baseMean): 2.8651639<br />gene: FBgn0039381","log2FoldChange: 0.3799733197<br />-log10(padj): 1.746073e+00<br />log10(baseMean): 2.8620769<br />gene: FBgn0039385","log2FoldChange: -0.4180019550<br />-log10(padj): 9.212457e-02<br />log10(baseMean): 1.0951633<br />gene: FBgn0039386","log2FoldChange: 0.1262281103<br />-log10(padj): 1.423391e-02<br />log10(baseMean): 0.8102911<br />gene: FBgn0039387","log2FoldChange: -0.0444595558<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 3.0764848<br />gene: FBgn0039402","log2FoldChange: 0.0278249258<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 2.3089818<br />gene: FBgn0039404","log2FoldChange: 0.0756699066<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 3.5059306<br />gene: FBgn0039406","log2FoldChange: -0.0682426110<br />-log10(padj): 3.792700e-02<br />log10(baseMean): 2.1782470<br />gene: FBgn0039407","log2FoldChange: 0.3156143333<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 1.3681933<br />gene: FBgn0039415","log2FoldChange: -0.3214113505<br />-log10(padj): 5.626611e-01<br />log10(baseMean): 2.3214054<br />gene: FBgn0039417","log2FoldChange: -1.1603789307<br />-log10(padj): 2.932092e+01<br />log10(baseMean): 3.1943148<br />gene: FBgn0039419","log2FoldChange: 0.4253809576<br />-log10(padj): 2.286833e-01<br />log10(baseMean): 1.5059926<br />gene: FBgn0039420","log2FoldChange: -0.0055718955<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.7936663<br />gene: FBgn0039427","log2FoldChange: 0.5994504761<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 0.9492396<br />gene: FBgn0039429","log2FoldChange: -0.1256040034<br />-log10(padj): 1.599553e-01<br />log10(baseMean): 2.7055248<br />gene: FBgn0039430","log2FoldChange: -0.1741514208<br />-log10(padj): 2.258934e-01<br />log10(baseMean): 3.4155729<br />gene: FBgn0039431","log2FoldChange: -0.0183315158<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.4501653<br />gene: FBgn0039449","log2FoldChange: -0.0132347244<br />-log10(padj): 8.910680e-03<br />log10(baseMean): 3.1356322<br />gene: FBgn0039450","log2FoldChange: -0.0996111689<br />-log10(padj): 6.868318e-02<br />log10(baseMean): 3.2000049<br />gene: FBgn0039451","log2FoldChange: 0.0456341293<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 1.0287557<br />gene: FBgn0039454","log2FoldChange: 0.0195237946<br />-log10(padj): 9.009111e-03<br />log10(baseMean): 2.3033644<br />gene: FBgn0039459","log2FoldChange: -0.0033097960<br />-log10(padj): 1.611385e-03<br />log10(baseMean): 2.3238523<br />gene: FBgn0039461","log2FoldChange: 0.3543150686<br />-log10(padj): 3.549557e-01<br />log10(baseMean): 1.9231967<br />gene: FBgn0039462","log2FoldChange: 1.1262212061<br />-log10(padj): 8.017504e+00<br />log10(baseMean): 2.9140993<br />gene: FBgn0039464","log2FoldChange: 0.2846381232<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 2.6717110<br />gene: FBgn0039465","log2FoldChange: -0.1575302511<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 3.2520792<br />gene: FBgn0039466","log2FoldChange: -0.4618249141<br />-log10(padj): 2.221655e+00<br />log10(baseMean): 3.7362225<br />gene: FBgn0039467","log2FoldChange: -0.3088632261<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.4844929<br />gene: FBgn0039478","log2FoldChange: 1.5494362861<br />-log10(padj): 1.215634e+01<br />log10(baseMean): 2.2447254<br />gene: FBgn0039479","log2FoldChange: 1.3256183484<br />-log10(padj): 4.950056e+00<br />log10(baseMean): 1.7676756<br />gene: FBgn0039485","log2FoldChange: 0.1368918981<br />-log10(padj): 1.253921e-01<br />log10(baseMean): 3.0334793<br />gene: FBgn0039487","log2FoldChange: -0.0372059246<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.6117612<br />gene: FBgn0039488","log2FoldChange: 0.1907730697<br />-log10(padj): 1.061781e-01<br />log10(baseMean): 2.2073567<br />gene: FBgn0039489","log2FoldChange: -0.3302235006<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 0.9550211<br />gene: FBgn0039490","log2FoldChange: 0.0738108191<br />-log10(padj): 4.192583e-02<br />log10(baseMean): 3.1629184<br />gene: FBgn0039492","log2FoldChange: -0.3700361063<br />-log10(padj): 7.833850e-01<br />log10(baseMean): 3.3444223<br />gene: FBgn0039494","log2FoldChange: -0.2226524738<br />-log10(padj): 2.809186e-02<br />log10(baseMean): 0.9697058<br />gene: FBgn0039498","log2FoldChange: -0.9790632949<br />-log10(padj): 6.004532e-01<br />log10(baseMean): 1.3073443<br />gene: FBgn0039500","log2FoldChange: -0.1146827561<br />-log10(padj): 7.847011e-02<br />log10(baseMean): 2.2335736<br />gene: FBgn0039501","log2FoldChange: 0.1519358559<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 1.1837218<br />gene: FBgn0039503","log2FoldChange: -0.2598519763<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.1727420<br />gene: FBgn0039504","log2FoldChange: 0.3044030644<br />-log10(padj): 6.774474e-01<br />log10(baseMean): 2.4620040<br />gene: FBgn0039505","log2FoldChange: -0.1554999406<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 3.1173807<br />gene: FBgn0039507","log2FoldChange: -0.0393701416<br />-log10(padj): 2.803547e-02<br />log10(baseMean): 3.3336373<br />gene: FBgn0039508","log2FoldChange: -0.2479270416<br />-log10(padj): 3.921646e-01<br />log10(baseMean): 2.6988240<br />gene: FBgn0039509","log2FoldChange: 0.1605213302<br />-log10(padj): 4.614145e-02<br />log10(baseMean): 1.4985073<br />gene: FBgn0039510","log2FoldChange: 0.4791408522<br />-log10(padj): 1.799779e+00<br />log10(baseMean): 2.5375573<br />gene: FBgn0039525","log2FoldChange: -0.7305267274<br />-log10(padj): 7.239618e-01<br />log10(baseMean): 1.5380413<br />gene: FBgn0039527","log2FoldChange: -0.1597017841<br />-log10(padj): 2.543856e-01<br />log10(baseMean): 3.5371515<br />gene: FBgn0039528","log2FoldChange: -0.1524179200<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.8485789<br />gene: FBgn0039530","log2FoldChange: 0.1532460629<br />-log10(padj): 1.180403e-01<br />log10(baseMean): 2.2263700<br />gene: FBgn0039531","log2FoldChange: 0.1185688148<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 2.9558145<br />gene: FBgn0039532","log2FoldChange: -0.6372716031<br />-log10(padj): 1.404418e-01<br />log10(baseMean): 0.9201708<br />gene: FBgn0039536","log2FoldChange: 0.0670619975<br />-log10(padj): 4.967206e-02<br />log10(baseMean): 2.9418274<br />gene: FBgn0039537","log2FoldChange: 0.8202953753<br />-log10(padj): 3.096008e+00<br />log10(baseMean): 2.0196629<br />gene: FBgn0039538","log2FoldChange: 0.3595509978<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.0367474<br />gene: FBgn0039540","log2FoldChange: 0.0874360388<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 2.5700034<br />gene: FBgn0039543","log2FoldChange: 0.3082411020<br />-log10(padj): 1.077568e+00<br />log10(baseMean): 3.0605187<br />gene: FBgn0039544","log2FoldChange: 0.0054472617<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.9444923<br />gene: FBgn0039554","log2FoldChange: -0.0365896222<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 3.1121251<br />gene: FBgn0039555","log2FoldChange: 0.0563583230<br />-log10(padj): 4.218202e-02<br />log10(baseMean): 2.7260194<br />gene: FBgn0039557","log2FoldChange: 0.2176347375<br />-log10(padj): 3.765035e-01<br />log10(baseMean): 2.5093923<br />gene: FBgn0039558","log2FoldChange: -0.2226479615<br />-log10(padj): 7.463906e-01<br />log10(baseMean): 3.3312570<br />gene: FBgn0039559","log2FoldChange: -0.0165498333<br />-log10(padj): 8.011960e-03<br />log10(baseMean): 3.1637007<br />gene: FBgn0039560","log2FoldChange: -0.0913568096<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 3.1007805<br />gene: FBgn0039561","log2FoldChange: 0.2650599162<br />-log10(padj): 5.630377e-01<br />log10(baseMean): 4.2706923<br />gene: FBgn0039562","log2FoldChange: -0.0245984129<br />-log10(padj): 1.315175e-02<br />log10(baseMean): 2.8380093<br />gene: FBgn0039563","log2FoldChange: -0.4145501294<br />-log10(padj): 2.545995e-01<br />log10(baseMean): 1.6662133<br />gene: FBgn0039564","log2FoldChange: 0.2168123698<br />-log10(padj): 2.303311e-01<br />log10(baseMean): 2.2611422<br />gene: FBgn0039565","log2FoldChange: -0.0023244928<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 3.0068525<br />gene: FBgn0039566","log2FoldChange: -0.3214048261<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 3.3866161<br />gene: FBgn0039580","log2FoldChange: -0.0446238626<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 3.3495599<br />gene: FBgn0039581","log2FoldChange: 0.0843431220<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 2.2123015<br />gene: FBgn0039588","log2FoldChange: 0.1307815497<br />-log10(padj): 1.170642e-01<br />log10(baseMean): 2.4364925<br />gene: FBgn0039589","log2FoldChange: 0.4217646459<br />-log10(padj): 2.312069e+00<br />log10(baseMean): 3.1629847<br />gene: FBgn0039590","log2FoldChange: 1.9379546653<br />-log10(padj): 7.462704e+00<br />log10(baseMean): 1.6283392<br />gene: FBgn0039593","log2FoldChange: 0.0927998181<br />-log10(padj): 1.528174e-02<br />log10(baseMean): 1.0727589<br />gene: FBgn0039594","log2FoldChange: -0.1873269968<br />-log10(padj): 5.222391e-01<br />log10(baseMean): 3.3215049<br />gene: FBgn0039600","log2FoldChange: 0.1084203386<br />-log10(padj): 1.084626e-01<br />log10(baseMean): 2.9271032<br />gene: FBgn0039601","log2FoldChange: 0.1108805464<br />-log10(padj): 1.081876e-01<br />log10(baseMean): 2.6682752<br />gene: FBgn0039602","log2FoldChange: 0.2014239400<br />-log10(padj): 2.508241e-02<br />log10(baseMean): 0.9398943<br />gene: FBgn0039611","log2FoldChange: -0.2990744922<br />-log10(padj): 9.684093e-01<br />log10(baseMean): 3.7608057<br />gene: FBgn0039612","log2FoldChange: -0.5339331001<br />-log10(padj): 3.379265e+00<br />log10(baseMean): 3.4522267<br />gene: FBgn0039613","log2FoldChange: -0.3405290491<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.4751403<br />gene: FBgn0039620","log2FoldChange: -0.1826859404<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 0.8810462<br />gene: FBgn0039621","log2FoldChange: 0.2853480838<br />-log10(padj): 2.073298e-01<br />log10(baseMean): 2.0083998<br />gene: FBgn0039622","log2FoldChange: -0.1535854453<br />-log10(padj): 3.169933e-01<br />log10(baseMean): 2.9555224<br />gene: FBgn0039623","log2FoldChange: 0.0826514277<br />-log10(padj): 5.444957e-02<br />log10(baseMean): 2.7288043<br />gene: FBgn0039625","log2FoldChange: -0.1896348174<br />-log10(padj): 3.398300e-01<br />log10(baseMean): 2.7644431<br />gene: FBgn0039626","log2FoldChange: -0.0814299820<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 2.2468475<br />gene: FBgn0039627","log2FoldChange: -0.0527988849<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 0.9702587<br />gene: FBgn0039630","log2FoldChange: 0.1916282385<br />-log10(padj): 1.692444e-01<br />log10(baseMean): 2.1762885<br />gene: FBgn0039631","log2FoldChange: 0.0262819477<br />-log10(padj): 2.116949e-02<br />log10(baseMean): 3.0586423<br />gene: FBgn0039632","log2FoldChange: -0.1501690469<br />-log10(padj): 1.224566e-01<br />log10(baseMean): 3.4843640<br />gene: FBgn0039633","log2FoldChange: -0.0771431151<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.0779642<br />gene: FBgn0039634","log2FoldChange: -0.0842722602<br />-log10(padj): 5.961065e-02<br />log10(baseMean): 3.2662489<br />gene: FBgn0039635","log2FoldChange: -0.2451466377<br />-log10(padj): 6.322214e-01<br />log10(baseMean): 2.9074506<br />gene: FBgn0039636","log2FoldChange: -0.5090031237<br />-log10(padj): 4.864486e+00<br />log10(baseMean): 3.4824982<br />gene: FBgn0039637","log2FoldChange: 0.1175875548<br />-log10(padj): 1.768229e-01<br />log10(baseMean): 2.8328844<br />gene: FBgn0039638","log2FoldChange: -0.0509914845<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.5904748<br />gene: FBgn0039639","log2FoldChange: -1.7987126567<br />-log10(padj): 9.240146e+00<br />log10(baseMean): 1.8574702<br />gene: FBgn0039640","log2FoldChange: 0.1641886416<br />-log10(padj): 1.052752e-01<br />log10(baseMean): 2.0722311<br />gene: FBgn0039641","log2FoldChange: 0.1932786944<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 2.2525244<br />gene: FBgn0039642","log2FoldChange: 0.4149973838<br />-log10(padj): 2.146238e+00<br />log10(baseMean): 3.0205470<br />gene: FBgn0039644","log2FoldChange: 1.1271947371<br />-log10(padj): 3.810681e-01<br />log10(baseMean): 0.8716666<br />gene: FBgn0039647","log2FoldChange: 0.4805060887<br />-log10(padj): 2.480212e-01<br />log10(baseMean): 1.7145136<br />gene: FBgn0039650","log2FoldChange: 1.1225110938<br />-log10(padj): 3.623814e+00<br />log10(baseMean): 1.8705763<br />gene: FBgn0039651","log2FoldChange: 0.2914982863<br />-log10(padj): 7.318301e-01<br />log10(baseMean): 2.8882217<br />gene: FBgn0039654","log2FoldChange: -0.3910218600<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 0.9666660<br />gene: FBgn0039655","log2FoldChange: 0.0166909717<br />-log10(padj): 5.649266e-03<br />log10(baseMean): 2.3290306<br />gene: FBgn0039663","log2FoldChange: 0.1161970208<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.9171002<br />gene: FBgn0039664","log2FoldChange: 0.2808363435<br />-log10(padj): 2.303742e-01<br />log10(baseMean): 3.3082039<br />gene: FBgn0039665","log2FoldChange: -0.2882788631<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 1.1564057<br />gene: FBgn0039667","log2FoldChange: -0.1367454884<br />-log10(padj): 1.331016e-01<br />log10(baseMean): 3.3701469<br />gene: FBgn0039668","log2FoldChange: -0.3427187984<br />-log10(padj): 4.517363e-02<br />log10(baseMean): 0.7892870<br />gene: FBgn0039673","log2FoldChange: -0.0308068084<br />-log10(padj): 2.426530e-02<br />log10(baseMean): 2.8073738<br />gene: FBgn0039674","log2FoldChange: -0.2536306316<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 1.2154298<br />gene: FBgn0039679","log2FoldChange: 0.2082329151<br />-log10(padj): 5.929130e-01<br />log10(baseMean): 3.1497934<br />gene: FBgn0039680","log2FoldChange: -0.0654063167<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 2.1296288<br />gene: FBgn0039682","log2FoldChange: -0.1339400071<br />-log10(padj): 9.902178e-02<br />log10(baseMean): 2.7721779<br />gene: FBgn0039687","log2FoldChange: 0.2099095745<br />-log10(padj): 3.366415e-02<br />log10(baseMean): 0.9734891<br />gene: FBgn0039688","log2FoldChange: 0.2478370477<br />-log10(padj): 4.756075e-01<br />log10(baseMean): 2.7936472<br />gene: FBgn0039689","log2FoldChange: 0.2093420065<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 2.4400639<br />gene: FBgn0039690","log2FoldChange: 0.0029641262<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 3.4196198<br />gene: FBgn0039691","log2FoldChange: -0.4165536403<br />-log10(padj): 5.739650e-01<br />log10(baseMean): 1.9765317<br />gene: FBgn0039694","log2FoldChange: 0.3056858444<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 1.1882747<br />gene: FBgn0039695","log2FoldChange: 0.3394261798<br />-log10(padj): 1.415959e+00<br />log10(baseMean): 2.8563665<br />gene: FBgn0039696","log2FoldChange: 0.1135308172<br />-log10(padj): 1.474974e-01<br />log10(baseMean): 3.3552024<br />gene: FBgn0039697","log2FoldChange: -0.0429993320<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.7089522<br />gene: FBgn0039698","log2FoldChange: 0.0369071997<br />-log10(padj): 2.373932e-02<br />log10(baseMean): 2.5895593<br />gene: FBgn0039702","log2FoldChange: -0.2829049869<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 1.0018993<br />gene: FBgn0039704","log2FoldChange: -0.0189759040<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 3.0586346<br />gene: FBgn0039705","log2FoldChange: -0.1171073097<br />-log10(padj): 9.409299e-02<br />log10(baseMean): 2.9301995<br />gene: FBgn0039710","log2FoldChange: 0.1450967394<br />-log10(padj): 2.158958e-01<br />log10(baseMean): 2.6812929<br />gene: FBgn0039711","log2FoldChange: 0.0155403049<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.9637524<br />gene: FBgn0039712","log2FoldChange: -0.1492229598<br />-log10(padj): 1.105617e-01<br />log10(baseMean): 4.9030193<br />gene: FBgn0039713","log2FoldChange: -0.6133026013<br />-log10(padj): 5.030836e+00<br />log10(baseMean): 3.3635896<br />gene: FBgn0039714","log2FoldChange: 0.1279083434<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.5240070<br />gene: FBgn0039726","log2FoldChange: 0.0074129371<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 3.0900472<br />gene: FBgn0039727","log2FoldChange: -0.0197503550<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.6238833<br />gene: FBgn0039730","log2FoldChange: 0.3803517910<br />-log10(padj): 6.092959e-01<br />log10(baseMean): 2.0530159<br />gene: FBgn0039731","log2FoldChange: -0.0107065038<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.1188864<br />gene: FBgn0039732","log2FoldChange: -0.1302371287<br />-log10(padj): 1.331873e-01<br />log10(baseMean): 2.8146926<br />gene: FBgn0039733","log2FoldChange: 0.3689045568<br />-log10(padj): 1.467019e+00<br />log10(baseMean): 3.1167552<br />gene: FBgn0039735","log2FoldChange: -0.2268774986<br />-log10(padj): 7.856885e-02<br />log10(baseMean): 1.4536137<br />gene: FBgn0039736","log2FoldChange: -0.1399746426<br />-log10(padj): 1.136128e-01<br />log10(baseMean): 3.6040762<br />gene: FBgn0039737","log2FoldChange: 0.2798944358<br />-log10(padj): 7.144529e-01<br />log10(baseMean): 2.7993012<br />gene: FBgn0039738","log2FoldChange: 0.0228167151<br />-log10(padj): 1.300138e-02<br />log10(baseMean): 2.6069431<br />gene: FBgn0039740","log2FoldChange: -0.2685627642<br />-log10(padj): 7.163575e-01<br />log10(baseMean): 2.7170597<br />gene: FBgn0039741","log2FoldChange: 0.3298835532<br />-log10(padj): 7.238624e-02<br />log10(baseMean): 1.1690192<br />gene: FBgn0039742","log2FoldChange: -0.0666098953<br />-log10(padj): 6.881408e-02<br />log10(baseMean): 3.0362445<br />gene: FBgn0039743","log2FoldChange: 0.3969083397<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 0.8659796<br />gene: FBgn0039747","log2FoldChange: 0.1238440397<br />-log10(padj): 1.075092e-01<br />log10(baseMean): 2.3257655<br />gene: FBgn0039751","log2FoldChange: 1.0255418143<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 1.0300548<br />gene: FBgn0039755","log2FoldChange: 0.4241693057<br />-log10(padj): 1.509222e+00<br />log10(baseMean): 3.2372015<br />gene: FBgn0039756","log2FoldChange: -0.1272727794<br />-log10(padj): 1.096547e-01<br />log10(baseMean): 4.6054650<br />gene: FBgn0039757","log2FoldChange: 1.9657085094<br />-log10(padj): 2.301617e+00<br />log10(baseMean): 1.1538402<br />gene: FBgn0039759","log2FoldChange: -0.2484180029<br />-log10(padj): 8.388118e-02<br />log10(baseMean): 2.0021906<br />gene: FBgn0039764","log2FoldChange: 0.1893499757<br />-log10(padj): 1.007396e-01<br />log10(baseMean): 2.0228947<br />gene: FBgn0039765","log2FoldChange: 0.3371147067<br />-log10(padj): 1.198375e-01<br />log10(baseMean): 1.8139228<br />gene: FBgn0039766","log2FoldChange: 0.0319502294<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 3.1118235<br />gene: FBgn0039767","log2FoldChange: 0.0357310709<br />-log10(padj): 1.685188e-02<br />log10(baseMean): 2.5489486<br />gene: FBgn0039773","log2FoldChange: 0.1065861414<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 2.6961911<br />gene: FBgn0039774","log2FoldChange: -0.4670956321<br />-log10(padj): 2.106054e+00<br />log10(baseMean): 3.5084766<br />gene: FBgn0039776","log2FoldChange: 1.7947849752<br />-log10(padj): 2.574824e+00<br />log10(baseMean): 1.2360562<br />gene: FBgn0039788","log2FoldChange: 0.0658623717<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.2988770<br />gene: FBgn0039789","log2FoldChange: -0.1126959264<br />-log10(padj): 1.250693e-01<br />log10(baseMean): 2.8158265<br />gene: FBgn0039790","log2FoldChange: -0.4779156519<br />-log10(padj): 1.643916e+00<br />log10(baseMean): 2.3882628<br />gene: FBgn0039797","log2FoldChange: -0.0074229025<br />-log10(padj): 1.989760e-03<br />log10(baseMean): 1.5476639<br />gene: FBgn0039798","log2FoldChange: -0.6797186144<br />-log10(padj): 7.135583e-01<br />log10(baseMean): 1.8176466<br />gene: FBgn0039800","log2FoldChange: -0.5455993028<br />-log10(padj): 9.543967e-01<br />log10(baseMean): 2.1626236<br />gene: FBgn0039801","log2FoldChange: -0.0406672628<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.9847426<br />gene: FBgn0039802","log2FoldChange: -0.4048177293<br />-log10(padj): 1.687576e+00<br />log10(baseMean): 2.9892318<br />gene: FBgn0039804","log2FoldChange: -0.9693242164<br />-log10(padj): 1.310368e+00<br />log10(baseMean): 1.5827673<br />gene: FBgn0039808","log2FoldChange: -0.8772921548<br />-log10(padj): 7.020595e+00<br />log10(baseMean): 2.7656579<br />gene: FBgn0039809","log2FoldChange: -4.1624252815<br />-log10(padj): 6.774940e+01<br />log10(baseMean): 2.4181540<br />gene: FBgn0039827","log2FoldChange: -0.1988490804<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.9150329<br />gene: FBgn0039828","log2FoldChange: -0.3201354480<br />-log10(padj): 3.258814e-01<br />log10(baseMean): 2.1654573<br />gene: FBgn0039829","log2FoldChange: -0.0587379553<br />-log10(padj): 4.328987e-02<br />log10(baseMean): 4.0098439<br />gene: FBgn0039830","log2FoldChange: -0.0497599006<br />-log10(padj): 2.451126e-02<br />log10(baseMean): 3.2521699<br />gene: FBgn0039831","log2FoldChange: -0.0092121013<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 0.9152092<br />gene: FBgn0039833","log2FoldChange: 0.3767082485<br />-log10(padj): 2.903845e-01<br />log10(baseMean): 2.2849934<br />gene: FBgn0039835","log2FoldChange: 0.0811745100<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 1.7083396<br />gene: FBgn0039836","log2FoldChange: 0.4001024141<br />-log10(padj): 6.196345e-01<br />log10(baseMean): 2.7653381<br />gene: FBgn0039844","log2FoldChange: -0.2502955002<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 2.7600204<br />gene: FBgn0039846","log2FoldChange: -0.5111919798<br />-log10(padj): 3.051412e-01<br />log10(baseMean): 1.4637729<br />gene: FBgn0039848","log2FoldChange: -0.0235778383<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 2.9737279<br />gene: FBgn0039849","log2FoldChange: 0.0698665783<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.3682915<br />gene: FBgn0039854","log2FoldChange: 0.1862901926<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 1.0246046<br />gene: FBgn0039855","log2FoldChange: -0.4700886262<br />-log10(padj): 1.243327e-01<br />log10(baseMean): 1.1842038<br />gene: FBgn0039856","log2FoldChange: -0.2320886485<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 4.6198978<br />gene: FBgn0039857","log2FoldChange: -0.0472486639<br />-log10(padj): 3.409843e-02<br />log10(baseMean): 4.2562435<br />gene: FBgn0039858","log2FoldChange: 0.2899753129<br />-log10(padj): 1.251486e-01<br />log10(baseMean): 1.8944565<br />gene: FBgn0039859","log2FoldChange: 0.0728674651<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.1781041<br />gene: FBgn0039860","log2FoldChange: -0.2535607453<br />-log10(padj): 5.276647e-01<br />log10(baseMean): 2.4800471<br />gene: FBgn0039861","log2FoldChange: 0.2465733849<br />-log10(padj): 8.286788e-02<br />log10(baseMean): 1.4787447<br />gene: FBgn0039862","log2FoldChange: 0.0563756517<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 3.3562339<br />gene: FBgn0039863","log2FoldChange: -0.0768059137<br />-log10(padj): 6.470652e-02<br />log10(baseMean): 2.5252275<br />gene: FBgn0039867","log2FoldChange: -0.1368288715<br />-log10(padj): 7.442140e-02<br />log10(baseMean): 2.4351443<br />gene: FBgn0039868","log2FoldChange: 0.3868158222<br />-log10(padj): 1.209635e+00<br />log10(baseMean): 3.2207692<br />gene: FBgn0039869","log2FoldChange: 0.3609910756<br />-log10(padj): 5.641826e-01<br />log10(baseMean): 2.3662694<br />gene: FBgn0039870","log2FoldChange: -0.3516691996<br />-log10(padj): 1.865885e+00<br />log10(baseMean): 2.9455054<br />gene: FBgn0039874","log2FoldChange: -0.0712617234<br />-log10(padj): 7.943526e-02<br />log10(baseMean): 3.1907569<br />gene: FBgn0039875","log2FoldChange: 0.5186004038<br />-log10(padj): 2.206135e+00<br />log10(baseMean): 2.4434308<br />gene: FBgn0039876","log2FoldChange: -0.0149812318<br />-log10(padj): 7.540631e-03<br />log10(baseMean): 2.7514966<br />gene: FBgn0039877","log2FoldChange: 0.5062941626<br />-log10(padj): 5.503417e-01<br />log10(baseMean): 1.7536682<br />gene: FBgn0039881","log2FoldChange: -0.0354463656<br />-log10(padj): 1.816628e-02<br />log10(baseMean): 2.3738130<br />gene: FBgn0039882","log2FoldChange: 0.5138153601<br />-log10(padj): 7.142616e-01<br />log10(baseMean): 1.9455467<br />gene: FBgn0039883","log2FoldChange: -0.6326075575<br />-log10(padj): 6.531488e+00<br />log10(baseMean): 2.9539323<br />gene: FBgn0039886","log2FoldChange: -0.0358559499<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.5566579<br />gene: FBgn0039889","log2FoldChange: -0.2202584301<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 3.0800078<br />gene: FBgn0039890","log2FoldChange: -0.2327319118<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 2.3596655<br />gene: FBgn0039896","log2FoldChange: -0.2814384109<br />-log10(padj): 2.158135e-01<br />log10(baseMean): 2.8684321<br />gene: FBgn0039897","log2FoldChange: -0.1289368696<br />-log10(padj): 5.262197e-02<br />log10(baseMean): 2.8402173<br />gene: FBgn0039902","log2FoldChange: -0.1446931412<br />-log10(padj): 7.848184e-02<br />log10(baseMean): 3.2703664<br />gene: FBgn0039904","log2FoldChange: 0.1889815217<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 1.6583586<br />gene: FBgn0039905","log2FoldChange: -0.1647816172<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 3.0749144<br />gene: FBgn0039907","log2FoldChange: 0.1766937360<br />-log10(padj): 8.584195e-02<br />log10(baseMean): 2.2126157<br />gene: FBgn0039908","log2FoldChange: 0.0283142218<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 3.2202460<br />gene: FBgn0039909","log2FoldChange: 0.3229159746<br />-log10(padj): 1.788166e-01<br />log10(baseMean): 1.7279704<br />gene: FBgn0039911","log2FoldChange: -0.0722004832<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 2.7733071<br />gene: FBgn0039914","log2FoldChange: -0.6328895588<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 1.0929687<br />gene: FBgn0039915","log2FoldChange: -0.1243938412<br />-log10(padj): 1.797597e-02<br />log10(baseMean): 1.2545906<br />gene: FBgn0039916","log2FoldChange: -0.0336988926<br />-log10(padj): 1.347426e-02<br />log10(baseMean): 2.8054736<br />gene: FBgn0039920","log2FoldChange: -0.1192830612<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.9423783<br />gene: FBgn0039923","log2FoldChange: -0.0742564704<br />-log10(padj): 2.197671e-02<br />log10(baseMean): 2.5561760<br />gene: FBgn0039924","log2FoldChange: 0.2374980931<br />-log10(padj): 3.330261e-02<br />log10(baseMean): 1.0152877<br />gene: FBgn0039925","log2FoldChange: 0.4610582408<br />-log10(padj): 3.169933e-01<br />log10(baseMean): 2.4444451<br />gene: FBgn0039927","log2FoldChange: 0.2567550785<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.1095895<br />gene: FBgn0039928","log2FoldChange: -0.2047603346<br />-log10(padj): 1.546995e-01<br />log10(baseMean): 2.6853612<br />gene: FBgn0039929","log2FoldChange: -0.2902629466<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 2.6119499<br />gene: FBgn0039930","log2FoldChange: 0.0798791841<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.2794617<br />gene: FBgn0039932","log2FoldChange: -0.0798145612<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 3.4674278<br />gene: FBgn0039936","log2FoldChange: -0.1400000407<br />-log10(padj): 8.535439e-02<br />log10(baseMean): 2.4795419<br />gene: FBgn0039938","log2FoldChange: -0.0711164607<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 2.5554440<br />gene: FBgn0039965","log2FoldChange: -0.3662429724<br />-log10(padj): 4.050933e-01<br />log10(baseMean): 1.9689112<br />gene: FBgn0039966","log2FoldChange: 0.0580455233<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.6256942<br />gene: FBgn0039969","log2FoldChange: -0.1723101026<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.8734163<br />gene: FBgn0039970","log2FoldChange: -0.1202648523<br />-log10(padj): 6.229977e-02<br />log10(baseMean): 3.7518938<br />gene: FBgn0039972","log2FoldChange: -0.1327563233<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.5500003<br />gene: FBgn0039977","log2FoldChange: 0.2080344995<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 1.4239017<br />gene: FBgn0039979","log2FoldChange: -0.0006087661<br />-log10(padj): 3.453894e-04<br />log10(baseMean): 2.7992878<br />gene: FBgn0039994","log2FoldChange: -0.0563862807<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.8941406<br />gene: FBgn0039997","log2FoldChange: -0.0462792635<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.5714901<br />gene: FBgn0040002","log2FoldChange: -0.0512778226<br />-log10(padj): 8.011960e-03<br />log10(baseMean): 2.6673903<br />gene: FBgn0040005","log2FoldChange: 0.1225413730<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 3.5412888<br />gene: FBgn0040007","log2FoldChange: -0.6051955506<br />-log10(padj): 8.928500e-01<br />log10(baseMean): 2.6749804<br />gene: FBgn0040009","log2FoldChange: 0.1060301681<br />-log10(padj): 4.369314e-02<br />log10(baseMean): 3.0148316<br />gene: FBgn0040010","log2FoldChange: -0.0269968481<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.4249840<br />gene: FBgn0040011","log2FoldChange: -0.0099273453<br />-log10(padj): 1.414129e-03<br />log10(baseMean): 1.0214168<br />gene: FBgn0040045","log2FoldChange: -0.1899022442<br />-log10(padj): 3.919386e-02<br />log10(baseMean): 2.6701205<br />gene: FBgn0040056","log2FoldChange: 0.0016004898<br />-log10(padj): 1.362130e-03<br />log10(baseMean): 3.1584339<br />gene: FBgn0040064","log2FoldChange: -0.1957238739<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 2.9324726<br />gene: FBgn0040066","log2FoldChange: -0.0464601080<br />-log10(padj): 4.225299e-02<br />log10(baseMean): 3.0787065<br />gene: FBgn0040068","log2FoldChange: 0.3113783489<br />-log10(padj): 1.359958e+00<br />log10(baseMean): 3.5651616<br />gene: FBgn0040070","log2FoldChange: -0.3031695508<br />-log10(padj): 5.461124e-01<br />log10(baseMean): 3.0979786<br />gene: FBgn0040071","log2FoldChange: 0.0569891778<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 2.9328804<br />gene: FBgn0040075","log2FoldChange: 0.1459453813<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 2.9466785<br />gene: FBgn0040078","log2FoldChange: -0.1723248181<br />-log10(padj): 3.589419e-01<br />log10(baseMean): 2.7666770<br />gene: FBgn0040079","log2FoldChange: 0.1781191588<br />-log10(padj): 5.884366e-01<br />log10(baseMean): 3.2979180<br />gene: FBgn0040080","log2FoldChange: 0.0445773907<br />-log10(padj): 2.957270e-02<br />log10(baseMean): 3.1172243<br />gene: FBgn0040087","log2FoldChange: 0.0691682917<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 3.1407773<br />gene: FBgn0040089","log2FoldChange: -1.5752915538<br />-log10(padj): 3.487438e+01<br />log10(baseMean): 3.0439549<br />gene: FBgn0040091","log2FoldChange: -1.7811399613<br />-log10(padj): 1.785701e+01<br />log10(baseMean): 2.9590808<br />gene: FBgn0040099","log2FoldChange: -0.4926967549<br />-log10(padj): 7.943526e-02<br />log10(baseMean): 0.8285531<br />gene: FBgn0040104","log2FoldChange: 0.4612907985<br />-log10(padj): 2.942091e-01<br />log10(baseMean): 1.9613803<br />gene: FBgn0040153","log2FoldChange: -0.0654934371<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.9397930<br />gene: FBgn0040206","log2FoldChange: -0.1210218060<br />-log10(padj): 2.205829e-01<br />log10(baseMean): 3.0091323<br />gene: FBgn0040207","log2FoldChange: -0.0261325923<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.9832274<br />gene: FBgn0040208","log2FoldChange: 0.1827000197<br />-log10(padj): 2.326510e-01<br />log10(baseMean): 2.7313232<br />gene: FBgn0040212","log2FoldChange: -0.1292993919<br />-log10(padj): 1.672272e-01<br />log10(baseMean): 3.5931293<br />gene: FBgn0040227","log2FoldChange: 0.0445843229<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.3306149<br />gene: FBgn0040228","log2FoldChange: 0.0052042203<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 2.8461015<br />gene: FBgn0040230","log2FoldChange: -0.0723485731<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.3610108<br />gene: FBgn0040232","log2FoldChange: -0.1215206906<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 2.7388416<br />gene: FBgn0040234","log2FoldChange: -0.2077764900<br />-log10(padj): 2.726663e-01<br />log10(baseMean): 2.5998145<br />gene: FBgn0040235","log2FoldChange: 0.1294349566<br />-log10(padj): 1.937941e-01<br />log10(baseMean): 3.4778099<br />gene: FBgn0040236","log2FoldChange: -0.0743288549<br />-log10(padj): 6.106439e-02<br />log10(baseMean): 3.3077809<br />gene: FBgn0040237","log2FoldChange: 0.2632978340<br />-log10(padj): 7.927105e-01<br />log10(baseMean): 3.2944249<br />gene: FBgn0040238","log2FoldChange: -0.1007628164<br />-log10(padj): 4.403512e-02<br />log10(baseMean): 2.6031015<br />gene: FBgn0040239","log2FoldChange: 0.0516884032<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 2.0537339<br />gene: FBgn0040250","log2FoldChange: 0.3782554023<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 2.5752590<br />gene: FBgn0040251","log2FoldChange: 0.0022546545<br />-log10(padj): 1.068987e-03<br />log10(baseMean): 1.4767257<br />gene: FBgn0040252","log2FoldChange: -0.0781359098<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 1.9865855<br />gene: FBgn0040256","log2FoldChange: -0.7821082839<br />-log10(padj): 3.032358e-01<br />log10(baseMean): 1.1460344<br />gene: FBgn0040257","log2FoldChange: 0.1137418651<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 3.2951437<br />gene: FBgn0040259","log2FoldChange: -0.2544690641<br />-log10(padj): 4.186728e-01<br />log10(baseMean): 2.5349851<br />gene: FBgn0040260","log2FoldChange: -0.0088013974<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.7922700<br />gene: FBgn0040262","log2FoldChange: -0.1089213944<br />-log10(padj): 2.331077e-01<br />log10(baseMean): 3.3022933<br />gene: FBgn0040268","log2FoldChange: 0.9168350730<br />-log10(padj): 1.457307e+01<br />log10(baseMean): 3.0043410<br />gene: FBgn0040271","log2FoldChange: -0.0007804569<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.2051767<br />gene: FBgn0040273","log2FoldChange: 0.6551702658<br />-log10(padj): 2.891206e-01<br />log10(baseMean): 1.1972080<br />gene: FBgn0040281","log2FoldChange: -0.2025514547<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.4931267<br />gene: FBgn0040282","log2FoldChange: -0.1736981256<br />-log10(padj): 3.566542e-01<br />log10(baseMean): 3.1856196<br />gene: FBgn0040283","log2FoldChange: 0.1608058324<br />-log10(padj): 3.046922e-01<br />log10(baseMean): 3.2866259<br />gene: FBgn0040284","log2FoldChange: 0.0382378053<br />-log10(padj): 2.180150e-02<br />log10(baseMean): 3.3669160<br />gene: FBgn0040285","log2FoldChange: 0.0826812788<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.0926723<br />gene: FBgn0040286","log2FoldChange: -0.5595974998<br />-log10(padj): 5.006462e+00<br />log10(baseMean): 2.8521756<br />gene: FBgn0040290","log2FoldChange: 0.1955051499<br />-log10(padj): 4.764795e-01<br />log10(baseMean): 2.8658474<br />gene: FBgn0040294","log2FoldChange: -0.2169133745<br />-log10(padj): 5.123301e-01<br />log10(baseMean): 3.1713491<br />gene: FBgn0040297","log2FoldChange: -0.0441231411<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 2.4470067<br />gene: FBgn0040298","log2FoldChange: -0.2004266157<br />-log10(padj): 1.182302e-01<br />log10(baseMean): 2.5536228<br />gene: FBgn0040299","log2FoldChange: -0.2195844519<br />-log10(padj): 4.456353e-01<br />log10(baseMean): 2.7490951<br />gene: FBgn0040305","log2FoldChange: 0.3946662579<br />-log10(padj): 1.747302e+00<br />log10(baseMean): 2.8623401<br />gene: FBgn0040308","log2FoldChange: 0.1012402128<br />-log10(padj): 1.300740e-01<br />log10(baseMean): 4.1022485<br />gene: FBgn0040309","log2FoldChange: -0.1863070544<br />-log10(padj): 4.836802e-01<br />log10(baseMean): 3.2366231<br />gene: FBgn0040319","log2FoldChange: -0.0831087876<br />-log10(padj): 1.141528e-01<br />log10(baseMean): 3.2588784<br />gene: FBgn0040321","log2FoldChange: -0.2483543852<br />-log10(padj): 6.438294e-01<br />log10(baseMean): 2.8183946<br />gene: FBgn0040322","log2FoldChange: 0.2265194031<br />-log10(padj): 5.319589e-01<br />log10(baseMean): 2.5662339<br />gene: FBgn0040323","log2FoldChange: -0.1604228078<br />-log10(padj): 6.819536e-02<br />log10(baseMean): 2.9452849<br />gene: FBgn0040324","log2FoldChange: 0.1424645677<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 1.6789959<br />gene: FBgn0040333","log2FoldChange: -0.0092859261<br />-log10(padj): 4.478921e-03<br />log10(baseMean): 2.8981298<br />gene: FBgn0040334","log2FoldChange: -0.0557222851<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 3.5761539<br />gene: FBgn0040335","log2FoldChange: -0.1382086801<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.5734492<br />gene: FBgn0040336","log2FoldChange: 0.2755411218<br />-log10(padj): 4.062885e-01<br />log10(baseMean): 2.2726934<br />gene: FBgn0040337","log2FoldChange: 0.0065054823<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.0542460<br />gene: FBgn0040339","log2FoldChange: 0.1451643343<br />-log10(padj): 1.475887e-01<br />log10(baseMean): 2.9594370<br />gene: FBgn0040340","log2FoldChange: -0.4030220659<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 0.9554560<br />gene: FBgn0040342","log2FoldChange: -0.1117866151<br />-log10(padj): 1.105678e-01<br />log10(baseMean): 2.6494184<br />gene: FBgn0040344","log2FoldChange: 0.0223626696<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 2.4494389<br />gene: FBgn0040346","log2FoldChange: -0.0307564834<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 1.8810702<br />gene: FBgn0040347","log2FoldChange: 0.3041858350<br />-log10(padj): 2.073298e-01<br />log10(baseMean): 1.7163794<br />gene: FBgn0040348","log2FoldChange: -0.6605116638<br />-log10(padj): 4.619223e-01<br />log10(baseMean): 1.4232703<br />gene: FBgn0040351","log2FoldChange: 0.4921813007<br />-log10(padj): 5.522168e-01<br />log10(baseMean): 1.9300973<br />gene: FBgn0040364","log2FoldChange: 0.0507053646<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 1.6782983<br />gene: FBgn0040366","log2FoldChange: -0.6200196460<br />-log10(padj): 1.526116e-01<br />log10(baseMean): 0.9528452<br />gene: FBgn0040367","log2FoldChange: -0.1741648375<br />-log10(padj): 1.590182e-01<br />log10(baseMean): 2.8099501<br />gene: FBgn0040372","log2FoldChange: 0.7222308867<br />-log10(padj): 1.561096e+00<br />log10(baseMean): 1.9231931<br />gene: FBgn0040373","log2FoldChange: -0.0533766486<br />-log10(padj): 3.666423e-02<br />log10(baseMean): 2.5868193<br />gene: FBgn0040375","log2FoldChange: 0.0162193371<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.0264681<br />gene: FBgn0040376","log2FoldChange: 0.4077376088<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 1.1961997<br />gene: FBgn0040377","log2FoldChange: 0.2436223623<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 2.5805022<br />gene: FBgn0040382","log2FoldChange: -0.0211649386<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.5668421<br />gene: FBgn0040383","log2FoldChange: -0.4546667301<br />-log10(padj): 1.650052e+00<br />log10(baseMean): 2.3427951<br />gene: FBgn0040384","log2FoldChange: -1.0343224166<br />-log10(padj): 8.250130e+00<br />log10(baseMean): 3.1698007<br />gene: FBgn0040388","log2FoldChange: 0.2297234345<br />-log10(padj): 3.628149e-01<br />log10(baseMean): 2.5121699<br />gene: FBgn0040389","log2FoldChange: -1.2249986323<br />-log10(padj): 1.152375e+00<br />log10(baseMean): 1.3369969<br />gene: FBgn0040391","log2FoldChange: -0.4092053536<br />-log10(padj): 1.181581e-01<br />log10(baseMean): 1.1917109<br />gene: FBgn0040392","log2FoldChange: 0.0990763002<br />-log10(padj): 6.534919e-02<br />log10(baseMean): 3.0824323<br />gene: FBgn0040394","log2FoldChange: 0.0645250399<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 3.3843499<br />gene: FBgn0040395","log2FoldChange: 0.2528978632<br />-log10(padj): 2.764490e-01<br />log10(baseMean): 2.2123422<br />gene: FBgn0040396","log2FoldChange: 0.0366238012<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 2.2705269<br />gene: FBgn0040397","log2FoldChange: -0.7959581045<br />-log10(padj): 6.387195e+00<br />log10(baseMean): 3.6228609<br />gene: FBgn0040398","log2FoldChange: 0.0618566749<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 1.6246971<br />gene: FBgn0040465","log2FoldChange: 0.4317036148<br />-log10(padj): 4.324058e-01<br />log10(baseMean): 2.1782919<br />gene: FBgn0040466","log2FoldChange: 0.1751830666<br />-log10(padj): 5.304070e-02<br />log10(baseMean): 1.5965647<br />gene: FBgn0040467","log2FoldChange: -0.1348790441<br />-log10(padj): 1.290286e-01<br />log10(baseMean): 3.2995504<br />gene: FBgn0040475","log2FoldChange: -0.5754672353<br />-log10(padj): 1.720633e+00<br />log10(baseMean): 2.1906209<br />gene: FBgn0040477","log2FoldChange: 0.6281266363<br />-log10(padj): 1.408800e-01<br />log10(baseMean): 0.9352107<br />gene: FBgn0040491","log2FoldChange: -0.2310290124<br />-log10(padj): 5.129201e-01<br />log10(baseMean): 3.3335309<br />gene: FBgn0040493","log2FoldChange: -1.3066125632<br />-log10(padj): 4.911461e-01<br />log10(baseMean): 0.8168123<br />gene: FBgn0040503","log2FoldChange: -0.7768267576<br />-log10(padj): 4.961244e-01<br />log10(baseMean): 1.5185836<br />gene: FBgn0040505","log2FoldChange: 0.0705303948<br />-log10(padj): 5.532509e-02<br />log10(baseMean): 3.0313339<br />gene: FBgn0040512","log2FoldChange: 0.3316656440<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.0085823<br />gene: FBgn0040528","log2FoldChange: 0.2469788061<br />-log10(padj): 3.661991e-01<br />log10(baseMean): 3.1351208<br />gene: FBgn0040529","log2FoldChange: -0.5242204403<br />-log10(padj): 1.401391e-01<br />log10(baseMean): 1.0860747<br />gene: FBgn0040532","log2FoldChange: 0.1252228700<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.1743725<br />gene: FBgn0040534","log2FoldChange: 0.3707856265<br />-log10(padj): 1.300466e-01<br />log10(baseMean): 1.3968096<br />gene: FBgn0040551","log2FoldChange: 0.1747359675<br />-log10(padj): 4.007536e-02<br />log10(baseMean): 1.3106852<br />gene: FBgn0040554","log2FoldChange: 0.4779368663<br />-log10(padj): 1.143540e-01<br />log10(baseMean): 1.0548043<br />gene: FBgn0040571","log2FoldChange: 0.3129288973<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 1.3495588<br />gene: FBgn0040575","log2FoldChange: -0.1078840302<br />-log10(padj): 1.452132e-02<br />log10(baseMean): 0.9969676<br />gene: FBgn0040588","log2FoldChange: 0.1143507269<br />-log10(padj): 6.238437e-02<br />log10(baseMean): 2.7093819<br />gene: FBgn0040600","log2FoldChange: 0.3510557038<br />-log10(padj): 4.463829e-01<br />log10(baseMean): 2.5443520<br />gene: FBgn0040602","log2FoldChange: -0.2121417812<br />-log10(padj): 2.898402e-01<br />log10(baseMean): 2.3790816<br />gene: FBgn0040609","log2FoldChange: -0.2597935640<br />-log10(padj): 1.114791e-01<br />log10(baseMean): 1.6383847<br />gene: FBgn0040636","log2FoldChange: 0.4138668449<br />-log10(padj): 1.679081e-01<br />log10(baseMean): 1.5031710<br />gene: FBgn0040650","log2FoldChange: -0.5187637106<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 1.1388286<br />gene: FBgn0040658","log2FoldChange: -0.2925786914<br />-log10(padj): 1.130030e+00<br />log10(baseMean): 3.0765414<br />gene: FBgn0040660","log2FoldChange: 0.2521750953<br />-log10(padj): 3.939281e-01<br />log10(baseMean): 2.4223096<br />gene: FBgn0040666","log2FoldChange: 0.0974776532<br />-log10(padj): 3.091232e-02<br />log10(baseMean): 1.6352136<br />gene: FBgn0040670","log2FoldChange: -0.1917742261<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 1.7614885<br />gene: FBgn0040687","log2FoldChange: 0.5586795890<br />-log10(padj): 6.017031e-01<br />log10(baseMean): 1.6640948<br />gene: FBgn0040751","log2FoldChange: -0.8217568274<br />-log10(padj): 4.841651e+00<br />log10(baseMean): 3.6862021<br />gene: FBgn0040752","log2FoldChange: -0.1174662553<br />-log10(padj): 3.727526e-02<br />log10(baseMean): 2.1163004<br />gene: FBgn0040754","log2FoldChange: 0.2747788772<br />-log10(padj): 5.680556e-02<br />log10(baseMean): 1.1117768<br />gene: FBgn0040763","log2FoldChange: 0.4045600022<br />-log10(padj): 1.144949e+00<br />log10(baseMean): 3.1419123<br />gene: FBgn0040765","log2FoldChange: -0.0763614461<br />-log10(padj): 9.466923e-03<br />log10(baseMean): 1.1752938<br />gene: FBgn0040766","log2FoldChange: 0.3060725981<br />-log10(padj): 6.632754e-01<br />log10(baseMean): 2.9455000<br />gene: FBgn0040773","log2FoldChange: -0.0789922350<br />-log10(padj): 8.401040e-02<br />log10(baseMean): 3.7229717<br />gene: FBgn0040777","log2FoldChange: 0.2578311997<br />-log10(padj): 6.187511e-02<br />log10(baseMean): 1.3959632<br />gene: FBgn0040778","log2FoldChange: -0.1748419279<br />-log10(padj): 4.881401e-02<br />log10(baseMean): 1.5389120<br />gene: FBgn0040780","log2FoldChange: 0.7889488753<br />-log10(padj): 4.329783e-01<br />log10(baseMean): 1.3327817<br />gene: FBgn0040784","log2FoldChange: -0.4283946287<br />-log10(padj): 8.740408e-02<br />log10(baseMean): 1.2681794<br />gene: FBgn0040786","log2FoldChange: 0.1484927361<br />-log10(padj): 1.949321e-01<br />log10(baseMean): 3.0816524<br />gene: FBgn0040793","log2FoldChange: -0.9644706054<br />-log10(padj): 3.090547e+00<br />log10(baseMean): 4.6449568<br />gene: FBgn0040813","log2FoldChange: -1.9898460092<br />-log10(padj): 2.607078e+01<br />log10(baseMean): 2.3344133<br />gene: FBgn0040827","log2FoldChange: -0.1373868031<br />-log10(padj): 7.629605e-02<br />log10(baseMean): 2.2910112<br />gene: FBgn0040837","log2FoldChange: 0.1090494572<br />-log10(padj): 2.311688e-02<br />log10(baseMean): 1.4270967<br />gene: FBgn0040850","log2FoldChange: 0.2856122656<br />-log10(padj): 2.056518e-01<br />log10(baseMean): 2.0743719<br />gene: FBgn0040862","log2FoldChange: 0.0800638975<br />-log10(padj): 4.355667e-02<br />log10(baseMean): 2.2652448<br />gene: FBgn0040890","log2FoldChange: 0.2329100349<br />-log10(padj): 2.803235e-01<br />log10(baseMean): 2.3760654<br />gene: FBgn0040899","log2FoldChange: 0.9566855568<br />-log10(padj): 4.430721e-01<br />log10(baseMean): 1.0416955<br />gene: FBgn0040906","log2FoldChange: 0.0087131954<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.1215914<br />gene: FBgn0040907","log2FoldChange: 0.0187121319<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.6145395<br />gene: FBgn0040918","log2FoldChange: -0.0548349201<br />-log10(padj): 1.254009e-02<br />log10(baseMean): 1.6497509<br />gene: FBgn0040928","log2FoldChange: 0.0036640479<br />-log10(padj): 1.483411e-03<br />log10(baseMean): 1.9573010<br />gene: FBgn0040929","log2FoldChange: -0.0017257979<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 2.4247852<br />gene: FBgn0040931","log2FoldChange: 0.2997947258<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 2.5961151<br />gene: FBgn0040942","log2FoldChange: -0.0163304176<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 2.9284893<br />gene: FBgn0040954","log2FoldChange: 0.3307045389<br />-log10(padj): 5.491635e-01<br />log10(baseMean): 2.1102592<br />gene: FBgn0040964","log2FoldChange: 0.1545858992<br />-log10(padj): 1.526116e-01<br />log10(baseMean): 3.1581423<br />gene: FBgn0040985","log2FoldChange: 0.0193233302<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.9658338<br />gene: FBgn0041004","log2FoldChange: 0.2151201717<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.5770176<br />gene: FBgn0041005","log2FoldChange: 0.6742663316<br />-log10(padj): 4.393303e-01<br />log10(baseMean): 3.1462476<br />gene: FBgn0041087","log2FoldChange: -0.1576996313<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.7739681<br />gene: FBgn0041092","log2FoldChange: -0.2708471594<br />-log10(padj): 3.281711e-01<br />log10(baseMean): 3.2878671<br />gene: FBgn0041094","log2FoldChange: 0.2669811086<br />-log10(padj): 1.223780e-01<br />log10(baseMean): 1.6927831<br />gene: FBgn0041096","log2FoldChange: 0.0602224438<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.1385998<br />gene: FBgn0041100","log2FoldChange: -0.1489001495<br />-log10(padj): 1.511863e-01<br />log10(baseMean): 3.3797370<br />gene: FBgn0041111","log2FoldChange: 0.3384811220<br />-log10(padj): 4.432190e-01<br />log10(baseMean): 2.1623324<br />gene: FBgn0041147","log2FoldChange: 0.4998325485<br />-log10(padj): 1.182827e-01<br />log10(baseMean): 1.1014341<br />gene: FBgn0041150","log2FoldChange: 0.1060767881<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 2.8403695<br />gene: FBgn0041161","log2FoldChange: -0.0009828091<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.3657581<br />gene: FBgn0041164","log2FoldChange: -0.2341249727<br />-log10(padj): 6.288571e-01<br />log10(baseMean): 3.3825307<br />gene: FBgn0041171","log2FoldChange: 0.1881484636<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 1.9140573<br />gene: FBgn0041174","log2FoldChange: -0.4574696995<br />-log10(padj): 3.340362e+00<br />log10(baseMean): 3.9484233<br />gene: FBgn0041180","log2FoldChange: 0.2146709517<br />-log10(padj): 3.817271e-01<br />log10(baseMean): 3.3739431<br />gene: FBgn0041182","log2FoldChange: -0.6847106751<br />-log10(padj): 2.557879e+00<br />log10(baseMean): 2.2098344<br />gene: FBgn0041183","log2FoldChange: 0.1196226618<br />-log10(padj): 1.136128e-01<br />log10(baseMean): 2.9089326<br />gene: FBgn0041184","log2FoldChange: -0.1908928872<br />-log10(padj): 3.648503e-01<br />log10(baseMean): 2.8192444<br />gene: FBgn0041186","log2FoldChange: -0.2788352810<br />-log10(padj): 4.741993e-01<br />log10(baseMean): 3.7109327<br />gene: FBgn0041188","log2FoldChange: -0.4214846145<br />-log10(padj): 1.367952e+00<br />log10(baseMean): 2.8838979<br />gene: FBgn0041191","log2FoldChange: -0.2511397417<br />-log10(padj): 5.929130e-01<br />log10(baseMean): 3.2753247<br />gene: FBgn0041203","log2FoldChange: 0.0083843563<br />-log10(padj): 4.609975e-03<br />log10(baseMean): 3.2090677<br />gene: FBgn0041205","log2FoldChange: -0.1754618374<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 3.9046269<br />gene: FBgn0041210","log2FoldChange: 0.5905933006<br />-log10(padj): 5.126792e-01<br />log10(baseMean): 1.5598376<br />gene: FBgn0041225","log2FoldChange: -0.0834644604<br />-log10(padj): 7.188587e-03<br />log10(baseMean): 0.8608936<br />gene: FBgn0041229","log2FoldChange: -0.1829841503<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 1.0873463<br />gene: FBgn0041241","log2FoldChange: 0.0885552577<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 0.8293331<br />gene: FBgn0041243","log2FoldChange: -0.2596339991<br />-log10(padj): 3.957681e-02<br />log10(baseMean): 1.0825388<br />gene: FBgn0041244","log2FoldChange: -0.5120239433<br />-log10(padj): 2.440775e+00<br />log10(baseMean): 3.5720734<br />gene: FBgn0041342","log2FoldChange: 0.3045119594<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 1.6710509<br />gene: FBgn0041581","log2FoldChange: 0.0372268924<br />-log10(padj): 2.532859e-02<br />log10(baseMean): 2.9453698<br />gene: FBgn0041582","log2FoldChange: 0.0074288655<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.0870952<br />gene: FBgn0041585","log2FoldChange: 0.3354570233<br />-log10(padj): 1.636443e+00<br />log10(baseMean): 2.9262327<br />gene: FBgn0041588","log2FoldChange: 0.5620880424<br />-log10(padj): 2.786470e+00<br />log10(baseMean): 2.5080231<br />gene: FBgn0041604","log2FoldChange: 1.0088054617<br />-log10(padj): 1.331442e+00<br />log10(baseMean): 1.6048530<br />gene: FBgn0041605","log2FoldChange: -0.4483260563<br />-log10(padj): 2.613509e+00<br />log10(baseMean): 2.8770344<br />gene: FBgn0041607","log2FoldChange: -0.2406737863<br />-log10(padj): 3.563915e-02<br />log10(baseMean): 0.9504573<br />gene: FBgn0041621","log2FoldChange: 0.4232014767<br />-log10(padj): 2.177100e+00<br />log10(baseMean): 2.7138133<br />gene: FBgn0041627","log2FoldChange: 0.6034076096<br />-log10(padj): 2.443435e+00<br />log10(baseMean): 4.1187730<br />gene: FBgn0041629","log2FoldChange: 0.1344325890<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.9704174<br />gene: FBgn0041630","log2FoldChange: 0.1503770064<br />-log10(padj): 1.307513e-01<br />log10(baseMean): 2.4097514<br />gene: FBgn0041702","log2FoldChange: 0.1110577072<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.6442168<br />gene: FBgn0041710","log2FoldChange: -0.2499621566<br />-log10(padj): 1.809581e-01<br />log10(baseMean): 2.0139641<br />gene: FBgn0041712","log2FoldChange: 0.6427897145<br />-log10(padj): 1.475558e-01<br />log10(baseMean): 1.3796629<br />gene: FBgn0041721","log2FoldChange: -0.2761977952<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 1.7231796<br />gene: FBgn0041723","log2FoldChange: -0.0934303156<br />-log10(padj): 9.874229e-02<br />log10(baseMean): 3.6060333<br />gene: FBgn0041775","log2FoldChange: -0.1576396756<br />-log10(padj): 2.869994e-01<br />log10(baseMean): 3.3016757<br />gene: FBgn0041781","log2FoldChange: 0.0617687786<br />-log10(padj): 6.715249e-02<br />log10(baseMean): 3.5325688<br />gene: FBgn0041789","log2FoldChange: 0.1513086388<br />-log10(padj): 2.249970e-01<br />log10(baseMean): 2.8443018<br />gene: FBgn0042083","log2FoldChange: -0.1360439189<br />-log10(padj): 2.159052e-01<br />log10(baseMean): 3.4256308<br />gene: FBgn0042085","log2FoldChange: -0.1885006163<br />-log10(padj): 1.125061e-01<br />log10(baseMean): 2.1931131<br />gene: FBgn0042092","log2FoldChange: -0.9123314286<br />-log10(padj): 3.513335e+00<br />log10(baseMean): 2.0530058<br />gene: FBgn0042094","log2FoldChange: 1.6825789946<br />-log10(padj): 1.418463e+00<br />log10(baseMean): 1.0115926<br />gene: FBgn0042102","log2FoldChange: 0.4953427231<br />-log10(padj): 5.584344e-01<br />log10(baseMean): 1.7802615<br />gene: FBgn0042104","log2FoldChange: -1.2192578680<br />-log10(padj): 1.260715e+00<br />log10(baseMean): 1.4225014<br />gene: FBgn0042106","log2FoldChange: -0.1614713044<br />-log10(padj): 2.666229e-01<br />log10(baseMean): 2.9473342<br />gene: FBgn0042111","log2FoldChange: 0.0918135888<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.3284996<br />gene: FBgn0042112","log2FoldChange: 0.2475070351<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 1.6610582<br />gene: FBgn0042125","log2FoldChange: -0.2960999500<br />-log10(padj): 5.490546e-02<br />log10(baseMean): 1.0325388<br />gene: FBgn0042126","log2FoldChange: -0.1784924196<br />-log10(padj): 3.572320e-02<br />log10(baseMean): 1.2663069<br />gene: FBgn0042127","log2FoldChange: -0.0807192061<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 2.5392736<br />gene: FBgn0042132","log2FoldChange: -0.3989601125<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 1.8103862<br />gene: FBgn0042133","log2FoldChange: 0.0373176520<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 3.6674961<br />gene: FBgn0042134","log2FoldChange: -0.0321941118<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.1748863<br />gene: FBgn0042135","log2FoldChange: -0.1914043264<br />-log10(padj): 2.775125e-01<br />log10(baseMean): 3.0118559<br />gene: FBgn0042138","log2FoldChange: 0.0612510613<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 3.5239806<br />gene: FBgn0042174","log2FoldChange: 0.3995839201<br />-log10(padj): 8.503333e-02<br />log10(baseMean): 2.1602450<br />gene: FBgn0042175","log2FoldChange: 0.2585052962<br />-log10(padj): 1.866713e-01<br />log10(baseMean): 2.6706549<br />gene: FBgn0042177","log2FoldChange: 0.0250190921<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.0336338<br />gene: FBgn0042178","log2FoldChange: -0.1204265881<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.4433392<br />gene: FBgn0042180","log2FoldChange: -0.2553870830<br />-log10(padj): 2.877029e-01<br />log10(baseMean): 2.4730754<br />gene: FBgn0042185","log2FoldChange: 0.1243340361<br />-log10(padj): 3.568450e-02<br />log10(baseMean): 1.7149635<br />gene: FBgn0042205","log2FoldChange: 0.0429341240<br />-log10(padj): 7.820026e-03<br />log10(baseMean): 1.5775566<br />gene: FBgn0042206","log2FoldChange: 0.3075299449<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 1.1266950<br />gene: FBgn0042207","log2FoldChange: 0.1659648984<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.5802188<br />gene: FBgn0042213","log2FoldChange: -0.3580950530<br />-log10(padj): 3.642698e-01<br />log10(baseMean): 1.9456050<br />gene: FBgn0042627","log2FoldChange: 0.2419956358<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 0.8656325<br />gene: FBgn0042650","log2FoldChange: 0.0232499652<br />-log10(padj): 1.821339e-02<br />log10(baseMean): 3.1548132<br />gene: FBgn0042693","log2FoldChange: -0.5435186039<br />-log10(padj): 2.182740e-01<br />log10(baseMean): 1.3620901<br />gene: FBgn0042696","log2FoldChange: -0.1355145994<br />-log10(padj): 6.044567e-02<br />log10(baseMean): 2.1748409<br />gene: FBgn0042701","log2FoldChange: 0.2323759496<br />-log10(padj): 4.130467e-01<br />log10(baseMean): 3.0137441<br />gene: FBgn0042712","log2FoldChange: 0.1206925522<br />-log10(padj): 9.164097e-02<br />log10(baseMean): 2.1828090<br />gene: FBgn0043001","log2FoldChange: 0.0201640896<br />-log10(padj): 1.300138e-02<br />log10(baseMean): 2.5709240<br />gene: FBgn0043010","log2FoldChange: -0.0731987231<br />-log10(padj): 3.537453e-02<br />log10(baseMean): 2.2312433<br />gene: FBgn0043012","log2FoldChange: -0.0797476827<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 1.6388882<br />gene: FBgn0043025","log2FoldChange: 0.2494186758<br />-log10(padj): 6.002928e-01<br />log10(baseMean): 3.0353846<br />gene: FBgn0043070","log2FoldChange: -0.3349499910<br />-log10(padj): 1.575637e+00<br />log10(baseMean): 3.1176546<br />gene: FBgn0043362","log2FoldChange: -1.2097404933<br />-log10(padj): 5.650360e+00<br />log10(baseMean): 3.0773452<br />gene: FBgn0043364","log2FoldChange: -0.2783363649<br />-log10(padj): 3.424323e-01<br />log10(baseMean): 2.3413389<br />gene: FBgn0043455","log2FoldChange: -0.1473895627<br />-log10(padj): 2.391993e-01<br />log10(baseMean): 3.4862140<br />gene: FBgn0043456","log2FoldChange: 0.0644406819<br />-log10(padj): 2.778407e-02<br />log10(baseMean): 2.2333499<br />gene: FBgn0043457","log2FoldChange: 0.0256053376<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 3.2503178<br />gene: FBgn0043458","log2FoldChange: 0.7899708868<br />-log10(padj): 3.332154e-01<br />log10(baseMean): 1.3528850<br />gene: FBgn0043575","log2FoldChange: 0.1783537699<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 1.3331269<br />gene: FBgn0043783","log2FoldChange: -0.0499080376<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.0911059<br />gene: FBgn0043791","log2FoldChange: 0.0647727162<br />-log10(padj): 4.418619e-02<br />log10(baseMean): 2.5760244<br />gene: FBgn0043796","log2FoldChange: -0.0135056593<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 1.8141813<br />gene: FBgn0043799","log2FoldChange: 0.2498427713<br />-log10(padj): 6.896834e-01<br />log10(baseMean): 3.1647264<br />gene: FBgn0043806","log2FoldChange: -0.6403160310<br />-log10(padj): 5.122050e+00<br />log10(baseMean): 4.1678456<br />gene: FBgn0043841","log2FoldChange: 0.6886898481<br />-log10(padj): 1.585935e-01<br />log10(baseMean): 1.0573521<br />gene: FBgn0043853","log2FoldChange: -0.2983413636<br />-log10(padj): 2.001008e-01<br />log10(baseMean): 2.2124999<br />gene: FBgn0043854","log2FoldChange: -0.1084688195<br />-log10(padj): 1.034432e-01<br />log10(baseMean): 4.0610251<br />gene: FBgn0043884","log2FoldChange: -0.1167299899<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.0444402<br />gene: FBgn0043900","log2FoldChange: 0.2537235671<br />-log10(padj): 4.091065e-01<br />log10(baseMean): 3.2601654<br />gene: FBgn0043903","log2FoldChange: 0.5802995002<br />-log10(padj): 3.753221e-01<br />log10(baseMean): 1.4906989<br />gene: FBgn0043904","log2FoldChange: 0.5417931099<br />-log10(padj): 1.074738e+00<br />log10(baseMean): 2.1861050<br />gene: FBgn0044020","log2FoldChange: -0.0175116827<br />-log10(padj): 1.006592e-02<br />log10(baseMean): 2.9541551<br />gene: FBgn0044028","log2FoldChange: 0.2398549814<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 2.1751893<br />gene: FBgn0044030","log2FoldChange: -0.8960935183<br />-log10(padj): 5.490843e+00<br />log10(baseMean): 2.4376647<br />gene: FBgn0044047","log2FoldChange: -0.1399154046<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 3.5607319<br />gene: FBgn0044323","log2FoldChange: -0.1585324032<br />-log10(padj): 2.427348e-01<br />log10(baseMean): 3.1858181<br />gene: FBgn0044324","log2FoldChange: -0.2061510541<br />-log10(padj): 7.986459e-01<br />log10(baseMean): 3.3628560<br />gene: FBgn0044452","log2FoldChange: -0.0125877216<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.3302853<br />gene: FBgn0044510","log2FoldChange: 0.2367765180<br />-log10(padj): 1.512365e-01<br />log10(baseMean): 2.2945277<br />gene: FBgn0044511","log2FoldChange: 0.1935794233<br />-log10(padj): 1.389551e-01<br />log10(baseMean): 2.2310271<br />gene: FBgn0044823","log2FoldChange: -0.0207338348<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.3842617<br />gene: FBgn0044826","log2FoldChange: 0.0458893439<br />-log10(padj): 2.344795e-02<br />log10(baseMean): 2.5237616<br />gene: FBgn0044871","log2FoldChange: 0.1321810462<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.5081854<br />gene: FBgn0044872","log2FoldChange: -0.1287065486<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 3.3204206<br />gene: FBgn0045035","log2FoldChange: -0.0373416784<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 3.4170439<br />gene: FBgn0045063","log2FoldChange: -0.1502019739<br />-log10(padj): 2.288921e-01<br />log10(baseMean): 2.8808080<br />gene: FBgn0045064","log2FoldChange: -0.1045397215<br />-log10(padj): 1.526116e-01<br />log10(baseMean): 3.2473274<br />gene: FBgn0045073","log2FoldChange: 0.0374222937<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.5234094<br />gene: FBgn0045468","log2FoldChange: 0.0749189956<br />-log10(padj): 1.533839e-02<br />log10(baseMean): 1.5248632<br />gene: FBgn0045469","log2FoldChange: 0.2810177219<br />-log10(padj): 9.713627e-02<br />log10(baseMean): 1.4431108<br />gene: FBgn0045470","log2FoldChange: 0.5164724349<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 0.9708382<br />gene: FBgn0045474","log2FoldChange: 0.1084644441<br />-log10(padj): 2.122202e-02<br />log10(baseMean): 1.4004062<br />gene: FBgn0045495","log2FoldChange: 0.0490379040<br />-log10(padj): 2.563528e-02<br />log10(baseMean): 2.8897561<br />gene: FBgn0045761","log2FoldChange: 0.4366762136<br />-log10(padj): 1.027917e-01<br />log10(baseMean): 1.1485595<br />gene: FBgn0045770","log2FoldChange: 0.3413471737<br />-log10(padj): 1.345004e-01<br />log10(baseMean): 1.9765130<br />gene: FBgn0045800","log2FoldChange: 0.0357566491<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 4.0631956<br />gene: FBgn0045823","log2FoldChange: -0.1904977346<br />-log10(padj): 5.784598e-01<br />log10(baseMean): 3.2740615<br />gene: FBgn0045842","log2FoldChange: -0.6722612501<br />-log10(padj): 4.193900e+00<br />log10(baseMean): 3.3072889<br />gene: FBgn0045852","log2FoldChange: -0.0190045787<br />-log10(padj): 7.598195e-03<br />log10(baseMean): 3.0386990<br />gene: FBgn0045862","log2FoldChange: 0.2971674483<br />-log10(padj): 7.931230e-01<br />log10(baseMean): 3.1671522<br />gene: FBgn0045866","log2FoldChange: 0.5926931466<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 0.9819660<br />gene: FBgn0045980","log2FoldChange: 0.0253759388<br />-log10(padj): 1.349861e-02<br />log10(baseMean): 2.6287474<br />gene: FBgn0046114","log2FoldChange: 0.3056094858<br />-log10(padj): 6.261658e-01<br />log10(baseMean): 3.5150387<br />gene: FBgn0046214","log2FoldChange: -0.0361936412<br />-log10(padj): 2.803547e-02<br />log10(baseMean): 2.8997919<br />gene: FBgn0046222","log2FoldChange: 0.4212317375<br />-log10(padj): 9.152652e-01<br />log10(baseMean): 2.3004534<br />gene: FBgn0046225","log2FoldChange: -0.1076307269<br />-log10(padj): 1.240953e-01<br />log10(baseMean): 2.6722749<br />gene: FBgn0046247","log2FoldChange: 2.1476341191<br />-log10(padj): 2.118152e+00<br />log10(baseMean): 0.9894360<br />gene: FBgn0046258","log2FoldChange: -0.0019806396<br />-log10(padj): 2.813947e-04<br />log10(baseMean): 0.8155982<br />gene: FBgn0046272","log2FoldChange: 0.2302466203<br />-log10(padj): 2.845705e-01<br />log10(baseMean): 2.3416072<br />gene: FBgn0046296","log2FoldChange: -0.0331309188<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.3471921<br />gene: FBgn0046301","log2FoldChange: -0.5544431380<br />-log10(padj): 1.275311e+00<br />log10(baseMean): 2.3002695<br />gene: FBgn0046322","log2FoldChange: 0.1748082214<br />-log10(padj): 2.356804e-01<br />log10(baseMean): 3.1011909<br />gene: FBgn0046685","log2FoldChange: -0.5303536469<br />-log10(padj): 1.478323e+00<br />log10(baseMean): 3.2398718<br />gene: FBgn0046687","log2FoldChange: 0.0167123685<br />-log10(padj): 2.853716e-03<br />log10(baseMean): 2.3163603<br />gene: FBgn0046692","log2FoldChange: -0.0653394894<br />-log10(padj): 1.873705e-02<br />log10(baseMean): 1.6425669<br />gene: FBgn0046696","log2FoldChange: -0.1175532632<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.3379877<br />gene: FBgn0046704","log2FoldChange: -0.4984514320<br />-log10(padj): 1.472591e+00<br />log10(baseMean): 3.3429975<br />gene: FBgn0046763","log2FoldChange: 0.0745266590<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 2.7085938<br />gene: FBgn0046776","log2FoldChange: -0.0505552200<br />-log10(padj): 3.085530e-02<br />log10(baseMean): 2.9136842<br />gene: FBgn0047038","log2FoldChange: -0.9080511508<br />-log10(padj): 2.303311e-01<br />log10(baseMean): 0.8274551<br />gene: FBgn0047095","log2FoldChange: -0.0895716553<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.1242940<br />gene: FBgn0047114","log2FoldChange: 0.1529839912<br />-log10(padj): 9.412115e-02<br />log10(baseMean): 3.2314790<br />gene: FBgn0047135","log2FoldChange: 0.0848061613<br />-log10(padj): 4.633512e-02<br />log10(baseMean): 2.2173948<br />gene: FBgn0047178","log2FoldChange: -0.3842642406<br />-log10(padj): 3.063752e-01<br />log10(baseMean): 2.8069199<br />gene: FBgn0047205","log2FoldChange: 0.2142160863<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 2.8050863<br />gene: FBgn0050000","log2FoldChange: 0.2366338048<br />-log10(padj): 1.277760e-01<br />log10(baseMean): 1.9401930<br />gene: FBgn0050001","log2FoldChange: 0.0109400983<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.3155229<br />gene: FBgn0050005","log2FoldChange: -0.1920350159<br />-log10(padj): 1.136128e-01<br />log10(baseMean): 2.7959075<br />gene: FBgn0050007","log2FoldChange: -0.4505009843<br />-log10(padj): 1.035391e-01<br />log10(baseMean): 1.0015809<br />gene: FBgn0050008","log2FoldChange: 0.0995446711<br />-log10(padj): 2.116118e-02<br />log10(baseMean): 1.4075596<br />gene: FBgn0050010","log2FoldChange: -0.3490018950<br />-log10(padj): 1.051477e+00<br />log10(baseMean): 3.6651765<br />gene: FBgn0050011","log2FoldChange: 0.0041788183<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 3.5297376<br />gene: FBgn0050015","log2FoldChange: -0.0679644797<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 3.1179771<br />gene: FBgn0050020","log2FoldChange: 0.0528126709<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.6380655<br />gene: FBgn0050022","log2FoldChange: 0.6564382425<br />-log10(padj): 2.588560e-01<br />log10(baseMean): 1.1775278<br />gene: FBgn0050033","log2FoldChange: 0.5250202624<br />-log10(padj): 3.336410e+00<br />log10(baseMean): 3.1148759<br />gene: FBgn0050035","log2FoldChange: 0.8538072675<br />-log10(padj): 1.917584e-01<br />log10(baseMean): 0.8681688<br />gene: FBgn0050043","log2FoldChange: -0.5973857688<br />-log10(padj): 6.553472e-01<br />log10(baseMean): 1.6116016<br />gene: FBgn0050044","log2FoldChange: -0.3064718027<br />-log10(padj): 1.315086e+00<br />log10(baseMean): 3.0343293<br />gene: FBgn0050046","log2FoldChange: 0.1933163184<br />-log10(padj): 3.919386e-02<br />log10(baseMean): 1.1801514<br />gene: FBgn0050047","log2FoldChange: -0.0126026848<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.8504920<br />gene: FBgn0050051","log2FoldChange: -0.0386941107<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.2586722<br />gene: FBgn0050052","log2FoldChange: -0.3511636542<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 2.2524249<br />gene: FBgn0050055","log2FoldChange: -0.0277465659<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 1.4115670<br />gene: FBgn0050062","log2FoldChange: -0.8694012910<br />-log10(padj): 7.230017e+00<br />log10(baseMean): 3.1913079<br />gene: FBgn0050069","log2FoldChange: 0.1007102511<br />-log10(padj): 5.961065e-02<br />log10(baseMean): 2.1504771<br />gene: FBgn0050077","log2FoldChange: -0.5929022615<br />-log10(padj): 3.067409e-01<br />log10(baseMean): 1.5363847<br />gene: FBgn0050081","log2FoldChange: -0.4330004005<br />-log10(padj): 3.257365e+00<br />log10(baseMean): 3.3925414<br />gene: FBgn0050085","log2FoldChange: -0.5873960028<br />-log10(padj): 1.638218e-01<br />log10(baseMean): 1.0873482<br />gene: FBgn0050087","log2FoldChange: -1.5161982731<br />-log10(padj): 1.443637e+00<br />log10(baseMean): 1.1586980<br />gene: FBgn0050088","log2FoldChange: 0.1123722931<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 2.7495640<br />gene: FBgn0050089","log2FoldChange: -0.1474693442<br />-log10(padj): 1.588723e-01<br />log10(baseMean): 2.6295579<br />gene: FBgn0050090","log2FoldChange: -0.2161689901<br />-log10(padj): 1.293714e-01<br />log10(baseMean): 1.9143737<br />gene: FBgn0050094","log2FoldChange: 0.1920459783<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 0.9122376<br />gene: FBgn0050095","log2FoldChange: 0.1045275347<br />-log10(padj): 2.579879e-02<br />log10(baseMean): 1.5862701<br />gene: FBgn0050096","log2FoldChange: -0.9962839453<br />-log10(padj): 5.743118e-01<br />log10(baseMean): 1.1712813<br />gene: FBgn0050104","log2FoldChange: -0.1526399411<br />-log10(padj): 4.490020e-02<br />log10(baseMean): 1.9253975<br />gene: FBgn0050105","log2FoldChange: -0.0314982549<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.2232827<br />gene: FBgn0050109","log2FoldChange: 0.2324238593<br />-log10(padj): 3.411494e-01<br />log10(baseMean): 2.9879828<br />gene: FBgn0050115","log2FoldChange: -0.0397022614<br />-log10(padj): 2.849528e-02<br />log10(baseMean): 2.9385980<br />gene: FBgn0050118","log2FoldChange: -0.0932383771<br />-log10(padj): 1.224566e-01<br />log10(baseMean): 3.8109077<br />gene: FBgn0050122","log2FoldChange: -0.9517786058<br />-log10(padj): 1.806840e+01<br />log10(baseMean): 2.9885204<br />gene: FBgn0050147","log2FoldChange: -1.0973586724<br />-log10(padj): 1.710147e+00<br />log10(baseMean): 1.7568203<br />gene: FBgn0050148","log2FoldChange: -0.0740357217<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.2015790<br />gene: FBgn0050156","log2FoldChange: 0.0137627746<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 1.6609526<br />gene: FBgn0050157","log2FoldChange: 0.0313090565<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 2.7656115<br />gene: FBgn0050159","log2FoldChange: -0.2179978867<br />-log10(padj): 1.313744e-01<br />log10(baseMean): 2.2910497<br />gene: FBgn0050169","log2FoldChange: -0.1959319962<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 0.8205766<br />gene: FBgn0050178","log2FoldChange: 0.0510224357<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.5329295<br />gene: FBgn0050183","log2FoldChange: 0.3470422444<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 1.6679825<br />gene: FBgn0050184","log2FoldChange: 0.4723150974<br />-log10(padj): 2.758380e+00<br />log10(baseMean): 2.6475634<br />gene: FBgn0050185","log2FoldChange: 0.0294078888<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 3.0493811<br />gene: FBgn0050187","log2FoldChange: 0.5199661257<br />-log10(padj): 9.336547e-02<br />log10(baseMean): 0.9615367<br />gene: FBgn0050196","log2FoldChange: 0.0493936640<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 1.6057020<br />gene: FBgn0050217","log2FoldChange: -0.7838894884<br />-log10(padj): 3.140329e+00<br />log10(baseMean): 2.1825625<br />gene: FBgn0050269","log2FoldChange: -0.7413910277<br />-log10(padj): 1.909681e+00<br />log10(baseMean): 2.1093429<br />gene: FBgn0050273","log2FoldChange: 0.9638183373<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 1.0298229<br />gene: FBgn0050281","log2FoldChange: -0.0268869151<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.1917050<br />gene: FBgn0050284","log2FoldChange: 0.6938766244<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 1.0025231<br />gene: FBgn0050285","log2FoldChange: -0.0967910325<br />-log10(padj): 7.485771e-02<br />log10(baseMean): 2.7115766<br />gene: FBgn0050291","log2FoldChange: -0.0926195097<br />-log10(padj): 6.453047e-02<br />log10(baseMean): 2.3100995<br />gene: FBgn0050295","log2FoldChange: -0.3753296449<br />-log10(padj): 3.909276e-01<br />log10(baseMean): 2.2253625<br />gene: FBgn0050323","log2FoldChange: -0.0920006967<br />-log10(padj): 2.847533e-02<br />log10(baseMean): 1.7219473<br />gene: FBgn0050338","log2FoldChange: 0.1450198327<br />-log10(padj): 2.180450e-01<br />log10(baseMean): 2.6466805<br />gene: FBgn0050339","log2FoldChange: -0.1103316267<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 2.1359349<br />gene: FBgn0050340","log2FoldChange: 0.0005153540<br />-log10(padj): 8.129480e-04<br />log10(baseMean): 2.6788495<br />gene: FBgn0050342","log2FoldChange: -0.3659094609<br />-log10(padj): 1.646593e+00<br />log10(baseMean): 2.8000334<br />gene: FBgn0050344","log2FoldChange: -0.4771416868<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 1.8115130<br />gene: FBgn0050345","log2FoldChange: -0.2980171839<br />-log10(padj): 1.240688e+00<br />log10(baseMean): 3.1225907<br />gene: FBgn0050349","log2FoldChange: 0.1590571654<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.9256350<br />gene: FBgn0050356","log2FoldChange: 0.0285986656<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 1.6171186<br />gene: FBgn0050357","log2FoldChange: 1.2165680924<br />-log10(padj): 2.119922e+00<br />log10(baseMean): 1.6603697<br />gene: FBgn0050359","log2FoldChange: 0.0656821256<br />-log10(padj): 7.608529e-02<br />log10(baseMean): 3.1920978<br />gene: FBgn0050372","log2FoldChange: -0.0708876375<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.3537332<br />gene: FBgn0050373","log2FoldChange: -0.3964641017<br />-log10(padj): 7.502956e-02<br />log10(baseMean): 1.1632637<br />gene: FBgn0050377","log2FoldChange: 0.1540510304<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 2.0931985<br />gene: FBgn0050381","log2FoldChange: 0.3835350739<br />-log10(padj): 2.435905e-01<br />log10(baseMean): 1.8309370<br />gene: FBgn0050383","log2FoldChange: 0.2224172481<br />-log10(padj): 5.122140e-01<br />log10(baseMean): 3.4706765<br />gene: FBgn0050389","log2FoldChange: -0.1197416669<br />-log10(padj): 6.284217e-02<br />log10(baseMean): 2.2417169<br />gene: FBgn0050390","log2FoldChange: -0.0050614577<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 2.3062688<br />gene: FBgn0050392","log2FoldChange: 0.0921502189<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.8411601<br />gene: FBgn0050394","log2FoldChange: 0.4037801272<br />-log10(padj): 5.532631e-02<br />log10(baseMean): 0.8371775<br />gene: FBgn0050398","log2FoldChange: 0.5365247963<br />-log10(padj): 4.387741e-01<br />log10(baseMean): 1.5629398<br />gene: FBgn0050403","log2FoldChange: -0.0135092346<br />-log10(padj): 7.188587e-03<br />log10(baseMean): 2.8191406<br />gene: FBgn0050404","log2FoldChange: 0.3030168032<br />-log10(padj): 3.895549e-01<br />log10(baseMean): 2.9873286<br />gene: FBgn0050410","log2FoldChange: 0.3261244742<br />-log10(padj): 3.311063e-01<br />log10(baseMean): 1.8902993<br />gene: FBgn0050414","log2FoldChange: -0.1927454709<br />-log10(padj): 2.254458e-01<br />log10(baseMean): 2.3373405<br />gene: FBgn0050420","log2FoldChange: 0.0606308664<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.2240734<br />gene: FBgn0050421","log2FoldChange: -0.1434070223<br />-log10(padj): 2.142935e-01<br />log10(baseMean): 3.0029514<br />gene: FBgn0050423","log2FoldChange: 0.8088481919<br />-log10(padj): 2.257992e-01<br />log10(baseMean): 0.9013975<br />gene: FBgn0050424","log2FoldChange: -0.1646656185<br />-log10(padj): 6.495454e-02<br />log10(baseMean): 2.0747134<br />gene: FBgn0050428","log2FoldChange: 0.0732224012<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.3355318<br />gene: FBgn0050438","log2FoldChange: -0.1593241293<br />-log10(padj): 5.090893e-02<br />log10(baseMean): 2.2694737<br />gene: FBgn0050440","log2FoldChange: 0.0550120021<br />-log10(padj): 3.442555e-02<br />log10(baseMean): 2.4189210<br />gene: FBgn0050443","log2FoldChange: -0.5710000717<br />-log10(padj): 1.290286e-01<br />log10(baseMean): 0.9346574<br />gene: FBgn0050446","log2FoldChange: 0.1068125557<br />-log10(padj): 1.106934e-01<br />log10(baseMean): 2.8881066<br />gene: FBgn0050456","log2FoldChange: 0.1167480639<br />-log10(padj): 6.536412e-02<br />log10(baseMean): 2.6659859<br />gene: FBgn0050460","log2FoldChange: 2.6181602285<br />-log10(padj): 1.137979e+01<br />log10(baseMean): 1.6242260<br />gene: FBgn0050463","log2FoldChange: 0.0259291727<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 1.8238536<br />gene: FBgn0050466","log2FoldChange: 0.1289337127<br />-log10(padj): 8.129830e-02<br />log10(baseMean): 2.3196120<br />gene: FBgn0050467","log2FoldChange: 0.0477952038<br />-log10(padj): 1.575319e-02<br />log10(baseMean): 2.0374406<br />gene: FBgn0050476","log2FoldChange: -0.1813452683<br />-log10(padj): 5.702578e-02<br />log10(baseMean): 1.6443127<br />gene: FBgn0050482","log2FoldChange: -0.4351797785<br />-log10(padj): 1.240841e-01<br />log10(baseMean): 1.3568822<br />gene: FBgn0050484","log2FoldChange: -0.5544908235<br />-log10(padj): 1.563473e-01<br />log10(baseMean): 1.1714121<br />gene: FBgn0050485","log2FoldChange: -0.2149307119<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.6022568<br />gene: FBgn0050489","log2FoldChange: -0.5842712463<br />-log10(padj): 1.105617e-01<br />log10(baseMean): 0.8836871<br />gene: FBgn0050490","log2FoldChange: 0.2222366889<br />-log10(padj): 8.699284e-02<br />log10(baseMean): 1.5960416<br />gene: FBgn0050491","log2FoldChange: -0.0871613896<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 3.1676372<br />gene: FBgn0050492","log2FoldChange: 0.2347964116<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 2.0022341<br />gene: FBgn0050493","log2FoldChange: 0.2074187285<br />-log10(padj): 3.468475e-01<br />log10(baseMean): 2.4537332<br />gene: FBgn0050496","log2FoldChange: -0.0430201386<br />-log10(padj): 1.885426e-02<br />log10(baseMean): 3.8290342<br />gene: FBgn0050497","log2FoldChange: 0.3167300226<br />-log10(padj): 7.217343e-01<br />log10(baseMean): 2.8620327<br />gene: FBgn0050499","log2FoldChange: 0.2534440119<br />-log10(padj): 1.744222e-01<br />log10(baseMean): 1.8240124<br />gene: FBgn0050502","log2FoldChange: 0.2037768527<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 0.9014408<br />gene: FBgn0051004","log2FoldChange: -0.5441455077<br />-log10(padj): 2.886149e+00<br />log10(baseMean): 2.4926463<br />gene: FBgn0051005","log2FoldChange: -0.6243768571<br />-log10(padj): 3.646662e-01<br />log10(baseMean): 1.3281344<br />gene: FBgn0051019","log2FoldChange: 0.2369187926<br />-log10(padj): 3.420011e-02<br />log10(baseMean): 0.9258218<br />gene: FBgn0051025","log2FoldChange: 0.2486916444<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 1.6266894<br />gene: FBgn0051029","log2FoldChange: 0.3520212749<br />-log10(padj): 1.578542e-01<br />log10(baseMean): 1.7604850<br />gene: FBgn0051032","log2FoldChange: -0.9450418962<br />-log10(padj): 6.929240e+00<br />log10(baseMean): 2.8531775<br />gene: FBgn0051038","log2FoldChange: 0.1320686112<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 2.7020208<br />gene: FBgn0051040","log2FoldChange: -0.0572933261<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.1336542<br />gene: FBgn0051044","log2FoldChange: -0.0447091904<br />-log10(padj): 3.957681e-02<br />log10(baseMean): 3.2387185<br />gene: FBgn0051048","log2FoldChange: 0.8543123183<br />-log10(padj): 7.297988e-01<br />log10(baseMean): 1.3774322<br />gene: FBgn0051051","log2FoldChange: -0.6860274443<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 0.8512589<br />gene: FBgn0051053","log2FoldChange: 0.2125161665<br />-log10(padj): 3.673804e-01<br />log10(baseMean): 2.8213027<br />gene: FBgn0051063","log2FoldChange: -0.0102662394<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.8930691<br />gene: FBgn0051064","log2FoldChange: 0.2136682406<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 1.0613564<br />gene: FBgn0051065","log2FoldChange: -0.1792312025<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 1.5771146<br />gene: FBgn0051068","log2FoldChange: -0.0396241338<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 2.8005769<br />gene: FBgn0051072","log2FoldChange: 0.1153467938<br />-log10(padj): 9.544956e-02<br />log10(baseMean): 3.2545780<br />gene: FBgn0051075","log2FoldChange: -0.9588228253<br />-log10(padj): 1.713009e+00<br />log10(baseMean): 1.6931181<br />gene: FBgn0051082","log2FoldChange: -0.3150153136<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 0.8050646<br />gene: FBgn0051088","log2FoldChange: 2.3279073282<br />-log10(padj): 3.576135e+01<br />log10(baseMean): 2.1848860<br />gene: FBgn0051092","log2FoldChange: -0.6806062717<br />-log10(padj): 2.616283e+00<br />log10(baseMean): 2.2224967<br />gene: FBgn0051098","log2FoldChange: -0.2184211886<br />-log10(padj): 1.871332e-01<br />log10(baseMean): 2.4934478<br />gene: FBgn0051108","log2FoldChange: 0.0218026366<br />-log10(padj): 5.777426e-03<br />log10(baseMean): 1.8628264<br />gene: FBgn0051109","log2FoldChange: 0.0260854166<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 1.6674112<br />gene: FBgn0051111","log2FoldChange: -0.0164307143<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.0386422<br />gene: FBgn0051115","log2FoldChange: 0.6983765451<br />-log10(padj): 3.607337e+00<br />log10(baseMean): 2.2290836<br />gene: FBgn0051116","log2FoldChange: -0.4440057032<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 1.6244089<br />gene: FBgn0051118","log2FoldChange: 0.5099172737<br />-log10(padj): 3.278556e-01<br />log10(baseMean): 1.4623348<br />gene: FBgn0051119","log2FoldChange: 0.0450540489<br />-log10(padj): 3.004220e-02<br />log10(baseMean): 2.8919932<br />gene: FBgn0051120","log2FoldChange: 0.0917933532<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 3.1970883<br />gene: FBgn0051122","log2FoldChange: 1.2530863393<br />-log10(padj): 2.267271e+00<br />log10(baseMean): 1.4332839<br />gene: FBgn0051125","log2FoldChange: -0.2241499583<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 1.7863414<br />gene: FBgn0051126","log2FoldChange: 0.3156672221<br />-log10(padj): 2.608363e-01<br />log10(baseMean): 1.9722323<br />gene: FBgn0051133","log2FoldChange: 0.0719004807<br />-log10(padj): 4.745188e-02<br />log10(baseMean): 2.6380207<br />gene: FBgn0051140","log2FoldChange: 0.0107771243<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.3592461<br />gene: FBgn0051145","log2FoldChange: -0.3591921297<br />-log10(padj): 2.507238e+00<br />log10(baseMean): 3.9458597<br />gene: FBgn0051150","log2FoldChange: -0.1085740612<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 3.3729459<br />gene: FBgn0051151","log2FoldChange: -0.3158897754<br />-log10(padj): 5.483165e-01<br />log10(baseMean): 2.4526462<br />gene: FBgn0051155","log2FoldChange: -0.0867203705<br />-log10(padj): 5.800266e-02<br />log10(baseMean): 2.3609186<br />gene: FBgn0051156","log2FoldChange: 0.0815221523<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 3.3880608<br />gene: FBgn0051158","log2FoldChange: -0.0108748246<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.1334886<br />gene: FBgn0051159","log2FoldChange: 0.3063190987<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 1.2902657<br />gene: FBgn0051161","log2FoldChange: 1.7044971948<br />-log10(padj): 1.880705e+00<br />log10(baseMean): 1.0853745<br />gene: FBgn0051163","log2FoldChange: 0.0989715513<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.4129755<br />gene: FBgn0051169","log2FoldChange: 0.1849241668<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.8565162<br />gene: FBgn0051174","log2FoldChange: 0.3106688345<br />-log10(padj): 6.318518e-01<br />log10(baseMean): 2.4842094<br />gene: FBgn0051184","log2FoldChange: 0.8012096732<br />-log10(padj): 1.337098e+01<br />log10(baseMean): 2.9661126<br />gene: FBgn0051195","log2FoldChange: -0.0308381196<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.1271121<br />gene: FBgn0051211","log2FoldChange: -0.1201885538<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.3335265<br />gene: FBgn0051213","log2FoldChange: 0.3242090899<br />-log10(padj): 1.500904e+00<br />log10(baseMean): 3.1487457<br />gene: FBgn0051216","log2FoldChange: -0.3495909686<br />-log10(padj): 3.115478e-01<br />log10(baseMean): 1.8183416<br />gene: FBgn0051217","log2FoldChange: 0.1913177560<br />-log10(padj): 9.536868e-02<br />log10(baseMean): 3.2945731<br />gene: FBgn0051219","log2FoldChange: 0.0781724550<br />-log10(padj): 4.675633e-02<br />log10(baseMean): 2.2580742<br />gene: FBgn0051223","log2FoldChange: -0.0195432500<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.3766172<br />gene: FBgn0051224","log2FoldChange: 0.0830981671<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.3381204<br />gene: FBgn0051229","log2FoldChange: 0.3179446814<br />-log10(padj): 5.940472e-02<br />log10(baseMean): 0.9667569<br />gene: FBgn0051230","log2FoldChange: 0.3886851945<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 0.9736892<br />gene: FBgn0051231","log2FoldChange: 0.1128002235<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 2.6214341<br />gene: FBgn0051232","log2FoldChange: -0.2273551196<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.3563638<br />gene: FBgn0051249","log2FoldChange: 0.1354714639<br />-log10(padj): 6.895106e-02<br />log10(baseMean): 2.0090910<br />gene: FBgn0051251","log2FoldChange: -0.6292738035<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 1.1793488<br />gene: FBgn0051262","log2FoldChange: 0.0602047596<br />-log10(padj): 2.125730e-02<br />log10(baseMean): 1.8465286<br />gene: FBgn0051278","log2FoldChange: 0.4087155769<br />-log10(padj): 1.165047e-01<br />log10(baseMean): 1.2477434<br />gene: FBgn0051296","log2FoldChange: -0.0520442807<br />-log10(padj): 4.160701e-02<br />log10(baseMean): 2.7708221<br />gene: FBgn0051301","log2FoldChange: 0.2193804066<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.5218959<br />gene: FBgn0051302","log2FoldChange: 0.2703450899<br />-log10(padj): 6.066753e-01<br />log10(baseMean): 2.9080826<br />gene: FBgn0051304","log2FoldChange: -0.1875716130<br />-log10(padj): 3.537453e-02<br />log10(baseMean): 1.1968306<br />gene: FBgn0051313","log2FoldChange: 0.3549292852<br />-log10(padj): 2.640034e+00<br />log10(baseMean): 3.7120494<br />gene: FBgn0051314","log2FoldChange: -0.0821018809<br />-log10(padj): 3.790338e-02<br />log10(baseMean): 2.5000752<br />gene: FBgn0051320","log2FoldChange: 0.3392576325<br />-log10(padj): 1.073889e+00<br />log10(baseMean): 2.9367357<br />gene: FBgn0051324","log2FoldChange: -0.8991162217<br />-log10(padj): 6.868720e-01<br />log10(baseMean): 1.3102489<br />gene: FBgn0051326","log2FoldChange: -0.9048647760<br />-log10(padj): 5.114039e+00<br />log10(baseMean): 2.9500753<br />gene: FBgn0051337","log2FoldChange: -0.0429077392<br />-log10(padj): 4.083628e-02<br />log10(baseMean): 3.3761642<br />gene: FBgn0051342","log2FoldChange: -0.1103067972<br />-log10(padj): 8.122516e-02<br />log10(baseMean): 2.5563533<br />gene: FBgn0051344","log2FoldChange: -0.1419668213<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 1.2257450<br />gene: FBgn0051347","log2FoldChange: 0.5189355739<br />-log10(padj): 5.773835e-01<br />log10(baseMean): 2.6556576<br />gene: FBgn0051352","log2FoldChange: -0.0006663070<br />-log10(padj): 8.760575e-04<br />log10(baseMean): 2.6437272<br />gene: FBgn0051357","log2FoldChange: 0.5563242032<br />-log10(padj): 3.889185e-01<br />log10(baseMean): 1.8775813<br />gene: FBgn0051360","log2FoldChange: 0.6339430408<br />-log10(padj): 2.547410e+00<br />log10(baseMean): 3.1285460<br />gene: FBgn0051361","log2FoldChange: 0.9184397665<br />-log10(padj): 8.961691e+00<br />log10(baseMean): 3.8550654<br />gene: FBgn0051363","log2FoldChange: -0.1215778280<br />-log10(padj): 1.180403e-01<br />log10(baseMean): 2.6769001<br />gene: FBgn0051365","log2FoldChange: -0.2526899238<br />-log10(padj): 4.806215e-01<br />log10(baseMean): 2.8107873<br />gene: FBgn0051367","log2FoldChange: 0.1539763504<br />-log10(padj): 2.942091e-01<br />log10(baseMean): 2.9670333<br />gene: FBgn0051368","log2FoldChange: -0.3694386793<br />-log10(padj): 3.329052e-01<br />log10(baseMean): 2.3918980<br />gene: FBgn0051370","log2FoldChange: 0.6450233562<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 1.0696983<br />gene: FBgn0051374","log2FoldChange: -0.0717907798<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.0694295<br />gene: FBgn0051388","log2FoldChange: 0.3241827452<br />-log10(padj): 8.439116e-02<br />log10(baseMean): 1.2631655<br />gene: FBgn0051389","log2FoldChange: 0.1401450542<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 2.3420347<br />gene: FBgn0051390","log2FoldChange: 0.0781579083<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.0935651<br />gene: FBgn0051392","log2FoldChange: -0.7605374349<br />-log10(padj): 3.353524e-01<br />log10(baseMean): 1.2024571<br />gene: FBgn0051415","log2FoldChange: -0.7325252188<br />-log10(padj): 1.004362e+01<br />log10(baseMean): 3.3586372<br />gene: FBgn0051431","log2FoldChange: 0.3574554184<br />-log10(padj): 5.490360e-01<br />log10(baseMean): 2.0789487<br />gene: FBgn0051436","log2FoldChange: -0.6614356292<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 0.9229055<br />gene: FBgn0051439","log2FoldChange: 0.1071863854<br />-log10(padj): 6.236166e-02<br />log10(baseMean): 2.1008365<br />gene: FBgn0051441","log2FoldChange: 0.0016861151<br />-log10(padj): 9.882284e-04<br />log10(baseMean): 1.6824881<br />gene: FBgn0051445","log2FoldChange: -0.0877015547<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.3413298<br />gene: FBgn0051450","log2FoldChange: 0.3441552074<br />-log10(padj): 1.842354e+00<br />log10(baseMean): 2.9052801<br />gene: FBgn0051453","log2FoldChange: 0.1630575497<br />-log10(padj): 1.094620e-01<br />log10(baseMean): 2.1427552<br />gene: FBgn0051457","log2FoldChange: 0.5087249455<br />-log10(padj): 1.125866e+00<br />log10(baseMean): 2.3732833<br />gene: FBgn0051460","log2FoldChange: 1.2399776671<br />-log10(padj): 2.636480e+00<br />log10(baseMean): 1.9562367<br />gene: FBgn0051462","log2FoldChange: -0.3727624557<br />-log10(padj): 1.425130e-01<br />log10(baseMean): 1.4513675<br />gene: FBgn0051467","log2FoldChange: 0.4674713575<br />-log10(padj): 4.603572e-01<br />log10(baseMean): 1.7706145<br />gene: FBgn0051472","log2FoldChange: 1.1033628047<br />-log10(padj): 2.086565e+00<br />log10(baseMean): 1.6361870<br />gene: FBgn0051477","log2FoldChange: 0.5420668997<br />-log10(padj): 3.932264e-01<br />log10(baseMean): 1.7582860<br />gene: FBgn0051495","log2FoldChange: -0.1621048136<br />-log10(padj): 6.528827e-02<br />log10(baseMean): 2.3488653<br />gene: FBgn0051510","log2FoldChange: -0.2308040929<br />-log10(padj): 5.623826e-02<br />log10(baseMean): 1.2045412<br />gene: FBgn0051516","log2FoldChange: 0.1616233060<br />-log10(padj): 2.510493e-01<br />log10(baseMean): 2.8784901<br />gene: FBgn0051522","log2FoldChange: -0.8217753817<br />-log10(padj): 5.090146e+00<br />log10(baseMean): 3.6623845<br />gene: FBgn0051523","log2FoldChange: 0.0651360299<br />-log10(padj): 1.466187e-02<br />log10(baseMean): 1.5534967<br />gene: FBgn0051525","log2FoldChange: 0.0978612255<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 0.9926220<br />gene: FBgn0051526","log2FoldChange: -0.0053221292<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 3.0581514<br />gene: FBgn0051531","log2FoldChange: 0.0692977352<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.9024962<br />gene: FBgn0051534","log2FoldChange: -0.0629952003<br />-log10(padj): 6.639638e-02<br />log10(baseMean): 3.5050503<br />gene: FBgn0051536","log2FoldChange: -0.5109393366<br />-log10(padj): 1.327685e-01<br />log10(baseMean): 1.0996475<br />gene: FBgn0051538","log2FoldChange: 0.5270949925<br />-log10(padj): 4.898668e-01<br />log10(baseMean): 1.6595160<br />gene: FBgn0051546","log2FoldChange: -0.2355135384<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 1.1186367<br />gene: FBgn0051547","log2FoldChange: -0.2623433769<br />-log10(padj): 3.917225e-01<br />log10(baseMean): 2.6392063<br />gene: FBgn0051548","log2FoldChange: 0.3611625357<br />-log10(padj): 6.285343e-01<br />log10(baseMean): 2.7336885<br />gene: FBgn0051549","log2FoldChange: 0.0562261323<br />-log10(padj): 3.243344e-02<br />log10(baseMean): 3.0847787<br />gene: FBgn0051550","log2FoldChange: 0.0006420842<br />-log10(padj): 3.176531e-04<br />log10(baseMean): 1.1954241<br />gene: FBgn0051551","log2FoldChange: 1.8106824779<br />-log10(padj): 1.348644e+00<br />log10(baseMean): 0.9539227<br />gene: FBgn0051555","log2FoldChange: 0.1208383585<br />-log10(padj): 3.561033e-02<br />log10(baseMean): 1.8182170<br />gene: FBgn0051600","log2FoldChange: -0.1856344909<br />-log10(padj): 4.603090e-02<br />log10(baseMean): 3.0585235<br />gene: FBgn0051601","log2FoldChange: 0.4399240273<br />-log10(padj): 4.932615e-01<br />log10(baseMean): 2.0396268<br />gene: FBgn0051607","log2FoldChange: 0.1289119038<br />-log10(padj): 6.472956e-02<br />log10(baseMean): 2.3788583<br />gene: FBgn0051612","log2FoldChange: 0.9476284010<br />-log10(padj): 2.659672e+00<br />log10(baseMean): 1.9686226<br />gene: FBgn0051614","log2FoldChange: 0.2269557209<br />-log10(padj): 6.349200e-01<br />log10(baseMean): 3.6015445<br />gene: FBgn0051619","log2FoldChange: 0.6251074231<br />-log10(padj): 4.547191e-01<br />log10(baseMean): 1.4916994<br />gene: FBgn0051624","log2FoldChange: 0.1481336751<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.0085702<br />gene: FBgn0051627","log2FoldChange: -0.5498657341<br />-log10(padj): 1.066951e+00<br />log10(baseMean): 1.8926860<br />gene: FBgn0051630","log2FoldChange: -0.0869130482<br />-log10(padj): 3.397990e-02<br />log10(baseMean): 2.0510279<br />gene: FBgn0051633","log2FoldChange: -0.0232911977<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 1.0802084<br />gene: FBgn0051634","log2FoldChange: -0.0988236021<br />-log10(padj): 8.747108e-02<br />log10(baseMean): 2.8383466<br />gene: FBgn0051635","log2FoldChange: -0.4162298960<br />-log10(padj): 2.431865e+00<br />log10(baseMean): 3.4275331<br />gene: FBgn0051637","log2FoldChange: 0.1458373428<br />-log10(padj): 2.424491e-01<br />log10(baseMean): 2.8783073<br />gene: FBgn0051638","log2FoldChange: -0.2776707614<br />-log10(padj): 2.863512e-01<br />log10(baseMean): 3.1590318<br />gene: FBgn0051641","log2FoldChange: 2.0152960767<br />-log10(padj): 1.406910e+01<br />log10(baseMean): 2.0435220<br />gene: FBgn0051642","log2FoldChange: 0.1442469648<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.3916982<br />gene: FBgn0051648","log2FoldChange: 0.3702905466<br />-log10(padj): 5.983789e-01<br />log10(baseMean): 2.2939519<br />gene: FBgn0051658","log2FoldChange: 1.6868831805<br />-log10(padj): 9.015875e-01<br />log10(baseMean): 0.8829317<br />gene: FBgn0051663","log2FoldChange: 0.1271766366<br />-log10(padj): 3.442555e-02<br />log10(baseMean): 1.4506532<br />gene: FBgn0051668","log2FoldChange: 0.1388305032<br />-log10(padj): 2.255230e-01<br />log10(baseMean): 3.1643647<br />gene: FBgn0051673","log2FoldChange: 0.2025377923<br />-log10(padj): 1.638394e-01<br />log10(baseMean): 2.0898787<br />gene: FBgn0051674","log2FoldChange: -0.7628188668<br />-log10(padj): 6.891340e-01<br />log10(baseMean): 1.5845407<br />gene: FBgn0051675","log2FoldChange: -0.0426189340<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 3.5268277<br />gene: FBgn0051678","log2FoldChange: -0.2134830522<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 1.2154604<br />gene: FBgn0051687","log2FoldChange: -0.2741085491<br />-log10(padj): 3.756419e-01<br />log10(baseMean): 2.9581190<br />gene: FBgn0051688","log2FoldChange: -0.0169130942<br />-log10(padj): 1.579496e-02<br />log10(baseMean): 3.2733389<br />gene: FBgn0051694","log2FoldChange: 0.1142020749<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 0.8274682<br />gene: FBgn0051697","log2FoldChange: -0.1160992814<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 2.0083687<br />gene: FBgn0051710","log2FoldChange: -0.0678246818<br />-log10(padj): 4.687994e-02<br />log10(baseMean): 2.7027875<br />gene: FBgn0051712","log2FoldChange: 0.0952622330<br />-log10(padj): 3.645199e-02<br />log10(baseMean): 1.8537260<br />gene: FBgn0051713","log2FoldChange: 0.1200034769<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 2.4480602<br />gene: FBgn0051715","log2FoldChange: -0.0698255245<br />-log10(padj): 7.084185e-02<br />log10(baseMean): 3.3991524<br />gene: FBgn0051716","log2FoldChange: -0.1087470874<br />-log10(padj): 1.083143e-01<br />log10(baseMean): 2.8022182<br />gene: FBgn0051717","log2FoldChange: -0.1235884363<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.0743428<br />gene: FBgn0051719","log2FoldChange: -0.0156839311<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.0981308<br />gene: FBgn0051720","log2FoldChange: -0.0653578826<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 3.4673043<br />gene: FBgn0051729","log2FoldChange: -0.0927094584<br />-log10(padj): 1.277462e-01<br />log10(baseMean): 3.3825347<br />gene: FBgn0051739","log2FoldChange: -0.1021873505<br />-log10(padj): 6.238437e-02<br />log10(baseMean): 2.5572976<br />gene: FBgn0051755","log2FoldChange: -0.0959360750<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.0193745<br />gene: FBgn0051759","log2FoldChange: -0.2490828260<br />-log10(padj): 3.120938e-02<br />log10(baseMean): 0.8047502<br />gene: FBgn0051763","log2FoldChange: -0.2496943687<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 1.8209850<br />gene: FBgn0051769","log2FoldChange: 0.4707561005<br />-log10(padj): 8.965997e-02<br />log10(baseMean): 0.9887725<br />gene: FBgn0051774","log2FoldChange: -0.2381670319<br />-log10(padj): 1.268350e-01<br />log10(baseMean): 2.5378448<br />gene: FBgn0051777","log2FoldChange: -1.0328009844<br />-log10(padj): 4.180558e-01<br />log10(baseMean): 1.0548156<br />gene: FBgn0051778","log2FoldChange: -0.1343295287<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.1312256<br />gene: FBgn0051782","log2FoldChange: 0.6257460633<br />-log10(padj): 2.200835e-01<br />log10(baseMean): 1.1154825<br />gene: FBgn0051789","log2FoldChange: 0.0783365853<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 1.6255000<br />gene: FBgn0051790","log2FoldChange: -0.0180294897<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 2.6113670<br />gene: FBgn0051793","log2FoldChange: -0.9769691875<br />-log10(padj): 9.904895e-01<br />log10(baseMean): 1.5547348<br />gene: FBgn0051798","log2FoldChange: -0.0368974670<br />-log10(padj): 1.079808e-02<br />log10(baseMean): 2.1516662<br />gene: FBgn0051800","log2FoldChange: 0.3367256288<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 0.8214545<br />gene: FBgn0051803","log2FoldChange: 0.0123146478<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.4369589<br />gene: FBgn0051812","log2FoldChange: 0.0617988657<br />-log10(padj): 8.153153e-03<br />log10(baseMean): 1.6239569<br />gene: FBgn0051815","log2FoldChange: 0.2063492266<br />-log10(padj): 4.355667e-02<br />log10(baseMean): 1.2383558<br />gene: FBgn0051816","log2FoldChange: 0.6930743448<br />-log10(padj): 1.146228e+00<br />log10(baseMean): 1.7600431<br />gene: FBgn0051849","log2FoldChange: 0.0916538727<br />-log10(padj): 7.638315e-02<br />log10(baseMean): 2.6784914<br />gene: FBgn0051852","log2FoldChange: 0.1346457048<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.5237259<br />gene: FBgn0051855","log2FoldChange: 1.0030303767<br />-log10(padj): 4.333929e+00<br />log10(baseMean): 1.9271883<br />gene: FBgn0051856","log2FoldChange: -0.3770123821<br />-log10(padj): 7.472126e-02<br />log10(baseMean): 1.1604952<br />gene: FBgn0051858","log2FoldChange: 0.6751238798<br />-log10(padj): 8.962574e-02<br />log10(baseMean): 1.6651376<br />gene: FBgn0051864","log2FoldChange: -0.4513764609<br />-log10(padj): 7.057462e-02<br />log10(baseMean): 1.1532654<br />gene: FBgn0051865","log2FoldChange: 0.4022725530<br />-log10(padj): 1.052083e-01<br />log10(baseMean): 1.4025244<br />gene: FBgn0051866","log2FoldChange: 0.2754365756<br />-log10(padj): 7.166896e-02<br />log10(baseMean): 1.2981774<br />gene: FBgn0051869","log2FoldChange: 0.4150045636<br />-log10(padj): 1.039305e-01<br />log10(baseMean): 1.0676206<br />gene: FBgn0051874","log2FoldChange: -0.0603850212<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.6409748<br />gene: FBgn0051875","log2FoldChange: -0.1110034742<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 1.1920249<br />gene: FBgn0051897","log2FoldChange: 0.0177259476<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.7946753<br />gene: FBgn0051898","log2FoldChange: -0.3222747446<br />-log10(padj): 7.964138e-02<br />log10(baseMean): 1.2371995<br />gene: FBgn0051900","log2FoldChange: -0.1751946951<br />-log10(padj): 3.621771e-01<br />log10(baseMean): 3.2079629<br />gene: FBgn0051908","log2FoldChange: 0.1667466463<br />-log10(padj): 2.482214e-01<br />log10(baseMean): 3.0324697<br />gene: FBgn0051915","log2FoldChange: 0.3932485768<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 1.5549832<br />gene: FBgn0051922","log2FoldChange: -0.1650093227<br />-log10(padj): 2.311790e-01<br />log10(baseMean): 2.5797539<br />gene: FBgn0051935","log2FoldChange: 0.0954465574<br />-log10(padj): 4.793149e-02<br />log10(baseMean): 2.0438370<br />gene: FBgn0051950","log2FoldChange: -0.5562991608<br />-log10(padj): 1.015151e+00<br />log10(baseMean): 2.6127577<br />gene: FBgn0051955","log2FoldChange: -0.1665085272<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 2.0480542<br />gene: FBgn0051957","log2FoldChange: -0.0666790805<br />-log10(padj): 4.517363e-02<br />log10(baseMean): 2.5486000<br />gene: FBgn0051961","log2FoldChange: -0.0256302591<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 1.7703730<br />gene: FBgn0051973","log2FoldChange: 0.2794406282<br />-log10(padj): 1.462327e-01<br />log10(baseMean): 1.6670901<br />gene: FBgn0051974","log2FoldChange: -0.2707331876<br />-log10(padj): 7.665496e-01<br />log10(baseMean): 2.7170417<br />gene: FBgn0051989","log2FoldChange: -0.0221303512<br />-log10(padj): 1.164540e-02<br />log10(baseMean): 3.7097947<br />gene: FBgn0051992","log2FoldChange: -0.0935293026<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 2.7782719<br />gene: FBgn0051997","log2FoldChange: -0.1373047621<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.0669844<br />gene: FBgn0051998","log2FoldChange: -0.2339275956<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 3.1171251<br />gene: FBgn0051999","log2FoldChange: -0.3277595307<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.3733718<br />gene: FBgn0052000","log2FoldChange: -0.5690681270<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.1562367<br />gene: FBgn0052011","log2FoldChange: -0.2319193652<br />-log10(padj): 1.184229e-01<br />log10(baseMean): 3.0957159<br />gene: FBgn0052016","log2FoldChange: 0.0769580594<br />-log10(padj): 9.356238e-03<br />log10(baseMean): 2.5962392<br />gene: FBgn0052017","log2FoldChange: 0.5509604966<br />-log10(padj): 2.341664e+00<br />log10(baseMean): 2.7041353<br />gene: FBgn0052021","log2FoldChange: 0.2849221950<br />-log10(padj): 3.701179e-01<br />log10(baseMean): 2.4210101<br />gene: FBgn0052022","log2FoldChange: 1.2206293608<br />-log10(padj): 7.297988e-01<br />log10(baseMean): 1.0722247<br />gene: FBgn0052024","log2FoldChange: 0.1583204565<br />-log10(padj): 4.130831e-02<br />log10(baseMean): 1.9914704<br />gene: FBgn0052026","log2FoldChange: 0.1485916366<br />-log10(padj): 1.047710e-01<br />log10(baseMean): 2.1224837<br />gene: FBgn0052027","log2FoldChange: 0.0453753236<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 1.3924024<br />gene: FBgn0052036","log2FoldChange: -0.0808819979<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.1231201<br />gene: FBgn0052037","log2FoldChange: -0.0969597676<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.4117127<br />gene: FBgn0052038","log2FoldChange: 0.0912567222<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.0471594<br />gene: FBgn0052039","log2FoldChange: -0.4357721855<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 1.0875544<br />gene: FBgn0052040","log2FoldChange: 0.1974485788<br />-log10(padj): 1.487293e-01<br />log10(baseMean): 2.1872729<br />gene: FBgn0052043","log2FoldChange: 0.4154087345<br />-log10(padj): 1.177805e+00<br />log10(baseMean): 3.0897965<br />gene: FBgn0052056","log2FoldChange: -0.9135309180<br />-log10(padj): 3.739047e-01<br />log10(baseMean): 1.1278260<br />gene: FBgn0052058","log2FoldChange: -0.2342177615<br />-log10(padj): 5.749248e-02<br />log10(baseMean): 1.4705205<br />gene: FBgn0052060","log2FoldChange: -0.0668318177<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.6159793<br />gene: FBgn0052062","log2FoldChange: -0.1500999449<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 0.9520784<br />gene: FBgn0052065","log2FoldChange: -0.1024294353<br />-log10(padj): 1.779246e-01<br />log10(baseMean): 3.4009980<br />gene: FBgn0052066","log2FoldChange: -0.3671133337<br />-log10(padj): 1.108934e+00<br />log10(baseMean): 2.7458006<br />gene: FBgn0052068","log2FoldChange: 0.4536554659<br />-log10(padj): 4.892476e-01<br />log10(baseMean): 1.9466635<br />gene: FBgn0052069","log2FoldChange: -0.0459994470<br />-log10(padj): 1.740744e-02<br />log10(baseMean): 2.3211384<br />gene: FBgn0052075","log2FoldChange: -0.0088024621<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.6005032<br />gene: FBgn0052076","log2FoldChange: 0.0126878275<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.4620951<br />gene: FBgn0052082","log2FoldChange: 0.1511101954<br />-log10(padj): 3.912983e-02<br />log10(baseMean): 1.8991677<br />gene: FBgn0052088","log2FoldChange: 0.0523727505<br />-log10(padj): 1.492365e-02<br />log10(baseMean): 1.6357036<br />gene: FBgn0052091","log2FoldChange: -0.0451894466<br />-log10(padj): 2.362377e-02<br />log10(baseMean): 2.3392690<br />gene: FBgn0052095","log2FoldChange: -0.1774799173<br />-log10(padj): 1.114791e-01<br />log10(baseMean): 2.1494301<br />gene: FBgn0052099","log2FoldChange: -0.0172906096<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.1406970<br />gene: FBgn0052100","log2FoldChange: -0.0757604747<br />-log10(padj): 6.232561e-02<br />log10(baseMean): 3.3214784<br />gene: FBgn0052103","log2FoldChange: 0.0341779426<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 2.2887088<br />gene: FBgn0052104","log2FoldChange: 0.2370273414<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 2.1494907<br />gene: FBgn0052109","log2FoldChange: -0.2594717143<br />-log10(padj): 2.700653e-01<br />log10(baseMean): 2.0944915<br />gene: FBgn0052112","log2FoldChange: -0.2081654384<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.9322042<br />gene: FBgn0052113","log2FoldChange: 0.2167257151<br />-log10(padj): 4.497479e-02<br />log10(baseMean): 2.5755638<br />gene: FBgn0052115","log2FoldChange: -0.3990247964<br />-log10(padj): 7.555807e-02<br />log10(baseMean): 1.3739171<br />gene: FBgn0052132","log2FoldChange: -0.3746267276<br />-log10(padj): 1.613335e+00<br />log10(baseMean): 3.2922754<br />gene: FBgn0052133","log2FoldChange: 1.3089730361<br />-log10(padj): 7.040355e+00<br />log10(baseMean): 2.3814934<br />gene: FBgn0052135","log2FoldChange: 0.0067208508<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 4.0765552<br />gene: FBgn0052138","log2FoldChange: 0.1954725899<br />-log10(padj): 4.162586e-02<br />log10(baseMean): 1.3487854<br />gene: FBgn0052152","log2FoldChange: 0.2511060939<br />-log10(padj): 1.287703e-01<br />log10(baseMean): 1.9161218<br />gene: FBgn0052155","log2FoldChange: 0.1543885974<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 0.8617574<br />gene: FBgn0052159","log2FoldChange: 0.0033736993<br />-log10(padj): 1.410214e-03<br />log10(baseMean): 2.0906772<br />gene: FBgn0052163","log2FoldChange: -0.5586423237<br />-log10(padj): 6.180418e-01<br />log10(baseMean): 1.8375614<br />gene: FBgn0052170","log2FoldChange: -0.0139736065<br />-log10(padj): 4.196143e-03<br />log10(baseMean): 2.4697837<br />gene: FBgn0052176","log2FoldChange: -0.2088664821<br />-log10(padj): 2.518947e-01<br />log10(baseMean): 2.6280987<br />gene: FBgn0052177","log2FoldChange: -0.2857890849<br />-log10(padj): 2.708725e-01<br />log10(baseMean): 2.0220667<br />gene: FBgn0052179","log2FoldChange: -0.1086786537<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 1.6359110<br />gene: FBgn0052181","log2FoldChange: -0.0836426197<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.5664300<br />gene: FBgn0052182","log2FoldChange: 1.2963505149<br />-log10(padj): 2.006347e+00<br />log10(baseMean): 1.3735445<br />gene: FBgn0052185","log2FoldChange: -0.0183018597<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 3.6700290<br />gene: FBgn0052190","log2FoldChange: -0.4383768417<br />-log10(padj): 5.183965e-02<br />log10(baseMean): 0.8047738<br />gene: FBgn0052193","log2FoldChange: 0.0594009332<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 1.7717013<br />gene: FBgn0052195","log2FoldChange: 0.4141470199<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.1972616<br />gene: FBgn0052196","log2FoldChange: -0.2746025192<br />-log10(padj): 1.686202e-01<br />log10(baseMean): 1.9175126<br />gene: FBgn0052202","log2FoldChange: -0.0588200202<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 1.9496980<br />gene: FBgn0052206","log2FoldChange: 0.3091301649<br />-log10(padj): 1.194533e-01<br />log10(baseMean): 1.8338918<br />gene: FBgn0052207","log2FoldChange: -0.2214717517<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 3.0410285<br />gene: FBgn0052210","log2FoldChange: 0.0499016330<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 2.2096627<br />gene: FBgn0052221","log2FoldChange: -0.0050056266<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 2.7217326<br />gene: FBgn0052226","log2FoldChange: 0.3220347587<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.0790056<br />gene: FBgn0052227","log2FoldChange: -0.0615511597<br />-log10(padj): 2.480451e-02<br />log10(baseMean): 2.7993068<br />gene: FBgn0052230","log2FoldChange: 0.0597580505<br />-log10(padj): 1.051577e-02<br />log10(baseMean): 2.7923876<br />gene: FBgn0052243","log2FoldChange: 0.0125663011<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.3290322<br />gene: FBgn0052250","log2FoldChange: -0.4225264272<br />-log10(padj): 8.191455e-01<br />log10(baseMean): 3.0496714<br />gene: FBgn0052251","log2FoldChange: 0.0897600529<br />-log10(padj): 4.421083e-02<br />log10(baseMean): 2.0517607<br />gene: FBgn0052262","log2FoldChange: -0.1771568234<br />-log10(padj): 1.337400e-01<br />log10(baseMean): 2.9920411<br />gene: FBgn0052264","log2FoldChange: 0.2572756330<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 2.3554406<br />gene: FBgn0052267","log2FoldChange: -0.3076378666<br />-log10(padj): 6.900827e-02<br />log10(baseMean): 1.1239537<br />gene: FBgn0052268","log2FoldChange: 0.3403342064<br />-log10(padj): 8.439116e-02<br />log10(baseMean): 1.2290450<br />gene: FBgn0052271","log2FoldChange: -0.1938357108<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 2.4385095<br />gene: FBgn0052278","log2FoldChange: 0.4710323291<br />-log10(padj): 1.518914e+00<br />log10(baseMean): 2.5623832<br />gene: FBgn0052280","log2FoldChange: -0.0859549057<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 2.2190250<br />gene: FBgn0052281","log2FoldChange: 0.1134168179<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.8463879<br />gene: FBgn0052296","log2FoldChange: -0.5528156303<br />-log10(padj): 1.062516e-01<br />log10(baseMean): 0.9354815<br />gene: FBgn0052302","log2FoldChange: -0.5753321080<br />-log10(padj): 2.386876e+00<br />log10(baseMean): 3.1583639<br />gene: FBgn0052311","log2FoldChange: -1.7771540336<br />-log10(padj): 1.307914e+00<br />log10(baseMean): 1.0979615<br />gene: FBgn0052313","log2FoldChange: -0.3437839498<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 0.8175554<br />gene: FBgn0052318","log2FoldChange: 0.1116332478<br />-log10(padj): 1.067740e-01<br />log10(baseMean): 2.6500142<br />gene: FBgn0052343","log2FoldChange: -0.1592810849<br />-log10(padj): 3.251174e-01<br />log10(baseMean): 3.0964849<br />gene: FBgn0052344","log2FoldChange: -0.0779055418<br />-log10(padj): 1.077674e-02<br />log10(baseMean): 2.3425031<br />gene: FBgn0052350","log2FoldChange: -0.1948774417<br />-log10(padj): 5.992028e-02<br />log10(baseMean): 1.4641783<br />gene: FBgn0052352","log2FoldChange: 0.4396075063<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 1.1174309<br />gene: FBgn0052353","log2FoldChange: 0.0800885554<br />-log10(padj): 1.353168e-02<br />log10(baseMean): 1.2022928<br />gene: FBgn0052354","log2FoldChange: 0.6359962458<br />-log10(padj): 1.346254e-01<br />log10(baseMean): 0.8997913<br />gene: FBgn0052355","log2FoldChange: 0.1209750898<br />-log10(padj): 9.517988e-02<br />log10(baseMean): 2.6154482<br />gene: FBgn0052364","log2FoldChange: -0.2263878727<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.0021874<br />gene: FBgn0052365","log2FoldChange: 0.3494325170<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 2.5659258<br />gene: FBgn0052368","log2FoldChange: 0.0362967575<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 3.3496578<br />gene: FBgn0052369","log2FoldChange: 0.4656850028<br />-log10(padj): 1.181581e-01<br />log10(baseMean): 1.0932193<br />gene: FBgn0052372","log2FoldChange: 0.0824384588<br />-log10(padj): 1.023213e-01<br />log10(baseMean): 3.0474578<br />gene: FBgn0052380","log2FoldChange: 0.0623601120<br />-log10(padj): 3.778552e-02<br />log10(baseMean): 2.6922542<br />gene: FBgn0052393","log2FoldChange: -0.1924646533<br />-log10(padj): 2.388550e-01<br />log10(baseMean): 3.4863781<br />gene: FBgn0052394","log2FoldChange: 0.6011797078<br />-log10(padj): 2.972161e-01<br />log10(baseMean): 1.4611322<br />gene: FBgn0052397","log2FoldChange: -2.2028708042<br />-log10(padj): 2.538102e+01<br />log10(baseMean): 2.2719581<br />gene: FBgn0052407","log2FoldChange: -0.1197114796<br />-log10(padj): 1.274928e-01<br />log10(baseMean): 2.8324916<br />gene: FBgn0052409","log2FoldChange: -0.6639999759<br />-log10(padj): 8.331324e+00<br />log10(baseMean): 3.0367406<br />gene: FBgn0052412","log2FoldChange: -0.2219495131<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 2.3904854<br />gene: FBgn0052418","log2FoldChange: -0.2738189263<br />-log10(padj): 2.567148e-01<br />log10(baseMean): 3.5800850<br />gene: FBgn0052423","log2FoldChange: -0.0293677963<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 2.9684647<br />gene: FBgn0052425","log2FoldChange: -0.2908800619<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 2.4729870<br />gene: FBgn0052428","log2FoldChange: 0.4595369635<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 2.6126966<br />gene: FBgn0052436","log2FoldChange: -0.0242522541<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.9335188<br />gene: FBgn0052438","log2FoldChange: 0.1306513463<br />-log10(padj): 1.685825e-01<br />log10(baseMean): 2.8926617<br />gene: FBgn0052441","log2FoldChange: 0.0433185433<br />-log10(padj): 1.060316e-02<br />log10(baseMean): 1.7787396<br />gene: FBgn0052442","log2FoldChange: 0.0865814709<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 0.8880093<br />gene: FBgn0052445","log2FoldChange: 0.3367069330<br />-log10(padj): 4.878643e-01<br />log10(baseMean): 2.1146150<br />gene: FBgn0052446","log2FoldChange: 0.3338889400<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 1.7686299<br />gene: FBgn0052448","log2FoldChange: -0.0966784650<br />-log10(padj): 1.467275e-01<br />log10(baseMean): 3.0931948<br />gene: FBgn0052451","log2FoldChange: 0.0273589563<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.3687565<br />gene: FBgn0052452","log2FoldChange: 0.6106669691<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 0.9203227<br />gene: FBgn0052457","log2FoldChange: -0.2978453230<br />-log10(padj): 1.598990e-01<br />log10(baseMean): 2.0850302<br />gene: FBgn0052458","log2FoldChange: -0.0161286922<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.7559944<br />gene: FBgn0052473","log2FoldChange: 0.6579553361<br />-log10(padj): 2.200835e-01<br />log10(baseMean): 1.0825532<br />gene: FBgn0052476","log2FoldChange: -0.0726897865<br />-log10(padj): 4.658288e-02<br />log10(baseMean): 4.1522419<br />gene: FBgn0052477","log2FoldChange: -0.0790831578<br />-log10(padj): 7.933913e-02<br />log10(baseMean): 3.8250742<br />gene: FBgn0052479","log2FoldChange: -0.0615256193<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 0.9848517<br />gene: FBgn0052480","log2FoldChange: 0.0716837721<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 1.6673485<br />gene: FBgn0052483","log2FoldChange: -0.1144559335<br />-log10(padj): 1.031211e-01<br />log10(baseMean): 2.9632440<br />gene: FBgn0052484","log2FoldChange: -0.4294492631<br />-log10(padj): 9.799692e-01<br />log10(baseMean): 2.3143887<br />gene: FBgn0052485","log2FoldChange: 0.2101712097<br />-log10(padj): 4.921742e-01<br />log10(baseMean): 3.2465629<br />gene: FBgn0052486","log2FoldChange: -0.6046342408<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.6195390<br />gene: FBgn0052495","log2FoldChange: 0.1312801611<br />-log10(padj): 1.829553e-02<br />log10(baseMean): 1.0871446<br />gene: FBgn0052499","log2FoldChange: -0.1115591353<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.9551712<br />gene: FBgn0052512","log2FoldChange: -0.9284932879<br />-log10(padj): 5.597151e+00<br />log10(baseMean): 3.4366110<br />gene: FBgn0052521","log2FoldChange: 0.1653842359<br />-log10(padj): 2.435905e-01<br />log10(baseMean): 3.2361660<br />gene: FBgn0052528","log2FoldChange: -0.3624083726<br />-log10(padj): 1.735633e+00<br />log10(baseMean): 4.0072096<br />gene: FBgn0052529","log2FoldChange: 0.1616854314<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.5945509<br />gene: FBgn0052532","log2FoldChange: 0.2922307346<br />-log10(padj): 2.755476e-01<br />log10(baseMean): 2.1887661<br />gene: FBgn0052533","log2FoldChange: 0.2347917027<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 1.5518567<br />gene: FBgn0052536","log2FoldChange: -0.0699654954<br />-log10(padj): 3.028478e-02<br />log10(baseMean): 2.3818896<br />gene: FBgn0052537","log2FoldChange: -0.0755591695<br />-log10(padj): 7.961053e-02<br />log10(baseMean): 3.4111473<br />gene: FBgn0052549","log2FoldChange: 0.1850012227<br />-log10(padj): 1.006262e-01<br />log10(baseMean): 1.8408977<br />gene: FBgn0052554","log2FoldChange: 0.4229793869<br />-log10(padj): 2.073298e-01<br />log10(baseMean): 1.5684694<br />gene: FBgn0052556","log2FoldChange: 0.0027056950<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.5791917<br />gene: FBgn0052557","log2FoldChange: 0.1557547178<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 1.6325856<br />gene: FBgn0052572","log2FoldChange: 0.1019463700<br />-log10(padj): 4.460811e-02<br />log10(baseMean): 2.4447108<br />gene: FBgn0052576","log2FoldChange: -0.0386666833<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.4383003<br />gene: FBgn0052579","log2FoldChange: 0.2289468047<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 2.4442456<br />gene: FBgn0052580","log2FoldChange: -0.4386751087<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.5109737<br />gene: FBgn0052581","log2FoldChange: 0.0663361436<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 1.8082598<br />gene: FBgn0052582","log2FoldChange: -0.2679940567<br />-log10(padj): 1.261444e-01<br />log10(baseMean): 1.9993263<br />gene: FBgn0052590","log2FoldChange: -0.1221531961<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 3.4436648<br />gene: FBgn0052594","log2FoldChange: -0.0699165650<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.2451459<br />gene: FBgn0052595","log2FoldChange: -1.1027675070<br />-log10(padj): 2.713173e-01<br />log10(baseMean): 0.9176589<br />gene: FBgn0052606","log2FoldChange: 0.2628918170<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.7198217<br />gene: FBgn0052613","log2FoldChange: 1.6035098891<br />-log10(padj): 8.781536e+00<br />log10(baseMean): 2.1925702<br />gene: FBgn0052625","log2FoldChange: -0.1654324300<br />-log10(padj): 2.200835e-01<br />log10(baseMean): 3.3485685<br />gene: FBgn0052626","log2FoldChange: -0.0536319781<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 2.1113839<br />gene: FBgn0052627","log2FoldChange: 0.1216194432<br />-log10(padj): 5.688062e-02<br />log10(baseMean): 1.8558714<br />gene: FBgn0052628","log2FoldChange: -0.2562687353<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 3.2019091<br />gene: FBgn0052638","log2FoldChange: -0.0060107793<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.2191993<br />gene: FBgn0052639","log2FoldChange: 0.3568171020<br />-log10(padj): 9.074436e-02<br />log10(baseMean): 1.2953216<br />gene: FBgn0052640","log2FoldChange: 0.3241022719<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 1.5423970<br />gene: FBgn0052641","log2FoldChange: 0.0419697173<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.8693095<br />gene: FBgn0052649","log2FoldChange: 0.0941513650<br />-log10(padj): 2.424616e-02<br />log10(baseMean): 1.7507307<br />gene: FBgn0052650","log2FoldChange: -0.1258324914<br />-log10(padj): 1.806168e-01<br />log10(baseMean): 3.4068951<br />gene: FBgn0052654","log2FoldChange: 0.5938096407<br />-log10(padj): 8.409693e-02<br />log10(baseMean): 0.9765790<br />gene: FBgn0052655","log2FoldChange: -0.0168791982<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 3.1574250<br />gene: FBgn0052662","log2FoldChange: -0.1827044653<br />-log10(padj): 1.042576e-01<br />log10(baseMean): 3.3067102<br />gene: FBgn0052663","log2FoldChange: 0.0627321408<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.3335660<br />gene: FBgn0052666","log2FoldChange: 0.3249585346<br />-log10(padj): 9.220293e-02<br />log10(baseMean): 1.2666126<br />gene: FBgn0052668","log2FoldChange: -0.2601653151<br />-log10(padj): 6.895106e-02<br />log10(baseMean): 1.2652083<br />gene: FBgn0052669","log2FoldChange: -0.2072180300<br />-log10(padj): 1.598990e-01<br />log10(baseMean): 3.3754978<br />gene: FBgn0052672","log2FoldChange: 0.0063293058<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 3.4305852<br />gene: FBgn0052675","log2FoldChange: 0.2088578897<br />-log10(padj): 2.901409e-01<br />log10(baseMean): 3.2718027<br />gene: FBgn0052676","log2FoldChange: 1.2699561590<br />-log10(padj): 3.753583e+00<br />log10(baseMean): 1.6641222<br />gene: FBgn0052677","log2FoldChange: -0.9988329853<br />-log10(padj): 6.111580e+00<br />log10(baseMean): 2.2056002<br />gene: FBgn0052681","log2FoldChange: -0.1512267673<br />-log10(padj): 1.737269e-01<br />log10(baseMean): 3.4780928<br />gene: FBgn0052685","log2FoldChange: -0.3484679792<br />-log10(padj): 1.311210e-01<br />log10(baseMean): 1.4288771<br />gene: FBgn0052686","log2FoldChange: -0.5608607118<br />-log10(padj): 3.532804e+00<br />log10(baseMean): 2.5148666<br />gene: FBgn0052687","log2FoldChange: 0.1187001700<br />-log10(padj): 1.441906e-01<br />log10(baseMean): 3.1556608<br />gene: FBgn0052699","log2FoldChange: 1.3156114730<br />-log10(padj): 2.083222e-01<br />log10(baseMean): 1.4314725<br />gene: FBgn0052700","log2FoldChange: 0.1201760562<br />-log10(padj): 1.658154e-01<br />log10(baseMean): 2.8442227<br />gene: FBgn0052702","log2FoldChange: -0.1799490683<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 0.8794184<br />gene: FBgn0052703","log2FoldChange: 0.1736906450<br />-log10(padj): 1.930828e-01<br />log10(baseMean): 2.4386880<br />gene: FBgn0052707","log2FoldChange: -0.2450755195<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 2.2153266<br />gene: FBgn0052708","log2FoldChange: 0.1358497722<br />-log10(padj): 5.048558e-02<br />log10(baseMean): 1.9052937<br />gene: FBgn0052711","log2FoldChange: -0.7527666503<br />-log10(padj): 5.509571e-01<br />log10(baseMean): 1.7626484<br />gene: FBgn0052719","log2FoldChange: -0.4569834617<br />-log10(padj): 7.995419e-02<br />log10(baseMean): 0.9256147<br />gene: FBgn0052720","log2FoldChange: 0.1161260505<br />-log10(padj): 1.175473e-01<br />log10(baseMean): 2.5253698<br />gene: FBgn0052732","log2FoldChange: -0.0523871886<br />-log10(padj): 3.434146e-02<br />log10(baseMean): 3.5186976<br />gene: FBgn0052743","log2FoldChange: 0.0996576583<br />-log10(padj): 1.124980e-02<br />log10(baseMean): 0.9866917<br />gene: FBgn0052745","log2FoldChange: 0.1652725779<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 1.7844097<br />gene: FBgn0052756","log2FoldChange: -0.0663807712<br />-log10(padj): 7.768911e-02<br />log10(baseMean): 3.3360266<br />gene: FBgn0052758","log2FoldChange: -0.0921526363<br />-log10(padj): 6.928401e-02<br />log10(baseMean): 3.3120535<br />gene: FBgn0052767","log2FoldChange: -0.2484749455<br />-log10(padj): 3.646662e-01<br />log10(baseMean): 2.7126715<br />gene: FBgn0052772","log2FoldChange: 0.3752932097<br />-log10(padj): 5.077932e-02<br />log10(baseMean): 0.7930539<br />gene: FBgn0052783","log2FoldChange: 0.0581506415<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.9473900<br />gene: FBgn0052791","log2FoldChange: 0.3889465492<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 1.5706842<br />gene: FBgn0052801","log2FoldChange: 0.4733686426<br />-log10(padj): 1.785218e+00<br />log10(baseMean): 2.4954846<br />gene: FBgn0052803","log2FoldChange: -0.0124823511<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 1.4639828<br />gene: FBgn0052808","log2FoldChange: -0.7371562658<br />-log10(padj): 2.953876e+00<br />log10(baseMean): 2.2202055<br />gene: FBgn0052813","log2FoldChange: -0.5736158451<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 0.8803930<br />gene: FBgn0052815","log2FoldChange: 0.2790800128<br />-log10(padj): 3.530073e-02<br />log10(baseMean): 0.8307437<br />gene: FBgn0052816","log2FoldChange: 0.0624692552<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 1.7512392<br />gene: FBgn0052823","log2FoldChange: -0.6195643580<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 0.8896704<br />gene: FBgn0052829","log2FoldChange: 1.5753051664<br />-log10(padj): 7.729714e-01<br />log10(baseMean): 0.8206410<br />gene: FBgn0052845","log2FoldChange: 0.1591035920<br />-log10(padj): 1.196377e-01<br />log10(baseMean): 2.4321341<br />gene: FBgn0052850","log2FoldChange: 0.2247513593<br />-log10(padj): 7.414978e-02<br />log10(baseMean): 1.8757213<br />gene: FBgn0052856","log2FoldChange: -0.4984863089<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 0.8221026<br />gene: FBgn0052865","log2FoldChange: -1.9599129814<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.2099972<br />gene: FBgn0052939","log2FoldChange: -0.0158766905<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.3563108<br />gene: FBgn0052944","log2FoldChange: 0.2821064046<br />-log10(padj): 5.092879e-01<br />log10(baseMean): 3.1406839<br />gene: FBgn0052982","log2FoldChange: -0.1964616344<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.0662689<br />gene: FBgn0052984","log2FoldChange: 0.8292644450<br />-log10(padj): 1.145334e+00<br />log10(baseMean): 1.8611712<br />gene: FBgn0052985","log2FoldChange: 0.2463307887<br />-log10(padj): 2.765362e-01<br />log10(baseMean): 2.4395043<br />gene: FBgn0053002","log2FoldChange: -0.7813656353<br />-log10(padj): 2.904404e+00<br />log10(baseMean): 2.0561821<br />gene: FBgn0053007","log2FoldChange: -0.1426133767<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.9734299<br />gene: FBgn0053017","log2FoldChange: 0.0840889803<br />-log10(padj): 3.696079e-02<br />log10(baseMean): 2.0610704<br />gene: FBgn0053051","log2FoldChange: 0.0327478153<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 1.7238331<br />gene: FBgn0053054","log2FoldChange: -0.4560807316<br />-log10(padj): 1.053160e+00<br />log10(baseMean): 2.5366352<br />gene: FBgn0053056","log2FoldChange: 0.1438934590<br />-log10(padj): 7.196422e-02<br />log10(baseMean): 2.0273268<br />gene: FBgn0053057","log2FoldChange: -0.2004887820<br />-log10(padj): 9.035501e-02<br />log10(baseMean): 1.7504895<br />gene: FBgn0053080","log2FoldChange: -0.0969646453<br />-log10(padj): 1.187329e-02<br />log10(baseMean): 1.0343657<br />gene: FBgn0053082","log2FoldChange: -0.0225228696<br />-log10(padj): 1.442589e-02<br />log10(baseMean): 3.7119361<br />gene: FBgn0053087","log2FoldChange: -0.2202752777<br />-log10(padj): 7.632928e-01<br />log10(baseMean): 3.1735286<br />gene: FBgn0053094","log2FoldChange: 0.1374876193<br />-log10(padj): 4.614145e-02<br />log10(baseMean): 2.7024684<br />gene: FBgn0053095","log2FoldChange: 0.2335279094<br />-log10(padj): 4.858187e-01<br />log10(baseMean): 2.5989439<br />gene: FBgn0053096","log2FoldChange: -0.2396063393<br />-log10(padj): 4.591891e-01<br />log10(baseMean): 3.0137853<br />gene: FBgn0053097","log2FoldChange: 0.5421663598<br />-log10(padj): 1.052021e-01<br />log10(baseMean): 0.9027459<br />gene: FBgn0053099","log2FoldChange: -0.0716516683<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 2.6711386<br />gene: FBgn0053100","log2FoldChange: 0.4141011798<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 2.4692361<br />gene: FBgn0053105","log2FoldChange: -0.0258497006<br />-log10(padj): 1.245357e-02<br />log10(baseMean): 2.2439015<br />gene: FBgn0053107","log2FoldChange: 0.8235875886<br />-log10(padj): 9.682114e-01<br />log10(baseMean): 1.4913873<br />gene: FBgn0053108","log2FoldChange: -0.3353033693<br />-log10(padj): 4.492750e-01<br />log10(baseMean): 2.2900026<br />gene: FBgn0053111","log2FoldChange: -0.0981409483<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 3.5124222<br />gene: FBgn0053113","log2FoldChange: 0.0439364219<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 2.9411357<br />gene: FBgn0053116","log2FoldChange: 0.0785423340<br />-log10(padj): 1.797597e-02<br />log10(baseMean): 1.5793590<br />gene: FBgn0053120","log2FoldChange: -0.1049045238<br />-log10(padj): 1.390945e-01<br />log10(baseMean): 3.3989265<br />gene: FBgn0053123","log2FoldChange: -0.0063825076<br />-log10(padj): 1.989760e-03<br />log10(baseMean): 1.6624697<br />gene: FBgn0053124","log2FoldChange: 0.6958034136<br />-log10(padj): 1.837508e-01<br />log10(baseMean): 0.9640121<br />gene: FBgn0053127","log2FoldChange: 0.0390826493<br />-log10(padj): 2.945997e-02<br />log10(baseMean): 3.4432641<br />gene: FBgn0053129","log2FoldChange: 1.1872885414<br />-log10(padj): 4.975627e-01<br />log10(baseMean): 0.9245824<br />gene: FBgn0053137","log2FoldChange: 0.4608202190<br />-log10(padj): 1.665850e+00<br />log10(baseMean): 3.1994914<br />gene: FBgn0053138","log2FoldChange: -0.0204384626<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.0912354<br />gene: FBgn0053139","log2FoldChange: 0.3136651112<br />-log10(padj): 4.866456e-01<br />log10(baseMean): 3.1188531<br />gene: FBgn0053144","log2FoldChange: 0.0171223320<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.6147762<br />gene: FBgn0053145","log2FoldChange: 0.2425218328<br />-log10(padj): 2.400870e-01<br />log10(baseMean): 2.2464309<br />gene: FBgn0053156","log2FoldChange: 0.0997059807<br />-log10(padj): 1.115347e-01<br />log10(baseMean): 2.7755263<br />gene: FBgn0053158","log2FoldChange: 0.1963988709<br />-log10(padj): 5.617499e-02<br />log10(baseMean): 1.4469376<br />gene: FBgn0053159","log2FoldChange: 0.4752274884<br />-log10(padj): 2.901409e-01<br />log10(baseMean): 1.6161996<br />gene: FBgn0053169","log2FoldChange: 0.2538631171<br />-log10(padj): 1.117342e-01<br />log10(baseMean): 2.2620028<br />gene: FBgn0053170","log2FoldChange: -0.1817260640<br />-log10(padj): 8.933533e-02<br />log10(baseMean): 2.0991622<br />gene: FBgn0053172","log2FoldChange: -0.2382978696<br />-log10(padj): 1.460573e-01<br />log10(baseMean): 1.8206514<br />gene: FBgn0053173","log2FoldChange: 0.1352354489<br />-log10(padj): 1.375785e-02<br />log10(baseMean): 0.8842869<br />gene: FBgn0053177","log2FoldChange: -0.1910281355<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 1.2009352<br />gene: FBgn0053178","log2FoldChange: -0.0861522471<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.9406702<br />gene: FBgn0053180","log2FoldChange: -0.5207137361<br />-log10(padj): 1.497830e+00<br />log10(baseMean): 2.4826213<br />gene: FBgn0053181","log2FoldChange: 0.0195879264<br />-log10(padj): 1.076328e-02<br />log10(baseMean): 2.7747249<br />gene: FBgn0053182","log2FoldChange: 0.5172264358<br />-log10(padj): 2.077428e+00<br />log10(baseMean): 2.5259512<br />gene: FBgn0053193","log2FoldChange: 0.3656144036<br />-log10(padj): 1.792604e-01<br />log10(baseMean): 1.7151688<br />gene: FBgn0053196","log2FoldChange: 0.2580927085<br />-log10(padj): 2.296369e-01<br />log10(baseMean): 3.2178205<br />gene: FBgn0053197","log2FoldChange: -0.0830063270<br />-log10(padj): 2.849528e-02<br />log10(baseMean): 2.1820537<br />gene: FBgn0053198","log2FoldChange: -0.1508829302<br />-log10(padj): 2.404269e-02<br />log10(baseMean): 1.1460804<br />gene: FBgn0053203","log2FoldChange: -0.1534361987<br />-log10(padj): 1.455059e-01<br />log10(baseMean): 2.8138419<br />gene: FBgn0053207","log2FoldChange: -0.0435008200<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 3.3394866<br />gene: FBgn0053208","log2FoldChange: -0.0315038820<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 2.2680112<br />gene: FBgn0053213","log2FoldChange: -0.0362508111<br />-log10(padj): 2.728113e-02<br />log10(baseMean): 3.4628838<br />gene: FBgn0053214","log2FoldChange: 0.3308386856<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 1.8686098<br />gene: FBgn0053217","log2FoldChange: -0.3582710126<br />-log10(padj): 2.690434e-01<br />log10(baseMean): 1.7263377<br />gene: FBgn0053225","log2FoldChange: -0.1076328012<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.5579246<br />gene: FBgn0053229","log2FoldChange: -0.2488915989<br />-log10(padj): 2.473805e-01<br />log10(baseMean): 2.5964876<br />gene: FBgn0053230","log2FoldChange: -0.0176469875<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.2249615<br />gene: FBgn0053260","log2FoldChange: -0.0163794246<br />-log10(padj): 2.985629e-03<br />log10(baseMean): 1.1913628<br />gene: FBgn0053267","log2FoldChange: -0.1343734685<br />-log10(padj): 2.957270e-02<br />log10(baseMean): 1.7265861<br />gene: FBgn0053276","log2FoldChange: 0.6688529791<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 0.8832859<br />gene: FBgn0053278","log2FoldChange: 0.0777202641<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 1.0076833<br />gene: FBgn0053288","log2FoldChange: 0.0450352699<br />-log10(padj): 9.053881e-03<br />log10(baseMean): 1.7720256<br />gene: FBgn0053294","log2FoldChange: -0.1795326699<br />-log10(padj): 1.789957e-02<br />log10(baseMean): 0.9231788<br />gene: FBgn0053300","log2FoldChange: 0.0955287687<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.7176832<br />gene: FBgn0053303","log2FoldChange: -1.4524710414<br />-log10(padj): 9.706516e+00<br />log10(baseMean): 3.0058523<br />gene: FBgn0053307","log2FoldChange: 0.3270965165<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.1432899<br />gene: FBgn0053317","log2FoldChange: -0.1880653614<br />-log10(padj): 2.426927e-01<br />log10(baseMean): 2.6477817<br />gene: FBgn0053324","log2FoldChange: -0.1578472148<br />-log10(padj): 3.436407e-02<br />log10(baseMean): 3.3995681<br />gene: FBgn0053328","log2FoldChange: -0.4997897128<br />-log10(padj): 1.011024e+00<br />log10(baseMean): 2.7799999<br />gene: FBgn0053329","log2FoldChange: -0.3977885502<br />-log10(padj): 7.252582e-01<br />log10(baseMean): 2.2154191<br />gene: FBgn0053330","log2FoldChange: 1.6963660258<br />-log10(padj): 1.325666e+00<br />log10(baseMean): 1.0888575<br />gene: FBgn0053346","log2FoldChange: -0.4166348796<br />-log10(padj): 1.327518e+00<br />log10(baseMean): 3.2640766<br />gene: FBgn0053460","log2FoldChange: 0.2782381065<br />-log10(padj): 1.157132e-01<br />log10(baseMean): 1.5889705<br />gene: FBgn0053461","log2FoldChange: -0.1086069389<br />-log10(padj): 6.938076e-02<br />log10(baseMean): 3.0479223<br />gene: FBgn0053464","log2FoldChange: -0.4792271125<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.4828895<br />gene: FBgn0053465","log2FoldChange: 1.6004597655<br />-log10(padj): 1.111444e+00<br />log10(baseMean): 1.0269705<br />gene: FBgn0053468","log2FoldChange: -0.2296095040<br />-log10(padj): 3.778552e-02<br />log10(baseMean): 1.1438503<br />gene: FBgn0053481","log2FoldChange: 0.1649602595<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.9017176<br />gene: FBgn0053493","log2FoldChange: -0.0170251388<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 3.0419443<br />gene: FBgn0053494","log2FoldChange: -5.9345609720<br />-log10(padj): 3.771626e-01<br />log10(baseMean): 0.8047323<br />gene: FBgn0053498","log2FoldChange: -0.4570501928<br />-log10(padj): 1.605136e-01<br />log10(baseMean): 1.7323492<br />gene: FBgn0053503","log2FoldChange: -0.1602971050<br />-log10(padj): 1.622135e-01<br />log10(baseMean): 2.4946457<br />gene: FBgn0053505","log2FoldChange: 0.1442749251<br />-log10(padj): 1.079031e-01<br />log10(baseMean): 2.2680115<br />gene: FBgn0053506","log2FoldChange: 0.9098876371<br />-log10(padj): 1.720633e+00<br />log10(baseMean): 1.7630264<br />gene: FBgn0053508","log2FoldChange: 0.5632496020<br />-log10(padj): 4.472881e-01<br />log10(baseMean): 1.8014774<br />gene: FBgn0053509","log2FoldChange: 0.0279840592<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.2960330<br />gene: FBgn0053510","log2FoldChange: 0.4866712992<br />-log10(padj): 6.744424e-02<br />log10(baseMean): 1.1272764<br />gene: FBgn0053511","log2FoldChange: -0.0019015264<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.7996783<br />gene: FBgn0053513","log2FoldChange: -0.7795501199<br />-log10(padj): 8.059840e+00<br />log10(baseMean): 2.8489479<br />gene: FBgn0053516","log2FoldChange: 0.2703126692<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 2.3559327<br />gene: FBgn0053519","log2FoldChange: -0.0449333172<br />-log10(padj): 2.770735e-02<br />log10(baseMean): 2.4390324<br />gene: FBgn0053520","log2FoldChange: -0.1475126594<br />-log10(padj): 1.820590e-01<br />log10(baseMean): 3.0328402<br />gene: FBgn0053522","log2FoldChange: -0.0261896949<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 2.9085877<br />gene: FBgn0053523","log2FoldChange: -0.1004130456<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 3.3327677<br />gene: FBgn0053526","log2FoldChange: 0.1237309782<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 0.8019363<br />gene: FBgn0053527","log2FoldChange: -0.0860395387<br />-log10(padj): 2.480727e-02<br />log10(baseMean): 1.6639489<br />gene: FBgn0053542","log2FoldChange: -0.4976819761<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 0.8413382<br />gene: FBgn0053548","log2FoldChange: -0.0340258060<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 3.4674426<br />gene: FBgn0053554","log2FoldChange: 0.7065753070<br />-log10(padj): 1.524123e+00<br />log10(baseMean): 1.8081344<br />gene: FBgn0053555","log2FoldChange: 1.0341507845<br />-log10(padj): 2.520382e+00<br />log10(baseMean): 2.3441043<br />gene: FBgn0053556","log2FoldChange: 0.5222622274<br />-log10(padj): 1.936628e+00<br />log10(baseMean): 3.4651222<br />gene: FBgn0053558","log2FoldChange: -0.0633255398<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.9050043<br />gene: FBgn0053635","log2FoldChange: 0.6499171716<br />-log10(padj): 2.113080e-01<br />log10(baseMean): 1.2189591<br />gene: FBgn0053640","log2FoldChange: 1.0250142929<br />-log10(padj): 6.763617e-01<br />log10(baseMean): 1.3380595<br />gene: FBgn0053641","log2FoldChange: 0.9531339480<br />-log10(padj): 6.157033e-01<br />log10(baseMean): 1.2907415<br />gene: FBgn0053642","log2FoldChange: 1.5084944238<br />-log10(padj): 9.418470e-01<br />log10(baseMean): 1.1557358<br />gene: FBgn0053643","log2FoldChange: 0.4619004217<br />-log10(padj): 1.201659e-01<br />log10(baseMean): 1.1787727<br />gene: FBgn0053644","log2FoldChange: 0.6530392788<br />-log10(padj): 1.399139e-01<br />log10(baseMean): 1.0536013<br />gene: FBgn0053645","log2FoldChange: 0.1405510698<br />-log10(padj): 4.503165e-02<br />log10(baseMean): 1.8721272<br />gene: FBgn0053653","log2FoldChange: -0.3295423659<br />-log10(padj): 4.020564e-02<br />log10(baseMean): 0.8103894<br />gene: FBgn0053696","log2FoldChange: -0.2407757874<br />-log10(padj): 2.818811e-02<br />log10(baseMean): 1.0124312<br />gene: FBgn0053757","log2FoldChange: 0.0235898522<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 1.9373680<br />gene: FBgn0053774","log2FoldChange: 0.3352759331<br />-log10(padj): 1.522534e-01<br />log10(baseMean): 1.7046501<br />gene: FBgn0053867","log2FoldChange: -1.3168719410<br />-log10(padj): 4.201799e-01<br />log10(baseMean): 1.1472776<br />gene: FBgn0053868","log2FoldChange: 0.3554706198<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.1444310<br />gene: FBgn0053909","log2FoldChange: 0.0228250809<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 0.8169490<br />gene: FBgn0053914","log2FoldChange: 0.6175017632<br />-log10(padj): 3.046453e-01<br />log10(baseMean): 2.8875764<br />gene: FBgn0053926","log2FoldChange: -0.1307448530<br />-log10(padj): 6.959715e-02<br />log10(baseMean): 3.8345831<br />gene: FBgn0053936","log2FoldChange: -0.1985917815<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 1.8076671<br />gene: FBgn0053937","log2FoldChange: -0.6985815193<br />-log10(padj): 4.353382e-01<br />log10(baseMean): 1.5383088<br />gene: FBgn0053963","log2FoldChange: -0.4586236309<br />-log10(padj): 1.720633e+00<br />log10(baseMean): 3.5163197<br />gene: FBgn0053967","log2FoldChange: 0.3258195585<br />-log10(padj): 5.794400e-01<br />log10(baseMean): 2.5396891<br />gene: FBgn0053969","log2FoldChange: 0.2611727709<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.2952303<br />gene: FBgn0053970","log2FoldChange: -0.3931664168<br />-log10(padj): 8.290431e-01<br />log10(baseMean): 2.2149125<br />gene: FBgn0053971","log2FoldChange: 0.0967576430<br />-log10(padj): 1.300138e-02<br />log10(baseMean): 2.0569674<br />gene: FBgn0053977","log2FoldChange: -0.0114586343<br />-log10(padj): 1.904030e-03<br />log10(baseMean): 2.3339642<br />gene: FBgn0053978","log2FoldChange: 0.8786426688<br />-log10(padj): 2.625609e-01<br />log10(baseMean): 0.8827624<br />gene: FBgn0053983","log2FoldChange: -0.2608846047<br />-log10(padj): 1.127675e-01<br />log10(baseMean): 1.9858489<br />gene: FBgn0053987","log2FoldChange: -0.2412684223<br />-log10(padj): 3.696516e-02<br />log10(baseMean): 1.0719983<br />gene: FBgn0053988","log2FoldChange: -0.0265095657<br />-log10(padj): 2.725201e-03<br />log10(baseMean): 1.5804810<br />gene: FBgn0053995","log2FoldChange: -0.1542354425<br />-log10(padj): 4.583018e-02<br />log10(baseMean): 1.5633460<br />gene: FBgn0054001","log2FoldChange: -0.2085820337<br />-log10(padj): 1.331016e-01<br />log10(baseMean): 2.3059128<br />gene: FBgn0054003","log2FoldChange: 0.5662527535<br />-log10(padj): 3.352636e-01<br />log10(baseMean): 1.5039284<br />gene: FBgn0054008","log2FoldChange: -0.1762371989<br />-log10(padj): 7.398062e-02<br />log10(baseMean): 1.8509715<br />gene: FBgn0054015","log2FoldChange: 0.0958414486<br />-log10(padj): 2.059298e-02<br />log10(baseMean): 1.6983735<br />gene: FBgn0054039","log2FoldChange: 0.2351484400<br />-log10(padj): 3.466159e-02<br />log10(baseMean): 0.9226768<br />gene: FBgn0054054","log2FoldChange: -0.8930802651<br />-log10(padj): 2.570095e+00<br />log10(baseMean): 1.8713121<br />gene: FBgn0054056","log2FoldChange: -1.0175007934<br />-log10(padj): 1.263262e+00<br />log10(baseMean): 1.4845008<br />gene: FBgn0054057","log2FoldChange: 0.6189540020<br />-log10(padj): 7.366068e-02<br />log10(baseMean): 0.8978209<br />gene: FBgn0058005","log2FoldChange: 0.2211878137<br />-log10(padj): 1.475887e-01<br />log10(baseMean): 1.9441254<br />gene: FBgn0058006","log2FoldChange: -0.1191569653<br />-log10(padj): 4.937499e-02<br />log10(baseMean): 3.1386991<br />gene: FBgn0058045","log2FoldChange: -0.3585103506<br />-log10(padj): 8.890809e-02<br />log10(baseMean): 2.6796089<br />gene: FBgn0058127","log2FoldChange: 0.1436059049<br />-log10(padj): 1.500149e-02<br />log10(baseMean): 1.0819690<br />gene: FBgn0058298","log2FoldChange: -0.1967003097<br />-log10(padj): 4.119705e-02<br />log10(baseMean): 2.7299855<br />gene: FBgn0058439","log2FoldChange: 0.0421713286<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.5558119<br />gene: FBgn0058470","log2FoldChange: 0.6247538030<br />-log10(padj): 1.285437e+00<br />log10(baseMean): 1.9842224<br />gene: FBgn0060296","log2FoldChange: 0.0592710743<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.8225004<br />gene: FBgn0061196","log2FoldChange: -0.2787543755<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.3820877<br />gene: FBgn0061198","log2FoldChange: -0.2085198572<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 3.6013221<br />gene: FBgn0061200","log2FoldChange: 0.1395619756<br />-log10(padj): 2.666229e-01<br />log10(baseMean): 3.0011827<br />gene: FBgn0061469","log2FoldChange: 0.1477098511<br />-log10(padj): 2.106661e-01<br />log10(baseMean): 2.6685191<br />gene: FBgn0061476","log2FoldChange: 0.2288513022<br />-log10(padj): 2.527712e-01<br />log10(baseMean): 2.6260045<br />gene: FBgn0061492","log2FoldChange: 0.1670748010<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 3.1157395<br />gene: FBgn0061515","log2FoldChange: -0.0284909484<br />-log10(padj): 1.977045e-02<br />log10(baseMean): 2.9148327<br />gene: FBgn0062413","log2FoldChange: -0.4077311135<br />-log10(padj): 1.054966e+00<br />log10(baseMean): 2.4652125<br />gene: FBgn0062440","log2FoldChange: -0.0049168002<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.9922624<br />gene: FBgn0062442","log2FoldChange: -0.2080703437<br />-log10(padj): 1.333676e-01<br />log10(baseMean): 2.2926941<br />gene: FBgn0062449","log2FoldChange: 0.0676439865<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.8284816<br />gene: FBgn0062463","log2FoldChange: -0.1770987085<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 1.7107021<br />gene: FBgn0062517","log2FoldChange: 1.2648590976<br />-log10(padj): 4.726491e-01<br />log10(baseMean): 0.8126827<br />gene: FBgn0062565","log2FoldChange: 0.4752625503<br />-log10(padj): 3.141135e-01<br />log10(baseMean): 1.7659617<br />gene: FBgn0062928","log2FoldChange: -1.6828983439<br />-log10(padj): 2.132647e+00<br />log10(baseMean): 1.2403448<br />gene: FBgn0062978","log2FoldChange: -0.2356578451<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 2.0169417<br />gene: FBgn0063449","log2FoldChange: -0.1771353323<br />-log10(padj): 3.549557e-01<br />log10(baseMean): 3.6295279<br />gene: FBgn0063485","log2FoldChange: 0.4087208922<br />-log10(padj): 1.560962e+00<br />log10(baseMean): 3.0301563<br />gene: FBgn0063491","log2FoldChange: 0.7962468702<br />-log10(padj): 9.251575e+00<br />log10(baseMean): 2.7691823<br />gene: FBgn0063492","log2FoldChange: 0.8642426708<br />-log10(padj): 4.233687e+00<br />log10(baseMean): 2.4801065<br />gene: FBgn0063493","log2FoldChange: 0.3209800150<br />-log10(padj): 6.209951e-01<br />log10(baseMean): 3.2225134<br />gene: FBgn0063494","log2FoldChange: 0.2758092472<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.0303192<br />gene: FBgn0063495","log2FoldChange: 0.0569281980<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 2.5003648<br />gene: FBgn0063497","log2FoldChange: -1.3707393029<br />-log10(padj): 1.360174e+01<br />log10(baseMean): 2.5363924<br />gene: FBgn0063498","log2FoldChange: -1.3093252868<br />-log10(padj): 2.107755e+01<br />log10(baseMean): 2.8103619<br />gene: FBgn0063649","log2FoldChange: 2.7993679360<br />-log10(padj): 4.447893e+00<br />log10(baseMean): 1.1156827<br />gene: FBgn0063667","log2FoldChange: -0.0183318894<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 5.0099214<br />gene: FBgn0064225","log2FoldChange: 0.0026776561<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 3.1428224<br />gene: FBgn0064766","log2FoldChange: -0.8729200009<br />-log10(padj): 2.510493e-01<br />log10(baseMean): 0.9283163<br />gene: FBgn0064912","log2FoldChange: 0.0774921591<br />-log10(padj): 3.684014e-02<br />log10(baseMean): 2.5952902<br />gene: FBgn0065032","log2FoldChange: -0.0893884718<br />-log10(padj): 2.063850e-02<br />log10(baseMean): 1.8345073<br />gene: FBgn0065035","log2FoldChange: -0.3665346321<br />-log10(padj): 2.891818e-02<br />log10(baseMean): 0.8757012<br />gene: FBgn0065053","log2FoldChange: 0.1392815473<br />-log10(padj): 2.185367e-02<br />log10(baseMean): 1.3886021<br />gene: FBgn0065064","log2FoldChange: -0.2707371464<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 1.6709498<br />gene: FBgn0065088","log2FoldChange: -0.8061926097<br />-log10(padj): 2.892640e-01<br />log10(baseMean): 1.2059628<br />gene: FBgn0065098","log2FoldChange: -0.0031428052<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.4391917<br />gene: FBgn0065099","log2FoldChange: -0.1827527665<br />-log10(padj): 1.250516e-01<br />log10(baseMean): 2.5161687<br />gene: FBgn0065104","log2FoldChange: 0.4459817929<br />-log10(padj): 5.693002e-01<br />log10(baseMean): 4.1650564<br />gene: FBgn0066084","log2FoldChange: 0.6665865339<br />-log10(padj): 1.865095e-01<br />log10(baseMean): 1.0026957<br />gene: FBgn0066101","log2FoldChange: -0.0761393563<br />-log10(padj): 6.500626e-02<br />log10(baseMean): 2.7712939<br />gene: FBgn0066114","log2FoldChange: 0.8171195131<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 1.0834213<br />gene: FBgn0066292","log2FoldChange: -0.2060680014<br />-log10(padj): 2.183150e-01<br />log10(baseMean): 2.8898067<br />gene: FBgn0067102","log2FoldChange: 0.1503772511<br />-log10(padj): 5.732811e-02<br />log10(baseMean): 1.6317993<br />gene: FBgn0067317","log2FoldChange: 0.3800921553<br />-log10(padj): 5.537464e-01<br />log10(baseMean): 2.0342696<br />gene: FBgn0067628","log2FoldChange: 0.2037370668<br />-log10(padj): 8.188638e-02<br />log10(baseMean): 1.7534305<br />gene: FBgn0067629","log2FoldChange: -0.1715283939<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 2.7680208<br />gene: FBgn0067779","log2FoldChange: -0.2384916036<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 1.7481785<br />gene: FBgn0067783","log2FoldChange: 0.4828454295<br />-log10(padj): 2.122202e-02<br />log10(baseMean): 1.5403095<br />gene: FBgn0067861","log2FoldChange: 0.1131317603<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 2.9739138<br />gene: FBgn0067864","log2FoldChange: -0.1350751609<br />-log10(padj): 5.794697e-02<br />log10(baseMean): 2.7452745<br />gene: FBgn0067895","log2FoldChange: 0.2821691596<br />-log10(padj): 3.513655e-01<br />log10(baseMean): 2.8223089<br />gene: FBgn0069242","log2FoldChange: -0.3492431115<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 1.2710288<br />gene: FBgn0069354","log2FoldChange: 0.5806023330<br />-log10(padj): 1.548746e-01<br />log10(baseMean): 1.0799333<br />gene: FBgn0069913","log2FoldChange: -0.1981874431<br />-log10(padj): 1.693673e-02<br />log10(baseMean): 0.7889934<br />gene: FBgn0069921","log2FoldChange: 0.1112728189<br />-log10(padj): 4.689734e-02<br />log10(baseMean): 2.1799662<br />gene: FBgn0069973","log2FoldChange: 0.0862727196<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 3.1021360<br />gene: FBgn0082582","log2FoldChange: 0.0741424939<br />-log10(padj): 3.627348e-02<br />log10(baseMean): 2.2836719<br />gene: FBgn0082585","log2FoldChange: 0.0248742867<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 3.4476573<br />gene: FBgn0082598","log2FoldChange: 0.0432887736<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 3.3112407<br />gene: FBgn0082831","log2FoldChange: -0.6040459809<br />-log10(padj): 8.439116e-02<br />log10(baseMean): 0.8785619<br />gene: FBgn0083047","log2FoldChange: -0.6226084464<br />-log10(padj): 8.182336e-02<br />log10(baseMean): 0.8895775<br />gene: FBgn0083048","log2FoldChange: -0.6091751611<br />-log10(padj): 9.650007e-02<br />log10(baseMean): 0.9079799<br />gene: FBgn0083050","log2FoldChange: 0.1651978552<br />-log10(padj): 2.523253e-02<br />log10(baseMean): 1.1046370<br />gene: FBgn0083068","log2FoldChange: 0.2030546496<br />-log10(padj): 3.290255e-01<br />log10(baseMean): 3.1951752<br />gene: FBgn0083077","log2FoldChange: 0.2061421477<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.2909770<br />gene: FBgn0083120","log2FoldChange: 0.1947231604<br />-log10(padj): 2.446854e-02<br />log10(baseMean): 1.0202572<br />gene: FBgn0083121","log2FoldChange: 0.0751285693<br />-log10(padj): 1.620599e-02<br />log10(baseMean): 1.9683449<br />gene: FBgn0083123","log2FoldChange: 0.1566567722<br />-log10(padj): 3.814593e-02<br />log10(baseMean): 1.7832011<br />gene: FBgn0083124","log2FoldChange: 0.1743579754<br />-log10(padj): 3.280476e-01<br />log10(baseMean): 2.7920303<br />gene: FBgn0083167","log2FoldChange: -0.2241783620<br />-log10(padj): 7.144873e-01<br />log10(baseMean): 3.5103152<br />gene: FBgn0083919","log2FoldChange: 0.7762247742<br />-log10(padj): 4.285157e-01<br />log10(baseMean): 1.2835401<br />gene: FBgn0083940","log2FoldChange: 0.0968880856<br />-log10(padj): 3.977426e-02<br />log10(baseMean): 2.2157455<br />gene: FBgn0083953","log2FoldChange: -0.4558538365<br />-log10(padj): 2.822951e+00<br />log10(baseMean): 3.1616207<br />gene: FBgn0083959","log2FoldChange: -0.1800917209<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 1.5569565<br />gene: FBgn0083960","log2FoldChange: 0.1557536642<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 2.4822334<br />gene: FBgn0083961","log2FoldChange: -0.0301819113<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 3.5392305<br />gene: FBgn0083962","log2FoldChange: 0.0364087822<br />-log10(padj): 1.180034e-02<br />log10(baseMean): 2.1214855<br />gene: FBgn0083968","log2FoldChange: -0.2169551869<br />-log10(padj): 6.671310e-01<br />log10(baseMean): 3.3157735<br />gene: FBgn0083969","log2FoldChange: 0.2480596531<br />-log10(padj): 2.155161e-01<br />log10(baseMean): 1.9815554<br />gene: FBgn0083970","log2FoldChange: 0.1880160404<br />-log10(padj): 4.164252e-02<br />log10(baseMean): 1.3049514<br />gene: FBgn0083972","log2FoldChange: -0.8082212443<br />-log10(padj): 4.130467e-01<br />log10(baseMean): 1.2141410<br />gene: FBgn0083973","log2FoldChange: -0.0071344418<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.0384471<br />gene: FBgn0083974","log2FoldChange: 0.1125936525<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.3664337<br />gene: FBgn0083977","log2FoldChange: 0.6795855233<br />-log10(padj): 8.111198e-01<br />log10(baseMean): 1.9538370<br />gene: FBgn0083983","log2FoldChange: 0.5589625300<br />-log10(padj): 4.925073e-01<br />log10(baseMean): 1.5619419<br />gene: FBgn0083985","log2FoldChange: 0.2471309741<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 1.9368340<br />gene: FBgn0083986","log2FoldChange: 0.1888440485<br />-log10(padj): 9.053881e-03<br />log10(baseMean): 1.7974047<br />gene: FBgn0083990","log2FoldChange: 0.8176032498<br />-log10(padj): 5.249561e-01<br />log10(baseMean): 1.2619088<br />gene: FBgn0084001","log2FoldChange: -0.0523117269<br />-log10(padj): 1.305418e-02<br />log10(baseMean): 1.6765439<br />gene: FBgn0084015","log2FoldChange: 0.0663103183<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 1.7005412<br />gene: FBgn0084049","log2FoldChange: -0.2376336161<br />-log10(padj): 2.348156e-01<br />log10(baseMean): 2.6869856<br />gene: FBgn0085188","log2FoldChange: -0.2323971097<br />-log10(padj): 4.814982e-02<br />log10(baseMean): 1.7979935<br />gene: FBgn0085192","log2FoldChange: -0.2402212339<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 1.6355455<br />gene: FBgn0085203","log2FoldChange: -0.0950158976<br />-log10(padj): 2.179021e-02<br />log10(baseMean): 1.7936733<br />gene: FBgn0085212","log2FoldChange: 0.1297850784<br />-log10(padj): 3.703734e-02<br />log10(baseMean): 2.3555366<br />gene: FBgn0085215","log2FoldChange: -0.2438981780<br />-log10(padj): 6.536412e-02<br />log10(baseMean): 1.3280914<br />gene: FBgn0085216","log2FoldChange: 0.2670049112<br />-log10(padj): 3.326566e-01<br />log10(baseMean): 2.2406936<br />gene: FBgn0085220","log2FoldChange: -0.4107057009<br />-log10(padj): 2.201481e-01<br />log10(baseMean): 1.6699816<br />gene: FBgn0085223","log2FoldChange: -0.2748281735<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.0527142<br />gene: FBgn0085224","log2FoldChange: 0.2291022017<br />-log10(padj): 2.001008e-01<br />log10(baseMean): 2.6501554<br />gene: FBgn0085226","log2FoldChange: 0.2157800802<br />-log10(padj): 3.434146e-02<br />log10(baseMean): 2.2934308<br />gene: FBgn0085229","log2FoldChange: 0.2333853626<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 1.6031112<br />gene: FBgn0085242","log2FoldChange: -0.3808740429<br />-log10(padj): 2.765185e-01<br />log10(baseMean): 2.3650538<br />gene: FBgn0085249","log2FoldChange: 0.1969475296<br />-log10(padj): 6.605863e-02<br />log10(baseMean): 1.7628093<br />gene: FBgn0085257","log2FoldChange: 0.1704387343<br />-log10(padj): 2.451126e-02<br />log10(baseMean): 1.1847933<br />gene: FBgn0085260","log2FoldChange: -0.6107766707<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.1756658<br />gene: FBgn0085261","log2FoldChange: 0.0943870801<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.5501460<br />gene: FBgn0085271","log2FoldChange: 0.3975606580<br />-log10(padj): 1.972791e-01<br />log10(baseMean): 1.7429867<br />gene: FBgn0085275","log2FoldChange: 0.0347050133<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 1.7761184<br />gene: FBgn0085279","log2FoldChange: 1.2117302579<br />-log10(padj): 4.494262e-01<br />log10(baseMean): 0.9715714<br />gene: FBgn0085281","log2FoldChange: -0.0783791574<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 1.4474427<br />gene: FBgn0085284","log2FoldChange: 0.1602563112<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 1.4401541<br />gene: FBgn0085290","log2FoldChange: -0.6609805456<br />-log10(padj): 4.728584e-01<br />log10(baseMean): 1.4069452<br />gene: FBgn0085292","log2FoldChange: -0.4394845563<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 1.5160561<br />gene: FBgn0085298","log2FoldChange: -0.2417615123<br />-log10(padj): 4.881401e-02<br />log10(baseMean): 1.1838915<br />gene: FBgn0085317","log2FoldChange: -0.0914656036<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.5223292<br />gene: FBgn0085322","log2FoldChange: -1.2980950730<br />-log10(padj): 1.682463e+00<br />log10(baseMean): 1.6361079<br />gene: FBgn0085334","log2FoldChange: -0.4115009175<br />-log10(padj): 4.472881e-01<br />log10(baseMean): 2.1627570<br />gene: FBgn0085335","log2FoldChange: 0.2189682990<br />-log10(padj): 5.119574e-02<br />log10(baseMean): 1.5369004<br />gene: FBgn0085339","log2FoldChange: 0.0911292119<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 2.0241925<br />gene: FBgn0085348","log2FoldChange: -0.3607424484<br />-log10(padj): 2.587641e-01<br />log10(baseMean): 1.7145392<br />gene: FBgn0085350","log2FoldChange: -0.2725574637<br />-log10(padj): 7.885919e-02<br />log10(baseMean): 1.4202925<br />gene: FBgn0085351","log2FoldChange: 0.5189255220<br />-log10(padj): 7.679941e-02<br />log10(baseMean): 0.7999539<br />gene: FBgn0085352","log2FoldChange: 0.0061590662<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 1.4315320<br />gene: FBgn0085353","log2FoldChange: 0.7768728144<br />-log10(padj): 2.822951e+00<br />log10(baseMean): 2.1609618<br />gene: FBgn0085354","log2FoldChange: 0.0762180814<br />-log10(padj): 8.211999e-03<br />log10(baseMean): 0.9204106<br />gene: FBgn0085357","log2FoldChange: -4.9177233040<br />-log10(padj): 2.010397e+01<br />log10(baseMean): 1.8363628<br />gene: FBgn0085359","log2FoldChange: -0.1505907095<br />-log10(padj): 8.042058e-02<br />log10(baseMean): 2.8463463<br />gene: FBgn0085360","log2FoldChange: -0.0003132129<br />-log10(padj): 4.855687e-04<br />log10(baseMean): 2.8207111<br />gene: FBgn0085363","log2FoldChange: 0.0815558568<br />-log10(padj): 7.628803e-02<br />log10(baseMean): 3.4851857<br />gene: FBgn0085370","log2FoldChange: -0.2396169066<br />-log10(padj): 2.178119e-01<br />log10(baseMean): 3.1401941<br />gene: FBgn0085375","log2FoldChange: -0.7075718806<br />-log10(padj): 7.647449e-01<br />log10(baseMean): 2.4638109<br />gene: FBgn0085376","log2FoldChange: -0.2009583166<br />-log10(padj): 1.351620e-01<br />log10(baseMean): 2.2728070<br />gene: FBgn0085377","log2FoldChange: -0.0143157485<br />-log10(padj): 4.598485e-03<br />log10(baseMean): 2.7688044<br />gene: FBgn0085378","log2FoldChange: -1.0430154454<br />-log10(padj): 9.934190e-01<br />log10(baseMean): 1.4297305<br />gene: FBgn0085379","log2FoldChange: 0.0785734473<br />-log10(padj): 1.952770e-02<br />log10(baseMean): 1.5910893<br />gene: FBgn0085384","log2FoldChange: 0.2629492578<br />-log10(padj): 2.762079e-01<br />log10(baseMean): 2.0784984<br />gene: FBgn0085386","log2FoldChange: 0.4633180906<br />-log10(padj): 1.419496e+00<br />log10(baseMean): 3.2889192<br />gene: FBgn0085388","log2FoldChange: -0.5167027265<br />-log10(padj): 4.834783e-01<br />log10(baseMean): 1.7470213<br />gene: FBgn0085390","log2FoldChange: -0.4614113974<br />-log10(padj): 3.294413e-01<br />log10(baseMean): 1.9619037<br />gene: FBgn0085395","log2FoldChange: 0.1015769825<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 1.1364435<br />gene: FBgn0085399","log2FoldChange: -0.5829635024<br />-log10(padj): 4.958183e+00<br />log10(baseMean): 3.7257846<br />gene: FBgn0085402","log2FoldChange: 0.9265280583<br />-log10(padj): 3.284245e+00<br />log10(baseMean): 2.3338840<br />gene: FBgn0085403","log2FoldChange: 0.0510276146<br />-log10(padj): 2.751499e-02<br />log10(baseMean): 2.4880543<br />gene: FBgn0085405","log2FoldChange: 0.9446184034<br />-log10(padj): 1.001202e+00<br />log10(baseMean): 1.3552671<br />gene: FBgn0085407","log2FoldChange: 0.0741047097<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.4889796<br />gene: FBgn0085408","log2FoldChange: 0.3254078913<br />-log10(padj): 5.001834e-02<br />log10(baseMean): 0.8339890<br />gene: FBgn0085411","log2FoldChange: -0.2282434081<br />-log10(padj): 8.004171e-02<br />log10(baseMean): 1.7936601<br />gene: FBgn0085412","log2FoldChange: -0.0787000238<br />-log10(padj): 9.014029e-03<br />log10(baseMean): 0.9542817<br />gene: FBgn0085413","log2FoldChange: 0.1145834432<br />-log10(padj): 1.410010e-02<br />log10(baseMean): 1.2439367<br />gene: FBgn0085417","log2FoldChange: 0.3734478437<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 0.9868227<br />gene: FBgn0085419","log2FoldChange: 0.3412915118<br />-log10(padj): 1.261043e-01<br />log10(baseMean): 1.6072951<br />gene: FBgn0085421","log2FoldChange: 0.1818132711<br />-log10(padj): 3.990713e-01<br />log10(baseMean): 3.2733570<br />gene: FBgn0085423","log2FoldChange: -0.2380146572<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 1.9165167<br />gene: FBgn0085425","log2FoldChange: 0.7074818487<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 1.1686004<br />gene: FBgn0085427","log2FoldChange: 0.0243807001<br />-log10(padj): 4.187748e-03<br />log10(baseMean): 1.4307103<br />gene: FBgn0085428","log2FoldChange: 1.1589826864<br />-log10(padj): 9.518441e-01<br />log10(baseMean): 1.3597331<br />gene: FBgn0085429","log2FoldChange: -0.0818747144<br />-log10(padj): 5.215772e-02<br />log10(baseMean): 3.3251081<br />gene: FBgn0085430","log2FoldChange: -0.0352140717<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.9556848<br />gene: FBgn0085432","log2FoldChange: -0.0209892528<br />-log10(padj): 1.393747e-02<br />log10(baseMean): 2.5254516<br />gene: FBgn0085433","log2FoldChange: -0.4400028579<br />-log10(padj): 1.927514e+00<br />log10(baseMean): 2.7124002<br />gene: FBgn0085434","log2FoldChange: 0.1793769126<br />-log10(padj): 6.151247e-02<br />log10(baseMean): 1.6770207<br />gene: FBgn0085435","log2FoldChange: -0.1660177973<br />-log10(padj): 3.382998e-01<br />log10(baseMean): 4.1647274<br />gene: FBgn0085436","log2FoldChange: -0.0505014329<br />-log10(padj): 3.163611e-02<br />log10(baseMean): 3.0687972<br />gene: FBgn0085437","log2FoldChange: -0.8687253999<br />-log10(padj): 1.629451e+00<br />log10(baseMean): 1.7734673<br />gene: FBgn0085438","log2FoldChange: 1.0135579176<br />-log10(padj): 5.065865e-01<br />log10(baseMean): 1.0767530<br />gene: FBgn0085440","log2FoldChange: -0.2258319897<br />-log10(padj): 9.735831e-01<br />log10(baseMean): 3.6615388<br />gene: FBgn0085442","log2FoldChange: 0.0400803357<br />-log10(padj): 2.041090e-02<br />log10(baseMean): 3.4510062<br />gene: FBgn0085443","log2FoldChange: -0.0899864789<br />-log10(padj): 6.600542e-02<br />log10(baseMean): 3.0961187<br />gene: FBgn0085444","log2FoldChange: 0.2530254224<br />-log10(padj): 5.455828e-02<br />log10(baseMean): 1.1882428<br />gene: FBgn0085445","log2FoldChange: 0.5810722954<br />-log10(padj): 3.439633e+00<br />log10(baseMean): 3.6012210<br />gene: FBgn0085446","log2FoldChange: -0.4232530845<br />-log10(padj): 1.013001e+00<br />log10(baseMean): 3.1483583<br />gene: FBgn0085450","log2FoldChange: -0.1095426054<br />-log10(padj): 8.809046e-02<br />log10(baseMean): 3.4948849<br />gene: FBgn0085451","log2FoldChange: 0.1995423552<br />-log10(padj): 4.793149e-02<br />log10(baseMean): 1.7473616<br />gene: FBgn0085452","log2FoldChange: 0.4116946544<br />-log10(padj): 6.417617e-01<br />log10(baseMean): 2.0985886<br />gene: FBgn0085453","log2FoldChange: 0.4984087160<br />-log10(padj): 1.230030e+00<br />log10(baseMean): 2.2889409<br />gene: FBgn0085459","log2FoldChange: 0.4859813953<br />-log10(padj): 2.638613e-01<br />log10(baseMean): 2.0844470<br />gene: FBgn0085460","log2FoldChange: -0.0629015813<br />-log10(padj): 7.951273e-03<br />log10(baseMean): 1.2036954<br />gene: FBgn0085465","log2FoldChange: -0.0824021851<br />-log10(padj): 1.182428e-02<br />log10(baseMean): 2.3852990<br />gene: FBgn0085466","log2FoldChange: 0.3788171267<br />-log10(padj): 1.599420e-01<br />log10(baseMean): 2.0802851<br />gene: FBgn0085468","log2FoldChange: 0.1259868530<br />-log10(padj): 7.339738e-02<br />log10(baseMean): 3.1832322<br />gene: FBgn0085478","log2FoldChange: -0.8354422667<br />-log10(padj): 8.703489e-01<br />log10(baseMean): 1.7156075<br />gene: FBgn0085483","log2FoldChange: 0.0036892356<br />-log10(padj): 2.353446e-03<br />log10(baseMean): 2.4167552<br />gene: FBgn0085484","log2FoldChange: -0.6825342471<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 0.8355418<br />gene: FBgn0085488","log2FoldChange: -0.7282632420<br />-log10(padj): 2.400870e-01<br />log10(baseMean): 1.0623960<br />gene: FBgn0085489","log2FoldChange: 0.6963936596<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.3940228<br />gene: FBgn0085787","log2FoldChange: 0.2080248263<br />-log10(padj): 5.290275e-02<br />log10(baseMean): 1.3825277<br />gene: FBgn0085789","log2FoldChange: -0.0291431506<br />-log10(padj): 3.291110e-03<br />log10(baseMean): 1.1967095<br />gene: FBgn0085792","log2FoldChange: -0.6294446031<br />-log10(padj): 3.829309e-01<br />log10(baseMean): 1.5740046<br />gene: FBgn0086031","log2FoldChange: -0.4645998118<br />-log10(padj): 1.165467e-01<br />log10(baseMean): 1.1654282<br />gene: FBgn0086038","log2FoldChange: -0.0853695275<br />-log10(padj): 6.609605e-02<br />log10(baseMean): 3.0294257<br />gene: FBgn0086129","log2FoldChange: 0.0821398001<br />-log10(padj): 6.153502e-02<br />log10(baseMean): 2.4579066<br />gene: FBgn0086130","log2FoldChange: -0.1972482078<br />-log10(padj): 3.443122e-01<br />log10(baseMean): 3.5024187<br />gene: FBgn0086133","log2FoldChange: 0.1274924014<br />-log10(padj): 1.766473e-01<br />log10(baseMean): 3.1927081<br />gene: FBgn0086134","log2FoldChange: 0.3007402318<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 1.8675290<br />gene: FBgn0086200","log2FoldChange: -0.0532953223<br />-log10(padj): 1.962272e-02<br />log10(baseMean): 3.0355447<br />gene: FBgn0086251","log2FoldChange: 0.3633538988<br />-log10(padj): 1.890702e+00<br />log10(baseMean): 2.8499355<br />gene: FBgn0086253","log2FoldChange: -0.0255132087<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 3.1447914<br />gene: FBgn0086254","log2FoldChange: -0.0790895151<br />-log10(padj): 8.903245e-02<br />log10(baseMean): 3.5937475<br />gene: FBgn0086346","log2FoldChange: -0.2010967866<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 3.7630596<br />gene: FBgn0086347","log2FoldChange: 0.0216822892<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 0.8871037<br />gene: FBgn0086348","log2FoldChange: 0.2269297750<br />-log10(padj): 9.945205e-02<br />log10(baseMean): 2.2908951<br />gene: FBgn0086350","log2FoldChange: 0.7316602161<br />-log10(padj): 3.584842e+00<br />log10(baseMean): 3.6167787<br />gene: FBgn0086355","log2FoldChange: 0.0926936068<br />-log10(padj): 9.921925e-02<br />log10(baseMean): 3.3052987<br />gene: FBgn0086356","log2FoldChange: 0.3676775397<br />-log10(padj): 1.630768e+00<br />log10(baseMean): 3.6882967<br />gene: FBgn0086357","log2FoldChange: -0.4338677853<br />-log10(padj): 1.727137e+00<br />log10(baseMean): 3.1619593<br />gene: FBgn0086358","log2FoldChange: 0.1540340707<br />-log10(padj): 1.265198e-01<br />log10(baseMean): 2.4316341<br />gene: FBgn0086359","log2FoldChange: -0.0076220974<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 3.4976947<br />gene: FBgn0086361","log2FoldChange: -0.1375064634<br />-log10(padj): 1.622799e-01<br />log10(baseMean): 2.5019737<br />gene: FBgn0086362","log2FoldChange: -0.2351058705<br />-log10(padj): 2.953616e-01<br />log10(baseMean): 3.0480992<br />gene: FBgn0086364","log2FoldChange: -0.5352078631<br />-log10(padj): 4.401837e+00<br />log10(baseMean): 3.5374284<br />gene: FBgn0086365","log2FoldChange: 1.3619337857<br />-log10(padj): 6.929979e-01<br />log10(baseMean): 1.0266207<br />gene: FBgn0086367","log2FoldChange: -0.1856837757<br />-log10(padj): 6.285343e-01<br />log10(baseMean): 3.1254641<br />gene: FBgn0086370","log2FoldChange: -0.0012920433<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 2.5986144<br />gene: FBgn0086371","log2FoldChange: -0.0187860022<br />-log10(padj): 7.055436e-03<br />log10(baseMean): 2.7450654<br />gene: FBgn0086372","log2FoldChange: -0.2722167298<br />-log10(padj): 6.554094e-01<br />log10(baseMean): 2.9556707<br />gene: FBgn0086377","log2FoldChange: -0.0375747032<br />-log10(padj): 2.749859e-02<br />log10(baseMean): 3.0068726<br />gene: FBgn0086384","log2FoldChange: -0.0838648647<br />-log10(padj): 8.721877e-02<br />log10(baseMean): 3.2203620<br />gene: FBgn0086441","log2FoldChange: 0.0039362826<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 3.4258771<br />gene: FBgn0086442","log2FoldChange: -0.1496024655<br />-log10(padj): 1.553941e-01<br />log10(baseMean): 3.6505359<br />gene: FBgn0086443","log2FoldChange: 0.0082572726<br />-log10(padj): 4.548884e-03<br />log10(baseMean): 2.8460443<br />gene: FBgn0086444","log2FoldChange: 0.1920235886<br />-log10(padj): 2.353998e-01<br />log10(baseMean): 2.4599790<br />gene: FBgn0086445","log2FoldChange: 0.1902293488<br />-log10(padj): 2.466545e-01<br />log10(baseMean): 2.6115223<br />gene: FBgn0086446","log2FoldChange: 0.1613878329<br />-log10(padj): 5.608230e-02<br />log10(baseMean): 2.0694923<br />gene: FBgn0086447","log2FoldChange: 0.2798517523<br />-log10(padj): 8.409693e-02<br />log10(baseMean): 1.8445765<br />gene: FBgn0086448","log2FoldChange: -0.5039799897<br />-log10(padj): 1.855718e+00<br />log10(baseMean): 3.7480072<br />gene: FBgn0086450","log2FoldChange: -0.2034601160<br />-log10(padj): 3.121768e-01<br />log10(baseMean): 3.2064136<br />gene: FBgn0086451","log2FoldChange: -0.0277504787<br />-log10(padj): 1.693673e-02<br />log10(baseMean): 4.4074681<br />gene: FBgn0086472","log2FoldChange: 0.0117587515<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.8896310<br />gene: FBgn0086475","log2FoldChange: 0.3303713577<br />-log10(padj): 1.060593e+00<br />log10(baseMean): 3.2218537<br />gene: FBgn0086532","log2FoldChange: 0.0659239632<br />-log10(padj): 1.056523e-02<br />log10(baseMean): 3.4397404<br />gene: FBgn0086558","log2FoldChange: -0.0436198955<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.7623157<br />gene: FBgn0086605","log2FoldChange: -0.2651032038<br />-log10(padj): 6.238437e-02<br />log10(baseMean): 1.2882808<br />gene: FBgn0086608","log2FoldChange: -0.1366202583<br />-log10(padj): 2.200835e-01<br />log10(baseMean): 3.0569723<br />gene: FBgn0086613","log2FoldChange: 0.0014235343<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 2.6524010<br />gene: FBgn0086655","log2FoldChange: -0.2434530234<br />-log10(padj): 2.872106e-01<br />log10(baseMean): 3.2702571<br />gene: FBgn0086656","log2FoldChange: 0.0625670346<br />-log10(padj): 5.365939e-02<br />log10(baseMean): 2.7049210<br />gene: FBgn0086657","log2FoldChange: -0.6513447117<br />-log10(padj): 1.253767e-01<br />log10(baseMean): 0.8173804<br />gene: FBgn0086661","log2FoldChange: -1.0465530235<br />-log10(padj): 3.876702e-01<br />log10(baseMean): 0.9048534<br />gene: FBgn0086664","log2FoldChange: 0.3904775537<br />-log10(padj): 5.936353e-02<br />log10(baseMean): 0.8300959<br />gene: FBgn0086666","log2FoldChange: -0.3177575652<br />-log10(padj): 1.991232e+00<br />log10(baseMean): 3.2153980<br />gene: FBgn0086674","log2FoldChange: 0.1330659807<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.7113839<br />gene: FBgn0086676","log2FoldChange: -0.1806403530<br />-log10(padj): 2.548905e-01<br />log10(baseMean): 2.8774036<br />gene: FBgn0086679","log2FoldChange: -0.5297538522<br />-log10(padj): 2.041448e-01<br />log10(baseMean): 1.3680219<br />gene: FBgn0086681","log2FoldChange: -0.1021491828<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 3.0401445<br />gene: FBgn0086683","log2FoldChange: -0.0860571750<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 3.1650736<br />gene: FBgn0086685","log2FoldChange: 0.0069285013<br />-log10(padj): 3.155619e-03<br />log10(baseMean): 3.4117035<br />gene: FBgn0086686","log2FoldChange: -0.3133061168<br />-log10(padj): 2.706395e+00<br />log10(baseMean): 3.9285507<br />gene: FBgn0086687","log2FoldChange: -0.1955227060<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 3.1591284<br />gene: FBgn0086689","log2FoldChange: -0.0187679877<br />-log10(padj): 1.473633e-02<br />log10(baseMean): 3.5586760<br />gene: FBgn0086690","log2FoldChange: -0.2072383264<br />-log10(padj): 1.476243e-01<br />log10(baseMean): 2.2486977<br />gene: FBgn0086691","log2FoldChange: 0.1647050388<br />-log10(padj): 4.792620e-02<br />log10(baseMean): 1.5377923<br />gene: FBgn0086693","log2FoldChange: -0.0088883094<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 3.1192607<br />gene: FBgn0086694","log2FoldChange: -0.2534575661<br />-log10(padj): 4.571600e-01<br />log10(baseMean): 2.5539867<br />gene: FBgn0086695","log2FoldChange: 0.0865594352<br />-log10(padj): 5.511886e-02<br />log10(baseMean): 2.9112901<br />gene: FBgn0086697","log2FoldChange: 0.0583822700<br />-log10(padj): 3.197179e-02<br />log10(baseMean): 2.6026303<br />gene: FBgn0086698","log2FoldChange: 0.2264406831<br />-log10(padj): 2.208791e-01<br />log10(baseMean): 2.1467320<br />gene: FBgn0086699","log2FoldChange: 0.7808722767<br />-log10(padj): 2.201481e-01<br />log10(baseMean): 0.8927182<br />gene: FBgn0086704","log2FoldChange: -0.2000504172<br />-log10(padj): 5.267132e-01<br />log10(baseMean): 3.8140286<br />gene: FBgn0086706","log2FoldChange: -0.0897487063<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 3.3457140<br />gene: FBgn0086707","log2FoldChange: -0.5297761553<br />-log10(padj): 1.375710e+00<br />log10(baseMean): 2.3838824<br />gene: FBgn0086708","log2FoldChange: -0.1526103797<br />-log10(padj): 1.341132e-01<br />log10(baseMean): 3.9548900<br />gene: FBgn0086710","log2FoldChange: -0.0362215626<br />-log10(padj): 2.187671e-02<br />log10(baseMean): 2.9294594<br />gene: FBgn0086712","log2FoldChange: -0.0382752377<br />-log10(padj): 2.498915e-02<br />log10(baseMean): 2.9607822<br />gene: FBgn0086736","log2FoldChange: -0.4172212758<br />-log10(padj): 1.791341e+00<br />log10(baseMean): 2.7848066<br />gene: FBgn0086757","log2FoldChange: -0.3970339547<br />-log10(padj): 5.940664e-01<br />log10(baseMean): 3.7433282<br />gene: FBgn0086758","log2FoldChange: 0.0591507216<br />-log10(padj): 3.653875e-02<br />log10(baseMean): 2.8878857<br />gene: FBgn0086768","log2FoldChange: -0.6385832690<br />-log10(padj): 1.399139e-01<br />log10(baseMean): 1.0242416<br />gene: FBgn0086778","log2FoldChange: 0.0072886794<br />-log10(padj): 3.692912e-03<br />log10(baseMean): 3.1335248<br />gene: FBgn0086779","log2FoldChange: -0.3987760680<br />-log10(padj): 2.611338e-01<br />log10(baseMean): 2.2973135<br />gene: FBgn0086782","log2FoldChange: -0.5392927100<br />-log10(padj): 2.558107e-01<br />log10(baseMean): 1.3837658<br />gene: FBgn0086783","log2FoldChange: -0.0971748726<br />-log10(padj): 9.036511e-02<br />log10(baseMean): 2.9627153<br />gene: FBgn0086784","log2FoldChange: 0.0563677264<br />-log10(padj): 3.802088e-02<br />log10(baseMean): 2.5644016<br />gene: FBgn0086785","log2FoldChange: 0.0154816419<br />-log10(padj): 7.066096e-03<br />log10(baseMean): 2.6978570<br />gene: FBgn0086855","log2FoldChange: -0.0067156051<br />-log10(padj): 1.967102e-03<br />log10(baseMean): 2.0992939<br />gene: FBgn0086856","log2FoldChange: 0.0669486355<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.8589828<br />gene: FBgn0086895","log2FoldChange: -0.2072712632<br />-log10(padj): 3.810279e-01<br />log10(baseMean): 3.8580484<br />gene: FBgn0086897","log2FoldChange: 0.2428605405<br />-log10(padj): 5.532631e-02<br />log10(baseMean): 1.2893749<br />gene: FBgn0086898","log2FoldChange: 0.1411745508<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.5759478<br />gene: FBgn0086899","log2FoldChange: 0.1942462186<br />-log10(padj): 2.927383e-01<br />log10(baseMean): 3.6391925<br />gene: FBgn0086901","log2FoldChange: -0.0918820566<br />-log10(padj): 5.794697e-02<br />log10(baseMean): 4.1205618<br />gene: FBgn0086902","log2FoldChange: -0.0217550352<br />-log10(padj): 1.353168e-02<br />log10(baseMean): 3.9454499<br />gene: FBgn0086904","log2FoldChange: 0.7670839885<br />-log10(padj): 4.868767e+00<br />log10(baseMean): 3.0523579<br />gene: FBgn0086906","log2FoldChange: 0.0301008625<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 3.1490616<br />gene: FBgn0086908","log2FoldChange: 0.0677774692<br />-log10(padj): 3.738336e-02<br />log10(baseMean): 2.3306261<br />gene: FBgn0086909","log2FoldChange: 1.8074466870<br />-log10(padj): 1.096546e+01<br />log10(baseMean): 1.9398606<br />gene: FBgn0086910","log2FoldChange: 0.2014587055<br />-log10(padj): 4.358114e-02<br />log10(baseMean): 2.4704714<br />gene: FBgn0086911","log2FoldChange: -0.2230360238<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 2.5701785<br />gene: FBgn0086912","log2FoldChange: -0.1628529933<br />-log10(padj): 3.245984e-02<br />log10(baseMean): 2.6387313<br />gene: FBgn0087002","log2FoldChange: 1.1234547341<br />-log10(padj): 4.565804e+00<br />log10(baseMean): 2.1247427<br />gene: FBgn0087007","log2FoldChange: -0.1374124073<br />-log10(padj): 1.155972e-01<br />log10(baseMean): 3.6443476<br />gene: FBgn0087008","log2FoldChange: -0.1325886838<br />-log10(padj): 2.187149e-01<br />log10(baseMean): 3.9960935<br />gene: FBgn0087013","log2FoldChange: 0.0684863858<br />-log10(padj): 3.520160e-02<br />log10(baseMean): 2.4574392<br />gene: FBgn0087021","log2FoldChange: -0.1896281218<br />-log10(padj): 1.542928e-01<br />log10(baseMean): 3.9588397<br />gene: FBgn0087035","log2FoldChange: 0.0489028849<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 2.4783501<br />gene: FBgn0087039","log2FoldChange: 0.1896378927<br />-log10(padj): 1.528909e-01<br />log10(baseMean): 3.3037612<br />gene: FBgn0243505","log2FoldChange: -0.0577786659<br />-log10(padj): 4.501543e-02<br />log10(baseMean): 3.1307285<br />gene: FBgn0243511","log2FoldChange: -0.4421803098<br />-log10(padj): 1.467019e+00<br />log10(baseMean): 3.3097835<br />gene: FBgn0243512","log2FoldChange: 0.1634520686<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 1.5142892<br />gene: FBgn0243513","log2FoldChange: 0.0235387184<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 3.7807762<br />gene: FBgn0243514","log2FoldChange: -0.1315242835<br />-log10(padj): 1.637484e-01<br />log10(baseMean): 3.1492181<br />gene: FBgn0243516","log2FoldChange: -0.1431649362<br />-log10(padj): 2.010943e-01<br />log10(baseMean): 2.6299678<br />gene: FBgn0243517","log2FoldChange: -0.2877053766<br />-log10(padj): 1.279208e+00<br />log10(baseMean): 3.0571643<br />gene: FBgn0250732","log2FoldChange: 0.2521309540<br />-log10(padj): 4.853377e-01<br />log10(baseMean): 3.2067220<br />gene: FBgn0250746","log2FoldChange: -0.3061027767<br />-log10(padj): 1.263777e+00<br />log10(baseMean): 3.0382370<br />gene: FBgn0250747","log2FoldChange: -0.0387255232<br />-log10(padj): 1.856410e-02<br />log10(baseMean): 3.7984403<br />gene: FBgn0250753","log2FoldChange: -0.0893180534<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 3.7239234<br />gene: FBgn0250754","log2FoldChange: 0.0498425650<br />-log10(padj): 2.216223e-02<br />log10(baseMean): 2.1774038<br />gene: FBgn0250755","log2FoldChange: 0.8958504119<br />-log10(padj): 2.431865e+00<br />log10(baseMean): 1.9133698<br />gene: FBgn0250757","log2FoldChange: 0.4317932645<br />-log10(padj): 1.837391e+00<br />log10(baseMean): 3.1104889<br />gene: FBgn0250785","log2FoldChange: -0.1230942532<br />-log10(padj): 1.489112e-01<br />log10(baseMean): 3.4781239<br />gene: FBgn0250786","log2FoldChange: 0.3032321594<br />-log10(padj): 8.226885e-01<br />log10(baseMean): 4.0649290<br />gene: FBgn0250788","log2FoldChange: 0.2057576890<br />-log10(padj): 3.169683e-01<br />log10(baseMean): 4.3487573<br />gene: FBgn0250789","log2FoldChange: 0.0019514472<br />-log10(padj): 1.734822e-03<br />log10(baseMean): 3.1945855<br />gene: FBgn0250791","log2FoldChange: -0.2053984711<br />-log10(padj): 1.520478e-01<br />log10(baseMean): 3.4915935<br />gene: FBgn0250814","log2FoldChange: 0.1181172256<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 2.2575566<br />gene: FBgn0250816","log2FoldChange: -0.5086525970<br />-log10(padj): 6.813664e-01<br />log10(baseMean): 3.0270887<br />gene: FBgn0250818","log2FoldChange: -0.4793880664<br />-log10(padj): 1.449700e-01<br />log10(baseMean): 1.5394115<br />gene: FBgn0250819","log2FoldChange: -0.0110070346<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.9133724<br />gene: FBgn0250820","log2FoldChange: 0.0805094638<br />-log10(padj): 2.363480e-02<br />log10(baseMean): 1.7378984<br />gene: FBgn0250821","log2FoldChange: -0.0661441632<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 3.3926534<br />gene: FBgn0250823","log2FoldChange: 0.2540226168<br />-log10(padj): 5.636755e-02<br />log10(baseMean): 1.3041640<br />gene: FBgn0250824","log2FoldChange: 1.2325992958<br />-log10(padj): 2.822951e+00<br />log10(baseMean): 1.8822658<br />gene: FBgn0250829","log2FoldChange: -0.2985109261<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 2.1866779<br />gene: FBgn0250830","log2FoldChange: 0.4286963819<br />-log10(padj): 1.914765e+00<br />log10(baseMean): 3.3457288<br />gene: FBgn0250837","log2FoldChange: 0.0131662049<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.1742697<br />gene: FBgn0250838","log2FoldChange: 0.1582615926<br />-log10(padj): 1.997256e-01<br />log10(baseMean): 3.3152016<br />gene: FBgn0250843","log2FoldChange: -0.1431797598<br />-log10(padj): 1.615018e-01<br />log10(baseMean): 4.0289505<br />gene: FBgn0250848","log2FoldChange: 0.0744482742<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.2380620<br />gene: FBgn0250850","log2FoldChange: -0.2096655437<br />-log10(padj): 4.975627e-01<br />log10(baseMean): 3.4531449<br />gene: FBgn0250861","log2FoldChange: 0.8442043936<br />-log10(padj): 4.646681e+00<br />log10(baseMean): 2.6394638<br />gene: FBgn0250867","log2FoldChange: -0.0017597523<br />-log10(padj): 3.176531e-04<br />log10(baseMean): 1.6873848<br />gene: FBgn0250868","log2FoldChange: -0.3366982804<br />-log10(padj): 7.743299e-01<br />log10(baseMean): 2.6565903<br />gene: FBgn0250869","log2FoldChange: -0.6820454596<br />-log10(padj): 1.133596e+00<br />log10(baseMean): 1.8885729<br />gene: FBgn0250870","log2FoldChange: -0.0428191979<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 1.2121792<br />gene: FBgn0250871","log2FoldChange: 0.0694340436<br />-log10(padj): 3.770620e-02<br />log10(baseMean): 3.3722502<br />gene: FBgn0250874","log2FoldChange: 0.5043201079<br />-log10(padj): 1.437043e+00<br />log10(baseMean): 2.4836001<br />gene: FBgn0250876","log2FoldChange: 1.8824685958<br />-log10(padj): 5.140899e+00<br />log10(baseMean): 1.4808566<br />gene: FBgn0250904","log2FoldChange: 0.5789009842<br />-log10(padj): 2.706395e+00<br />log10(baseMean): 3.4220159<br />gene: FBgn0250906","log2FoldChange: -0.1611012995<br />-log10(padj): 1.125750e-01<br />log10(baseMean): 2.5848441<br />gene: FBgn0250907","log2FoldChange: 0.2042021120<br />-log10(padj): 3.030837e-01<br />log10(baseMean): 2.5609953<br />gene: FBgn0259100","log2FoldChange: -0.1656866797<br />-log10(padj): 1.891881e-02<br />log10(baseMean): 0.8498590<br />gene: FBgn0259101","log2FoldChange: -0.2573546933<br />-log10(padj): 2.942522e-01<br />log10(baseMean): 2.5302022<br />gene: FBgn0259108","log2FoldChange: 0.5065520459<br />-log10(padj): 1.083796e+00<br />log10(baseMean): 2.1049076<br />gene: FBgn0259109","log2FoldChange: -0.5138721583<br />-log10(padj): 2.676539e+00<br />log10(baseMean): 2.8156406<br />gene: FBgn0259110","log2FoldChange: 0.6391603430<br />-log10(padj): 2.777385e+00<br />log10(baseMean): 3.3695566<br />gene: FBgn0259111","log2FoldChange: 0.2426897686<br />-log10(padj): 8.340349e-02<br />log10(baseMean): 1.6514395<br />gene: FBgn0259112","log2FoldChange: -0.2500878949<br />-log10(padj): 8.378512e-01<br />log10(baseMean): 3.2090319<br />gene: FBgn0259113","log2FoldChange: -0.0308310489<br />-log10(padj): 2.703040e-02<br />log10(baseMean): 3.3601330<br />gene: FBgn0259139","log2FoldChange: 1.4031891456<br />-log10(padj): 8.453524e-01<br />log10(baseMean): 1.0080439<br />gene: FBgn0259140","log2FoldChange: 0.1060068388<br />-log10(padj): 1.036168e-01<br />log10(baseMean): 2.5796116<br />gene: FBgn0259142","log2FoldChange: -0.3546000614<br />-log10(padj): 6.196999e-01<br />log10(baseMean): 2.6903744<br />gene: FBgn0259143","log2FoldChange: -0.7480249010<br />-log10(padj): 8.243211e-01<br />log10(baseMean): 1.9510909<br />gene: FBgn0259144","log2FoldChange: -0.6960395317<br />-log10(padj): 3.794101e-01<br />log10(baseMean): 1.2403474<br />gene: FBgn0259150","log2FoldChange: 1.7319081969<br />-log10(padj): 8.113146e-01<br />log10(baseMean): 0.8628373<br />gene: FBgn0259151","log2FoldChange: -0.0174257115<br />-log10(padj): 6.591161e-03<br />log10(baseMean): 2.6931818<br />gene: FBgn0259152","log2FoldChange: 0.0662901875<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 1.9962910<br />gene: FBgn0259163","log2FoldChange: -0.2127463629<br />-log10(padj): 9.970644e-02<br />log10(baseMean): 1.7764806<br />gene: FBgn0259164","log2FoldChange: 0.3716394376<br />-log10(padj): 7.093263e-01<br />log10(baseMean): 2.8699877<br />gene: FBgn0259166","log2FoldChange: -0.5289543976<br />-log10(padj): 9.247321e-01<br />log10(baseMean): 2.0325497<br />gene: FBgn0259167","log2FoldChange: -0.0624214430<br />-log10(padj): 4.501290e-02<br />log10(baseMean): 3.4185366<br />gene: FBgn0259168","log2FoldChange: -0.0014462461<br />-log10(padj): 1.175523e-03<br />log10(baseMean): 2.8127793<br />gene: FBgn0259170","log2FoldChange: 0.2592212003<br />-log10(padj): 3.494344e-01<br />log10(baseMean): 2.9032882<br />gene: FBgn0259171","log2FoldChange: 0.2885956520<br />-log10(padj): 5.349869e-01<br />log10(baseMean): 2.3371952<br />gene: FBgn0259173","log2FoldChange: -0.1848219387<br />-log10(padj): 4.770010e-01<br />log10(baseMean): 3.4343730<br />gene: FBgn0259174","log2FoldChange: 0.1610495267<br />-log10(padj): 9.921925e-02<br />log10(baseMean): 2.0010442<br />gene: FBgn0259175","log2FoldChange: -0.1157895616<br />-log10(padj): 1.307513e-01<br />log10(baseMean): 3.7946969<br />gene: FBgn0259176","log2FoldChange: -0.2553180440<br />-log10(padj): 1.799032e-01<br />log10(baseMean): 2.2528560<br />gene: FBgn0259178","log2FoldChange: -1.5983807717<br />-log10(padj): 3.904677e-01<br />log10(baseMean): 1.0026871<br />gene: FBgn0259183","log2FoldChange: -0.7198761888<br />-log10(padj): 1.395516e-01<br />log10(baseMean): 1.2464266<br />gene: FBgn0259184","log2FoldChange: -0.9592942622<br />-log10(padj): 1.255543e+00<br />log10(baseMean): 1.4662146<br />gene: FBgn0259188","log2FoldChange: 0.1557246495<br />-log10(padj): 5.118578e-02<br />log10(baseMean): 1.5835311<br />gene: FBgn0259209","log2FoldChange: 0.3070646629<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 1.4431018<br />gene: FBgn0259211","log2FoldChange: -0.1082210808<br />-log10(padj): 1.359133e-01<br />log10(baseMean): 3.5263222<br />gene: FBgn0259212","log2FoldChange: -0.2010875372<br />-log10(padj): 1.207488e-01<br />log10(baseMean): 3.4773812<br />gene: FBgn0259214","log2FoldChange: 0.0439755684<br />-log10(padj): 4.731748e-03<br />log10(baseMean): 1.0403139<br />gene: FBgn0259215","log2FoldChange: 0.1987046606<br />-log10(padj): 2.634694e-02<br />log10(baseMean): 1.2136771<br />gene: FBgn0259216","log2FoldChange: 0.3802805882<br />-log10(padj): 2.045996e+00<br />log10(baseMean): 3.8624389<br />gene: FBgn0259217","log2FoldChange: 0.0636252081<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.6452566<br />gene: FBgn0259220","log2FoldChange: -0.0436658657<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.5292197<br />gene: FBgn0259221","log2FoldChange: 1.1343254554<br />-log10(padj): 5.833167e+00<br />log10(baseMean): 2.4013098<br />gene: FBgn0259224","log2FoldChange: -0.0540631779<br />-log10(padj): 2.349338e-02<br />log10(baseMean): 2.2493024<br />gene: FBgn0259225","log2FoldChange: -0.8591596253<br />-log10(padj): 6.960757e-01<br />log10(baseMean): 1.3752757<br />gene: FBgn0259226","log2FoldChange: 0.0231388399<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.5416168<br />gene: FBgn0259227","log2FoldChange: -0.2463434906<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.8033347<br />gene: FBgn0259228","log2FoldChange: -0.0841887727<br />-log10(padj): 4.705987e-02<br />log10(baseMean): 2.5257886<br />gene: FBgn0259233","log2FoldChange: -0.0933866156<br />-log10(padj): 1.074640e-01<br />log10(baseMean): 2.8840868<br />gene: FBgn0259234","log2FoldChange: -2.9064152501<br />-log10(padj): 1.515343e+00<br />log10(baseMean): 0.8003000<br />gene: FBgn0259236","log2FoldChange: 0.1475551322<br />-log10(padj): 6.533645e-02<br />log10(baseMean): 2.5281458<br />gene: FBgn0259238","log2FoldChange: 0.1286377800<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.4477916<br />gene: FBgn0259240","log2FoldChange: -0.5772631592<br />-log10(padj): 1.771833e-01<br />log10(baseMean): 1.1090367<br />gene: FBgn0259242","log2FoldChange: -0.1721326452<br />-log10(padj): 3.090305e-01<br />log10(baseMean): 2.8492511<br />gene: FBgn0259243","log2FoldChange: 0.9353217746<br />-log10(padj): 2.257248e+00<br />log10(baseMean): 2.5395735<br />gene: FBgn0259246","log2FoldChange: -0.0678124925<br />-log10(padj): 4.202573e-02<br />log10(baseMean): 2.9385010<br />gene: FBgn0259247","log2FoldChange: -0.1784942170<br />-log10(padj): 2.395894e-01<br />log10(baseMean): 3.5316692<br />gene: FBgn0259481","log2FoldChange: -0.1643422601<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.2034751<br />gene: FBgn0259482","log2FoldChange: 0.0947362271<br />-log10(padj): 1.313744e-01<br />log10(baseMean): 3.1154665<br />gene: FBgn0259483","log2FoldChange: -0.2714777311<br />-log10(padj): 4.922819e-01<br />log10(baseMean): 2.5864035<br />gene: FBgn0259676","log2FoldChange: -0.0727260071<br />-log10(padj): 2.887343e-02<br />log10(baseMean): 2.0113366<br />gene: FBgn0259677","log2FoldChange: -0.4539315653<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 0.9627872<br />gene: FBgn0259678","log2FoldChange: -0.4256623441<br />-log10(padj): 1.114791e-01<br />log10(baseMean): 1.1579526<br />gene: FBgn0259682","log2FoldChange: 0.7019793951<br />-log10(padj): 1.036623e+00<br />log10(baseMean): 2.0605502<br />gene: FBgn0259683","log2FoldChange: -0.0192898730<br />-log10(padj): 9.475185e-03<br />log10(baseMean): 2.5744919<br />gene: FBgn0259685","log2FoldChange: -0.7649837706<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 0.9826452<br />gene: FBgn0259700","log2FoldChange: -0.1866233267<br />-log10(padj): 1.744222e-01<br />log10(baseMean): 2.4237359<br />gene: FBgn0259704","log2FoldChange: -0.2544405829<br />-log10(padj): 1.638394e-01<br />log10(baseMean): 1.9592691<br />gene: FBgn0259705","log2FoldChange: -0.1365476455<br />-log10(padj): 2.065888e-02<br />log10(baseMean): 1.0797408<br />gene: FBgn0259707","log2FoldChange: -0.2680808188<br />-log10(padj): 6.493917e-02<br />log10(baseMean): 1.3595990<br />gene: FBgn0259710","log2FoldChange: 0.1948680098<br />-log10(padj): 8.004060e-02<br />log10(baseMean): 2.0499916<br />gene: FBgn0259711","log2FoldChange: -0.5609416927<br />-log10(padj): 2.582533e-01<br />log10(baseMean): 1.4914085<br />gene: FBgn0259712","log2FoldChange: -1.0292455972<br />-log10(padj): 3.339965e+00<br />log10(baseMean): 2.0682651<br />gene: FBgn0259714","log2FoldChange: -1.7844407342<br />-log10(padj): 8.273456e+00<br />log10(baseMean): 2.9056012<br />gene: FBgn0259715","log2FoldChange: -0.8694718018<br />-log10(padj): 1.854187e-01<br />log10(baseMean): 0.7955084<br />gene: FBgn0259716","log2FoldChange: 0.2033962222<br />-log10(padj): 8.647306e-02<br />log10(baseMean): 1.7855070<br />gene: FBgn0259728","log2FoldChange: 0.6862167755<br />-log10(padj): 2.052620e+00<br />log10(baseMean): 2.4276096<br />gene: FBgn0259734","log2FoldChange: -0.1111693530<br />-log10(padj): 7.025723e-02<br />log10(baseMean): 3.9516825<br />gene: FBgn0259735","log2FoldChange: -0.3748099575<br />-log10(padj): 5.209222e-01<br />log10(baseMean): 2.3605594<br />gene: FBgn0259736","log2FoldChange: 0.5691867735<br />-log10(padj): 1.330892e-01<br />log10(baseMean): 1.3358421<br />gene: FBgn0259739","log2FoldChange: 0.9295138043<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 1.6676604<br />gene: FBgn0259740","log2FoldChange: 1.3061139905<br />-log10(padj): 2.706395e+00<br />log10(baseMean): 1.5206449<br />gene: FBgn0259741","log2FoldChange: 0.0109236043<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.2588851<br />gene: FBgn0259742","log2FoldChange: 0.0409846309<br />-log10(padj): 2.936411e-02<br />log10(baseMean): 3.3983835<br />gene: FBgn0259743","log2FoldChange: -0.1414679192<br />-log10(padj): 1.669236e-02<br />log10(baseMean): 0.8791587<br />gene: FBgn0259744","log2FoldChange: 0.0083358715<br />-log10(padj): 4.311641e-03<br />log10(baseMean): 3.1855241<br />gene: FBgn0259745","log2FoldChange: -0.6324693894<br />-log10(padj): 4.153561e+00<br />log10(baseMean): 3.1450031<br />gene: FBgn0259749","log2FoldChange: 0.0267048636<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 2.3739334<br />gene: FBgn0259784","log2FoldChange: -0.1287445183<br />-log10(padj): 1.821376e-01<br />log10(baseMean): 3.3852528<br />gene: FBgn0259785","log2FoldChange: 0.1851329311<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 1.4767366<br />gene: FBgn0259789","log2FoldChange: 0.1265965362<br />-log10(padj): 1.147664e-01<br />log10(baseMean): 2.4223459<br />gene: FBgn0259791","log2FoldChange: -0.1764740805<br />-log10(padj): 7.307603e-02<br />log10(baseMean): 1.7308315<br />gene: FBgn0259818","log2FoldChange: -0.0889327085<br />-log10(padj): 1.640678e-02<br />log10(baseMean): 1.3925590<br />gene: FBgn0259823","log2FoldChange: -0.0984761508<br />-log10(padj): 1.105723e-01<br />log10(baseMean): 3.1021888<br />gene: FBgn0259824","log2FoldChange: -0.2493149525<br />-log10(padj): 8.129830e-02<br />log10(baseMean): 1.6143714<br />gene: FBgn0259832","log2FoldChange: -0.2575259633<br />-log10(padj): 6.202592e-01<br />log10(baseMean): 2.5274810<br />gene: FBgn0259834","log2FoldChange: -0.0047467832<br />-log10(padj): 3.666988e-03<br />log10(baseMean): 3.2218901<br />gene: FBgn0259876","log2FoldChange: -0.2847598474<br />-log10(padj): 7.677419e-02<br />log10(baseMean): 1.3178587<br />gene: FBgn0259878","log2FoldChange: 0.0702703097<br />-log10(padj): 4.633512e-02<br />log10(baseMean): 3.0879442<br />gene: FBgn0259896","log2FoldChange: -0.1064226734<br />-log10(padj): 3.568450e-02<br />log10(baseMean): 1.9719126<br />gene: FBgn0259916","log2FoldChange: 0.0604426850<br />-log10(padj): 1.167738e-02<br />log10(baseMean): 1.4186397<br />gene: FBgn0259918","log2FoldChange: 0.4578373780<br />-log10(padj): 5.093919e-01<br />log10(baseMean): 1.8242789<br />gene: FBgn0259923","log2FoldChange: 0.4038671363<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 0.8447175<br />gene: FBgn0259926","log2FoldChange: 1.2184384509<br />-log10(padj): 3.030651e+00<br />log10(baseMean): 1.6522300<br />gene: FBgn0259935","log2FoldChange: -0.1587907308<br />-log10(padj): 8.049433e-02<br />log10(baseMean): 2.2892237<br />gene: FBgn0259936","log2FoldChange: 0.0348930790<br />-log10(padj): 2.474879e-02<br />log10(baseMean): 3.5598464<br />gene: FBgn0259937","log2FoldChange: -0.2999197065<br />-log10(padj): 6.288426e-01<br />log10(baseMean): 3.0276315<br />gene: FBgn0259938","log2FoldChange: 0.3495809129<br />-log10(padj): 2.170299e-01<br />log10(baseMean): 1.6442505<br />gene: FBgn0259977","log2FoldChange: -0.0510075885<br />-log10(padj): 3.211539e-02<br />log10(baseMean): 3.0135621<br />gene: FBgn0259978","log2FoldChange: -0.2064472522<br />-log10(padj): 2.556949e-01<br />log10(baseMean): 3.0556317<br />gene: FBgn0259979","log2FoldChange: 0.0957536306<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.1575958<br />gene: FBgn0259982","log2FoldChange: 0.2665634157<br />-log10(padj): 6.406388e-01<br />log10(baseMean): 3.8426190<br />gene: FBgn0259984","log2FoldChange: -0.0674502674<br />-log10(padj): 2.789379e-02<br />log10(baseMean): 2.1631807<br />gene: FBgn0259985","log2FoldChange: 0.0027679331<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 1.4789074<br />gene: FBgn0259990","log2FoldChange: -1.0350687528<br />-log10(padj): 1.327153e-01<br />log10(baseMean): 0.8713388<br />gene: FBgn0259991","log2FoldChange: -0.0497579715<br />-log10(padj): 1.650947e-02<br />log10(baseMean): 1.9141663<br />gene: FBgn0259993","log2FoldChange: 0.0895670769<br />-log10(padj): 2.814148e-02<br />log10(baseMean): 2.1710391<br />gene: FBgn0259994","log2FoldChange: -0.0863248556<br />-log10(padj): 9.921925e-02<br />log10(baseMean): 3.5437451<br />gene: FBgn0260003","log2FoldChange: 0.0186104829<br />-log10(padj): 6.069662e-03<br />log10(baseMean): 2.1228270<br />gene: FBgn0260004","log2FoldChange: -0.5082555716<br />-log10(padj): 4.661415e-01<br />log10(baseMean): 1.6582335<br />gene: FBgn0260005","log2FoldChange: 0.2243632591<br />-log10(padj): 1.805616e-01<br />log10(baseMean): 2.2815544<br />gene: FBgn0260006","log2FoldChange: 0.1328591164<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.5458899<br />gene: FBgn0260010","log2FoldChange: 2.3884003586<br />-log10(padj): 1.586979e+01<br />log10(baseMean): 1.9507510<br />gene: FBgn0260011","log2FoldChange: -0.1188894212<br />-log10(padj): 1.753139e-01<br />log10(baseMean): 3.2774133<br />gene: FBgn0260012","log2FoldChange: 0.2350654045<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 1.3326167<br />gene: FBgn0260026","log2FoldChange: 0.5325860227<br />-log10(padj): 1.171483e-01<br />log10(baseMean): 1.0908395<br />gene: FBgn0260027","log2FoldChange: 0.1704347272<br />-log10(padj): 2.762079e-01<br />log10(baseMean): 3.5196564<br />gene: FBgn0260049","log2FoldChange: 0.0657166744<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 2.6438629<br />gene: FBgn0260243","log2FoldChange: -0.1847832312<br />-log10(padj): 2.763200e-02<br />log10(baseMean): 1.1141638<br />gene: FBgn0260386","log2FoldChange: -0.1162437542<br />-log10(padj): 7.689075e-02<br />log10(baseMean): 2.6964598<br />gene: FBgn0260388","log2FoldChange: -0.1461628010<br />-log10(padj): 1.836853e-01<br />log10(baseMean): 2.7017214<br />gene: FBgn0260397","log2FoldChange: 0.0771972816<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.6785602<br />gene: FBgn0260399","log2FoldChange: 0.3454617469<br />-log10(padj): 6.700448e-01<br />log10(baseMean): 2.3270177<br />gene: FBgn0260400","log2FoldChange: -0.1449600635<br />-log10(padj): 7.115813e-02<br />log10(baseMean): 2.2639731<br />gene: FBgn0260407","log2FoldChange: -1.3813666703<br />-log10(padj): 2.078683e+00<br />log10(baseMean): 1.8346561<br />gene: FBgn0260429","log2FoldChange: 0.6852445869<br />-log10(padj): 4.961244e-01<br />log10(baseMean): 1.4648520<br />gene: FBgn0260431","log2FoldChange: 0.0219707592<br />-log10(padj): 2.009260e-02<br />log10(baseMean): 3.8098850<br />gene: FBgn0260439","log2FoldChange: -0.3284257108<br />-log10(padj): 1.395805e-01<br />log10(baseMean): 1.4813227<br />gene: FBgn0260440","log2FoldChange: -0.1891713146<br />-log10(padj): 2.262589e-01<br />log10(baseMean): 4.0003197<br />gene: FBgn0260441","log2FoldChange: -0.0826691217<br />-log10(padj): 6.532694e-02<br />log10(baseMean): 3.6027924<br />gene: FBgn0260442","log2FoldChange: 0.0522866221<br />-log10(padj): 2.774356e-02<br />log10(baseMean): 2.6806073<br />gene: FBgn0260444","log2FoldChange: 0.4662234926<br />-log10(padj): 1.260715e+00<br />log10(baseMean): 2.8724634<br />gene: FBgn0260450","log2FoldChange: -0.2135349858<br />-log10(padj): 2.466545e-01<br />log10(baseMean): 2.8132207<br />gene: FBgn0260456","log2FoldChange: -0.0973041489<br />-log10(padj): 8.547716e-02<br />log10(baseMean): 3.3498106<br />gene: FBgn0260457","log2FoldChange: 0.2468568150<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.5653993<br />gene: FBgn0260460","log2FoldChange: -0.0032324635<br />-log10(padj): 2.725201e-03<br />log10(baseMean): 3.7496317<br />gene: FBgn0260462","log2FoldChange: 0.4529987881<br />-log10(padj): 5.635371e-02<br />log10(baseMean): 1.9018694<br />gene: FBgn0260463","log2FoldChange: 0.0198214146<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 3.1475552<br />gene: FBgn0260465","log2FoldChange: 0.4819243790<br />-log10(padj): 1.418290e-01<br />log10(baseMean): 2.3736791<br />gene: FBgn0260475","log2FoldChange: -0.1707814155<br />-log10(padj): 1.767870e-02<br />log10(baseMean): 0.8243916<br />gene: FBgn0260477","log2FoldChange: 0.3244452637<br />-log10(padj): 1.244962e-01<br />log10(baseMean): 1.7651871<br />gene: FBgn0260478","log2FoldChange: -0.5268180418<br />-log10(padj): 1.437531e-01<br />log10(baseMean): 1.0953480<br />gene: FBgn0260481","log2FoldChange: 0.6171791852<br />-log10(padj): 6.554094e-01<br />log10(baseMean): 2.2218130<br />gene: FBgn0260484","log2FoldChange: -0.6289993014<br />-log10(padj): 3.325836e+00<br />log10(baseMean): 3.2981679<br />gene: FBgn0260486","log2FoldChange: -0.3115800091<br />-log10(padj): 1.281924e+00<br />log10(baseMean): 3.2979891<br />gene: FBgn0260632","log2FoldChange: 0.0243150075<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 3.3376829<br />gene: FBgn0260634","log2FoldChange: 0.0394290830<br />-log10(padj): 4.515497e-02<br />log10(baseMean): 3.9499092<br />gene: FBgn0260635","log2FoldChange: 0.1904238214<br />-log10(padj): 4.113010e-01<br />log10(baseMean): 2.9542462<br />gene: FBgn0260639","log2FoldChange: -0.1016326632<br />-log10(padj): 2.197671e-02<br />log10(baseMean): 1.4983837<br />gene: FBgn0260645","log2FoldChange: 0.0217629420<br />-log10(padj): 8.393752e-03<br />log10(baseMean): 2.1924066<br />gene: FBgn0260648","log2FoldChange: -0.0482191332<br />-log10(padj): 5.698177e-03<br />log10(baseMean): 0.9801854<br />gene: FBgn0260653","log2FoldChange: -0.2167668531<br />-log10(padj): 2.986472e-01<br />log10(baseMean): 2.9006544<br />gene: FBgn0260655","log2FoldChange: -0.1162361088<br />-log10(padj): 2.525758e-02<br />log10(baseMean): 1.4092717<br />gene: FBgn0260657","log2FoldChange: -0.3557692309<br />-log10(padj): 1.256470e-01<br />log10(baseMean): 1.7589445<br />gene: FBgn0260658","log2FoldChange: 0.0641634993<br />-log10(padj): 4.548636e-02<br />log10(baseMean): 2.6905534<br />gene: FBgn0260659","log2FoldChange: -0.8150120498<br />-log10(padj): 3.104823e-01<br />log10(baseMean): 1.0308035<br />gene: FBgn0260660","log2FoldChange: -0.1743358076<br />-log10(padj): 1.970482e-02<br />log10(baseMean): 0.9382235<br />gene: FBgn0260720","log2FoldChange: 0.0214533335<br />-log10(padj): 5.938280e-03<br />log10(baseMean): 2.5445619<br />gene: FBgn0260722","log2FoldChange: -0.0942323851<br />-log10(padj): 1.360272e-02<br />log10(baseMean): 1.6107850<br />gene: FBgn0260723","log2FoldChange: -0.1442871715<br />-log10(padj): 1.855676e-01<br />log10(baseMean): 4.0244595<br />gene: FBgn0260724","log2FoldChange: 0.1666675744<br />-log10(padj): 1.437531e-01<br />log10(baseMean): 2.2444764<br />gene: FBgn0260741","log2FoldChange: -0.0008361159<br />-log10(padj): 1.163804e-03<br />log10(baseMean): 3.2181808<br />gene: FBgn0260742","log2FoldChange: -0.1573904298<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.3966095<br />gene: FBgn0260743","log2FoldChange: 0.2024705424<br />-log10(padj): 2.066211e-01<br />log10(baseMean): 2.6977614<br />gene: FBgn0260744","log2FoldChange: -0.5586444975<br />-log10(padj): 3.584842e+00<br />log10(baseMean): 3.7832442<br />gene: FBgn0260745","log2FoldChange: 0.4189729420<br />-log10(padj): 1.124926e+00<br />log10(baseMean): 3.0570398<br />gene: FBgn0260746","log2FoldChange: -0.1426332193<br />-log10(padj): 1.940753e-01<br />log10(baseMean): 3.3557979<br />gene: FBgn0260747","log2FoldChange: -0.2112343141<br />-log10(padj): 8.899297e-01<br />log10(baseMean): 3.6533250<br />gene: FBgn0260748","log2FoldChange: -0.2398628484<br />-log10(padj): 3.158828e-01<br />log10(baseMean): 2.8513120<br />gene: FBgn0260749","log2FoldChange: 0.2406180074<br />-log10(padj): 1.954740e-01<br />log10(baseMean): 2.2645446<br />gene: FBgn0260750","log2FoldChange: 0.8592642314<br />-log10(padj): 7.810866e-01<br />log10(baseMean): 1.3355298<br />gene: FBgn0260753","log2FoldChange: 0.0695624942<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 2.5835891<br />gene: FBgn0260755","log2FoldChange: -0.3048239548<br />-log10(padj): 1.986805e-01<br />log10(baseMean): 1.9017055<br />gene: FBgn0260756","log2FoldChange: -0.1499718928<br />-log10(padj): 1.300740e-01<br />log10(baseMean): 3.0669736<br />gene: FBgn0260757","log2FoldChange: 0.0074672086<br />-log10(padj): 1.399186e-03<br />log10(baseMean): 1.0681353<br />gene: FBgn0260758","log2FoldChange: -0.2879237597<br />-log10(padj): 5.286181e-02<br />log10(baseMean): 1.0110005<br />gene: FBgn0260766","log2FoldChange: 0.1855790745<br />-log10(padj): 2.672681e-01<br />log10(baseMean): 2.4612051<br />gene: FBgn0260767","log2FoldChange: 0.5887233113<br />-log10(padj): 1.796917e+00<br />log10(baseMean): 2.0961654<br />gene: FBgn0260768","log2FoldChange: -0.1297915001<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 2.4104572<br />gene: FBgn0260771","log2FoldChange: 0.2880120346<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.5130255<br />gene: FBgn0260779","log2FoldChange: 0.2204277766<br />-log10(padj): 6.042879e-02<br />log10(baseMean): 1.3237882<br />gene: FBgn0260780","log2FoldChange: 0.1408634236<br />-log10(padj): 2.258934e-01<br />log10(baseMean): 3.0398006<br />gene: FBgn0260789","log2FoldChange: -0.2412469246<br />-log10(padj): 7.769992e-02<br />log10(baseMean): 1.4927109<br />gene: FBgn0260793","log2FoldChange: 0.1068748496<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 3.8150442<br />gene: FBgn0260794","log2FoldChange: 0.8259410492<br />-log10(padj): 2.903845e-01<br />log10(baseMean): 0.9995716<br />gene: FBgn0260795","log2FoldChange: -0.2241579679<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 2.4657765<br />gene: FBgn0260798","log2FoldChange: 0.0766265655<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 3.0275038<br />gene: FBgn0260799","log2FoldChange: 0.2480893814<br />-log10(padj): 3.886434e-01<br />log10(baseMean): 2.4490698<br />gene: FBgn0260817","log2FoldChange: 0.0824113934<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 2.7771957<br />gene: FBgn0260855","log2FoldChange: 0.0388366066<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.4619908<br />gene: FBgn0260856","log2FoldChange: 0.1164607352<br />-log10(padj): 1.078691e-01<br />log10(baseMean): 2.4446416<br />gene: FBgn0260857","log2FoldChange: -0.2154480139<br />-log10(padj): 3.418896e-01<br />log10(baseMean): 2.8573160<br />gene: FBgn0260858","log2FoldChange: 0.1207686499<br />-log10(padj): 6.146884e-02<br />log10(baseMean): 2.1115481<br />gene: FBgn0260859","log2FoldChange: 0.0108395606<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.3402559<br />gene: FBgn0260860","log2FoldChange: 0.0924939624<br />-log10(padj): 5.611394e-02<br />log10(baseMean): 2.3298723<br />gene: FBgn0260861","log2FoldChange: -0.1542890435<br />-log10(padj): 9.463495e-02<br />log10(baseMean): 2.0053371<br />gene: FBgn0260862","log2FoldChange: -0.4853196002<br />-log10(padj): 5.064937e-01<br />log10(baseMean): 1.7566214<br />gene: FBgn0260866","log2FoldChange: 0.2597743252<br />-log10(padj): 3.208521e-01<br />log10(baseMean): 2.2246079<br />gene: FBgn0260874","log2FoldChange: 0.2939575753<br />-log10(padj): 9.412115e-02<br />log10(baseMean): 1.4814713<br />gene: FBgn0260932","log2FoldChange: 0.6142198028<br />-log10(padj): 4.910926e+00<br />log10(baseMean): 2.8738112<br />gene: FBgn0260933","log2FoldChange: -0.2509376274<br />-log10(padj): 8.110079e-01<br />log10(baseMean): 3.7469563<br />gene: FBgn0260934","log2FoldChange: -0.0775059268<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.5819284<br />gene: FBgn0260935","log2FoldChange: -0.3215539834<br />-log10(padj): 1.750342e+00<br />log10(baseMean): 3.4237280<br />gene: FBgn0260936","log2FoldChange: -0.0717644072<br />-log10(padj): 5.385665e-02<br />log10(baseMean): 3.1692536<br />gene: FBgn0260937","log2FoldChange: -0.1794783717<br />-log10(padj): 3.390721e-01<br />log10(baseMean): 3.4369013<br />gene: FBgn0260938","log2FoldChange: 0.1063976375<br />-log10(padj): 8.045554e-02<br />log10(baseMean): 2.7906629<br />gene: FBgn0260939","log2FoldChange: 0.1972239000<br />-log10(padj): 2.227992e-01<br />log10(baseMean): 2.4045913<br />gene: FBgn0260940","log2FoldChange: -0.1736903146<br />-log10(padj): 4.329783e-01<br />log10(baseMean): 3.0628546<br />gene: FBgn0260941","log2FoldChange: 0.1550465635<br />-log10(padj): 1.274255e-01<br />log10(baseMean): 2.8251223<br />gene: FBgn0260944","log2FoldChange: 0.1561656104<br />-log10(padj): 1.717150e-01<br />log10(baseMean): 3.1461505<br />gene: FBgn0260945","log2FoldChange: -0.1805938343<br />-log10(padj): 4.253737e-01<br />log10(baseMean): 3.1220145<br />gene: FBgn0260946","log2FoldChange: -0.5280534339<br />-log10(padj): 9.990498e-01<br />log10(baseMean): 4.3268247<br />gene: FBgn0260952","log2FoldChange: -0.2087276167<br />-log10(padj): 4.101715e-01<br />log10(baseMean): 3.1373849<br />gene: FBgn0260959","log2FoldChange: -0.4589625536<br />-log10(padj): 1.914765e+00<br />log10(baseMean): 3.4811320<br />gene: FBgn0260960","log2FoldChange: -0.1033756507<br />-log10(padj): 1.696066e-01<br />log10(baseMean): 3.4575310<br />gene: FBgn0260962","log2FoldChange: 1.2808801107<br />-log10(padj): 7.629574e-01<br />log10(baseMean): 0.9762685<br />gene: FBgn0260963","log2FoldChange: -0.3644039019<br />-log10(padj): 4.407270e-01<br />log10(baseMean): 2.8877999<br />gene: FBgn0260965","log2FoldChange: -0.9370998973<br />-log10(padj): 4.143318e+00<br />log10(baseMean): 2.8728248<br />gene: FBgn0260966","log2FoldChange: -0.0123194450<br />-log10(padj): 7.543257e-03<br />log10(baseMean): 3.4577620<br />gene: FBgn0260970","log2FoldChange: -0.1241078361<br />-log10(padj): 1.245799e-01<br />log10(baseMean): 2.9036186<br />gene: FBgn0260972","log2FoldChange: -0.0058994145<br />-log10(padj): 3.856315e-03<br />log10(baseMean): 3.0632123<br />gene: FBgn0260985","log2FoldChange: 0.0076195600<br />-log10(padj): 4.079967e-03<br />log10(baseMean): 2.5712300<br />gene: FBgn0260986","log2FoldChange: -0.2117244234<br />-log10(padj): 6.772630e-01<br />log10(baseMean): 3.5934270<br />gene: FBgn0260990","log2FoldChange: 0.0332222545<br />-log10(padj): 2.122202e-02<br />log10(baseMean): 3.3699504<br />gene: FBgn0260991","log2FoldChange: -0.4854051198<br />-log10(padj): 1.991167e+00<br />log10(baseMean): 2.9677351<br />gene: FBgn0260992","log2FoldChange: 1.0727372465<br />-log10(padj): 5.078607e-01<br />log10(baseMean): 1.0504128<br />gene: FBgn0260993","log2FoldChange: 0.1534376770<br />-log10(padj): 2.619073e-01<br />log10(baseMean): 3.1643498<br />gene: FBgn0261004","log2FoldChange: 0.1941514758<br />-log10(padj): 2.683111e-01<br />log10(baseMean): 3.9339760<br />gene: FBgn0261014","log2FoldChange: -1.1431442164<br />-log10(padj): 5.781503e+00<br />log10(baseMean): 2.6380132<br />gene: FBgn0261015","log2FoldChange: -0.0942499633<br />-log10(padj): 2.763200e-02<br />log10(baseMean): 1.7024992<br />gene: FBgn0261016","log2FoldChange: 0.0699172190<br />-log10(padj): 3.750608e-02<br />log10(baseMean): 3.1098811<br />gene: FBgn0261020","log2FoldChange: -0.2258878820<br />-log10(padj): 4.203212e-01<br />log10(baseMean): 3.1493602<br />gene: FBgn0261041","log2FoldChange: -0.1001815823<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 2.4098753<br />gene: FBgn0261049","log2FoldChange: 0.5905961274<br />-log10(padj): 1.348044e+00<br />log10(baseMean): 1.9819978<br />gene: FBgn0261053","log2FoldChange: -0.1981958524<br />-log10(padj): 4.749053e-01<br />log10(baseMean): 2.8661384<br />gene: FBgn0261064","log2FoldChange: 0.0453799455<br />-log10(padj): 4.130831e-02<br />log10(baseMean): 2.8701010<br />gene: FBgn0261065","log2FoldChange: -0.3737230053<br />-log10(padj): 6.772263e-01<br />log10(baseMean): 2.2941687<br />gene: FBgn0261067","log2FoldChange: 0.1540299144<br />-log10(padj): 9.901479e-02<br />log10(baseMean): 2.6055905<br />gene: FBgn0261068","log2FoldChange: -0.1091467398<br />-log10(padj): 1.591860e-02<br />log10(baseMean): 1.0077539<br />gene: FBgn0261085","log2FoldChange: -0.3397190523<br />-log10(padj): 8.582465e-02<br />log10(baseMean): 1.1787630<br />gene: FBgn0261086","log2FoldChange: -0.0083136992<br />-log10(padj): 1.761089e-03<br />log10(baseMean): 1.3024372<br />gene: FBgn0261089","log2FoldChange: -0.3346192397<br />-log10(padj): 4.923107e-02<br />log10(baseMean): 1.0060404<br />gene: FBgn0261090","log2FoldChange: -0.0188311264<br />-log10(padj): 8.835079e-03<br />log10(baseMean): 2.7970031<br />gene: FBgn0261108","log2FoldChange: 0.5922979446<br />-log10(padj): 2.467643e+00<br />log10(baseMean): 2.5598556<br />gene: FBgn0261109","log2FoldChange: -0.0111071943<br />-log10(padj): 4.116838e-03<br />log10(baseMean): 2.3392647<br />gene: FBgn0261111","log2FoldChange: 0.2232772011<br />-log10(padj): 3.886434e-01<br />log10(baseMean): 2.6039181<br />gene: FBgn0261112","log2FoldChange: 0.1570590090<br />-log10(padj): 2.133430e-01<br />log10(baseMean): 4.1356836<br />gene: FBgn0261113","log2FoldChange: -0.1111150607<br />-log10(padj): 1.140666e-01<br />log10(baseMean): 3.0992612<br />gene: FBgn0261119","log2FoldChange: 0.7800846061<br />-log10(padj): 7.858370e+00<br />log10(baseMean): 2.9602850<br />gene: FBgn0261238","log2FoldChange: -0.1206439992<br />-log10(padj): 1.229447e-01<br />log10(baseMean): 2.8101926<br />gene: FBgn0261239","log2FoldChange: -0.0960790350<br />-log10(padj): 9.795206e-02<br />log10(baseMean): 3.1585096<br />gene: FBgn0261241","log2FoldChange: 0.0725814164<br />-log10(padj): 8.552398e-02<br />log10(baseMean): 3.6009015<br />gene: FBgn0261243","log2FoldChange: 0.2311477918<br />-log10(padj): 5.027731e-01<br />log10(baseMean): 3.1678018<br />gene: FBgn0261244","log2FoldChange: -0.2542667266<br />-log10(padj): 5.688062e-02<br />log10(baseMean): 1.1553687<br />gene: FBgn0261245","log2FoldChange: 0.5816329067<br />-log10(padj): 2.529423e+00<br />log10(baseMean): 3.2990806<br />gene: FBgn0261258","log2FoldChange: -0.2791651444<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 2.6187364<br />gene: FBgn0261259","log2FoldChange: 0.2287772979<br />-log10(padj): 4.883024e-01<br />log10(baseMean): 3.8039219<br />gene: FBgn0261260","log2FoldChange: -0.3626649679<br />-log10(padj): 1.263777e+00<br />log10(baseMean): 3.0673280<br />gene: FBgn0261261","log2FoldChange: 0.2471812083<br />-log10(padj): 7.161981e-01<br />log10(baseMean): 3.7291049<br />gene: FBgn0261263","log2FoldChange: -0.1415730446<br />-log10(padj): 7.502956e-02<br />log10(baseMean): 2.0525076<br />gene: FBgn0261266","log2FoldChange: -0.0934728772<br />-log10(padj): 8.987888e-02<br />log10(baseMean): 3.3046127<br />gene: FBgn0261268","log2FoldChange: 0.3104265052<br />-log10(padj): 2.823653e-01<br />log10(baseMean): 2.3337182<br />gene: FBgn0261269","log2FoldChange: -0.1926656338<br />-log10(padj): 2.471119e-01<br />log10(baseMean): 3.3154486<br />gene: FBgn0261270","log2FoldChange: -0.1711638494<br />-log10(padj): 3.229774e-01<br />log10(baseMean): 2.9151006<br />gene: FBgn0261274","log2FoldChange: 0.1252046011<br />-log10(padj): 2.844502e-01<br />log10(baseMean): 3.4471548<br />gene: FBgn0261276","log2FoldChange: 0.9044669528<br />-log10(padj): 3.370556e-01<br />log10(baseMean): 0.9621546<br />gene: FBgn0261277","log2FoldChange: 0.5011113339<br />-log10(padj): 4.761487e+00<br />log10(baseMean): 2.9996546<br />gene: FBgn0261278","log2FoldChange: 0.0349543608<br />-log10(padj): 2.719368e-02<br />log10(baseMean): 2.9297868<br />gene: FBgn0261279","log2FoldChange: -0.2369453963<br />-log10(padj): 4.094269e-01<br />log10(baseMean): 4.2208248<br />gene: FBgn0261283","log2FoldChange: 2.3108286695<br />-log10(padj): 1.248896e+01<br />log10(baseMean): 1.7076881<br />gene: FBgn0261284","log2FoldChange: -0.4343465166<br />-log10(padj): 1.348269e+00<br />log10(baseMean): 2.6350537<br />gene: FBgn0261285","log2FoldChange: -0.1903070458<br />-log10(padj): 2.326510e-01<br />log10(baseMean): 2.9569852<br />gene: FBgn0261286","log2FoldChange: -0.0906383651<br />-log10(padj): 9.466923e-03<br />log10(baseMean): 0.9748467<br />gene: FBgn0261287","log2FoldChange: -0.2247828178<br />-log10(padj): 4.640179e-02<br />log10(baseMean): 1.2821980<br />gene: FBgn0261349","log2FoldChange: 0.9755149564<br />-log10(padj): 3.237581e-02<br />log10(baseMean): 1.1435923<br />gene: FBgn0261359","log2FoldChange: 1.5196736298<br />-log10(padj): 1.415351e+01<br />log10(baseMean): 3.7631622<br />gene: FBgn0261362","log2FoldChange: -0.1212461946<br />-log10(padj): 7.802744e-02<br />log10(baseMean): 2.7915248<br />gene: FBgn0261363","log2FoldChange: -0.2673808505<br />-log10(padj): 1.032628e-01<br />log10(baseMean): 1.6419954<br />gene: FBgn0261373","log2FoldChange: -0.0686563358<br />-log10(padj): 4.442824e-02<br />log10(baseMean): 2.5270715<br />gene: FBgn0261380","log2FoldChange: -0.2208570709<br />-log10(padj): 4.896415e-01<br />log10(baseMean): 3.0580276<br />gene: FBgn0261383","log2FoldChange: 0.0969543588<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.6509936<br />gene: FBgn0261385","log2FoldChange: -0.0022057437<br />-log10(padj): 1.213041e-03<br />log10(baseMean): 3.0119696<br />gene: FBgn0261387","log2FoldChange: 1.1418465062<br />-log10(padj): 6.502280e-01<br />log10(baseMean): 1.0889158<br />gene: FBgn0261388","log2FoldChange: 0.1901999534<br />-log10(padj): 2.761376e-01<br />log10(baseMean): 3.2348501<br />gene: FBgn0261394","log2FoldChange: 0.1867548280<br />-log10(padj): 5.490181e-01<br />log10(baseMean): 3.3362123<br />gene: FBgn0261396","log2FoldChange: 0.0318198369<br />-log10(padj): 2.452371e-02<br />log10(baseMean): 3.2525050<br />gene: FBgn0261397","log2FoldChange: -0.0556731204<br />-log10(padj): 1.045264e-02<br />log10(baseMean): 3.2533403<br />gene: FBgn0261403","log2FoldChange: 0.5099061940<br />-log10(padj): 1.182302e-01<br />log10(baseMean): 0.9810496<br />gene: FBgn0261429","log2FoldChange: -1.1181830155<br />-log10(padj): 5.247652e-01<br />log10(baseMean): 1.0359563<br />gene: FBgn0261430","log2FoldChange: -0.0761881165<br />-log10(padj): 6.459380e-02<br />log10(baseMean): 2.8565748<br />gene: FBgn0261436","log2FoldChange: 0.0510703754<br />-log10(padj): 1.715502e-02<br />log10(baseMean): 2.2923427<br />gene: FBgn0261437","log2FoldChange: 0.0880909831<br />-log10(padj): 6.726744e-02<br />log10(baseMean): 3.4178320<br />gene: FBgn0261439","log2FoldChange: 0.6909402658<br />-log10(padj): 3.262004e+00<br />log10(baseMean): 2.8570015<br />gene: FBgn0261444","log2FoldChange: -0.2583603724<br />-log10(padj): 1.074738e+00<br />log10(baseMean): 3.8234400<br />gene: FBgn0261445","log2FoldChange: -0.5021411323<br />-log10(padj): 4.992606e-01<br />log10(baseMean): 1.7521716<br />gene: FBgn0261446","log2FoldChange: 0.1641336626<br />-log10(padj): 1.619769e-01<br />log10(baseMean): 4.0361575<br />gene: FBgn0261451","log2FoldChange: 0.1043148449<br />-log10(padj): 1.271125e-01<br />log10(baseMean): 2.9051317<br />gene: FBgn0261456","log2FoldChange: -0.0140241615<br />-log10(padj): 1.075565e-02<br />log10(baseMean): 3.7714527<br />gene: FBgn0261458","log2FoldChange: -0.2021012991<br />-log10(padj): 4.038476e-01<br />log10(baseMean): 3.3479621<br />gene: FBgn0261461","log2FoldChange: -0.1964320591<br />-log10(padj): 4.878643e-01<br />log10(baseMean): 3.0740034<br />gene: FBgn0261477","log2FoldChange: -0.0792331214<br />-log10(padj): 5.794697e-02<br />log10(baseMean): 2.6543002<br />gene: FBgn0261479","log2FoldChange: 0.0605190365<br />-log10(padj): 1.586349e-02<br />log10(baseMean): 1.5750392<br />gene: FBgn0261509","log2FoldChange: -0.1686387232<br />-log10(padj): 5.064448e-01<br />log10(baseMean): 3.2332882<br />gene: FBgn0261524","log2FoldChange: -0.0285623566<br />-log10(padj): 1.680948e-02<br />log10(baseMean): 2.6782135<br />gene: FBgn0261530","log2FoldChange: -0.0655511860<br />-log10(padj): 5.894869e-02<br />log10(baseMean): 3.0918688<br />gene: FBgn0261532","log2FoldChange: -0.1610013571<br />-log10(padj): 2.507520e-01<br />log10(baseMean): 2.8043209<br />gene: FBgn0261535","log2FoldChange: -0.5514666792<br />-log10(padj): 2.904404e+00<br />log10(baseMean): 3.2194830<br />gene: FBgn0261545","log2FoldChange: -1.8284426408<br />-log10(padj): 3.482989e+00<br />log10(baseMean): 1.4490359<br />gene: FBgn0261546","log2FoldChange: -0.5669879523<br />-log10(padj): 3.261169e+00<br />log10(baseMean): 2.8114156<br />gene: FBgn0261547","log2FoldChange: 0.1853298250<br />-log10(padj): 3.574581e-01<br />log10(baseMean): 3.2337406<br />gene: FBgn0261548","log2FoldChange: 0.1256825647<br />-log10(padj): 8.264702e-02<br />log10(baseMean): 2.7527035<br />gene: FBgn0261549","log2FoldChange: -0.3281275575<br />-log10(padj): 1.037479e+00<br />log10(baseMean): 3.9828472<br />gene: FBgn0261550","log2FoldChange: 0.0876675158<br />-log10(padj): 5.643673e-02<br />log10(baseMean): 3.7117288<br />gene: FBgn0261551","log2FoldChange: -1.8961198392<br />-log10(padj): 2.010669e+01<br />log10(baseMean): 3.7100002<br />gene: FBgn0261552","log2FoldChange: -0.2993250189<br />-log10(padj): 7.863513e-01<br />log10(baseMean): 3.1277340<br />gene: FBgn0261553","log2FoldChange: -0.0972283692<br />-log10(padj): 8.659630e-02<br />log10(baseMean): 3.0546592<br />gene: FBgn0261554","log2FoldChange: 0.6531355517<br />-log10(padj): 1.181582e+00<br />log10(baseMean): 1.8234522<br />gene: FBgn0261555","log2FoldChange: 0.2447191388<br />-log10(padj): 3.904677e-01<br />log10(baseMean): 3.4647364<br />gene: FBgn0261556","log2FoldChange: -0.8871763965<br />-log10(padj): 1.133741e+01<br />log10(baseMean): 3.2444811<br />gene: FBgn0261560","log2FoldChange: 0.6110778801<br />-log10(padj): 1.268749e+00<br />log10(baseMean): 1.8817263<br />gene: FBgn0261561","log2FoldChange: -0.1077122698<br />-log10(padj): 9.412115e-02<br />log10(baseMean): 2.5965171<br />gene: FBgn0261562","log2FoldChange: 0.6936276847<br />-log10(padj): 3.027504e+00<br />log10(baseMean): 3.2613175<br />gene: FBgn0261563","log2FoldChange: -0.0877384733<br />-log10(padj): 9.993745e-02<br />log10(baseMean): 3.0638458<br />gene: FBgn0261564","log2FoldChange: -0.2966975935<br />-log10(padj): 6.731530e-01<br />log10(baseMean): 2.7921503<br />gene: FBgn0261565","log2FoldChange: 0.2955328897<br />-log10(padj): 8.409143e-01<br />log10(baseMean): 3.5062870<br />gene: FBgn0261570","log2FoldChange: -0.9588229646<br />-log10(padj): 2.162051e-01<br />log10(baseMean): 0.7921947<br />gene: FBgn0261572","log2FoldChange: 0.0127194420<br />-log10(padj): 7.598195e-03<br />log10(baseMean): 3.3504379<br />gene: FBgn0261573","log2FoldChange: 0.0153924162<br />-log10(padj): 5.164278e-03<br />log10(baseMean): 3.6864289<br />gene: FBgn0261574","log2FoldChange: 0.1635705142<br />-log10(padj): 1.415695e-02<br />log10(baseMean): 1.0286737<br />gene: FBgn0261575"],"key":["FBgn0000008","FBgn0000017","FBgn0000018","FBgn0000024","FBgn0000032","FBgn0000037","FBgn0000042","FBgn0000043","FBgn0000044","FBgn0000045","FBgn0000046","FBgn0000047","FBgn0000052","FBgn0000053","FBgn0000054","FBgn0000057","FBgn0000063","FBgn0000064","FBgn0000071","FBgn0000077","FBgn0000078","FBgn0000079","FBgn0000083","FBgn0000084","FBgn0000092","FBgn0000094","FBgn0000097","FBgn0000100","FBgn0000108","FBgn0000109","FBgn0000115","FBgn0000116","FBgn0000117","FBgn0000119","FBgn0000121","FBgn0000139","FBgn0000140","FBgn0000142","FBgn0000146","FBgn0000147","FBgn0000150","FBgn0000152","FBgn0000153","FBgn0000163","FBgn0000166","FBgn0000171","FBgn0000173","FBgn0000180","FBgn0000181","FBgn0000183","FBgn0000210","FBgn0000212","FBgn0000221","FBgn0000228","FBgn0000229","FBgn0000239","FBgn0000241","FBgn0000242","FBgn0000244","FBgn0000246","FBgn0000247","FBgn0000250","FBgn0000253","FBgn0000256","FBgn0000257","FBgn0000258","FBgn0000259","FBgn0000261","FBgn0000273","FBgn0000276","FBgn0000277","FBgn0000278","FBgn0000279","FBgn0000283","FBgn0000286","FBgn0000289","FBgn0000299","FBgn0000307","FBgn0000308","FBgn0000313","FBgn0000317","FBgn0000318","FBgn0000319","FBgn0000326","FBgn0000330","FBgn0000337","FBgn0000338","FBgn0000339","FBgn0000346","FBgn0000351","FBgn0000352","FBgn0000370","FBgn0000376","FBgn0000377","FBgn0000382","FBgn0000384","FBgn0000392","FBgn0000394","FBgn0000395","FBgn0000404","FBgn0000405","FBgn0000406","FBgn0000409","FBgn0000412","FBgn0000413","FBgn0000414","FBgn0000416","FBgn0000422","FBgn0000426","FBgn0000447","FBgn0000448","FBgn0000454","FBgn0000455","FBgn0000464","FBgn0000472","FBgn0000473","FBgn0000479","FBgn0000482","FBgn0000489","FBgn0000490","FBgn0000499","FBgn0000504","FBgn0000520","FBgn0000524","FBgn0000527","FBgn0000533","FBgn0000535","FBgn0000536","FBgn0000541","FBgn0000542","FBgn0000543","FBgn0000546","FBgn0000547","FBgn0000556","FBgn0000557","FBgn0000559","FBgn0000562","FBgn0000565","FBgn0000566","FBgn0000567","FBgn0000568","FBgn0000575","FBgn0000578","FBgn0000579","FBgn0000581","FBgn0000588","FBgn0000594","FBgn0000611","FBgn0000615","FBgn0000617","FBgn0000618","FBgn0000629","FBgn0000630","FBgn0000633","FBgn0000634","FBgn0000635","FBgn0000636","FBgn0000662","FBgn0000667","FBgn0000709","FBgn0000711","FBgn0000719","FBgn0000721","FBgn0000723","FBgn0000808","FBgn0000810","FBgn0000826","FBgn0000927","FBgn0000928","FBgn0000986","FBgn0000996","FBgn0001075","FBgn0001078","FBgn0001079","FBgn0001083","FBgn0001084","FBgn0001085","FBgn0001086","FBgn0001087","FBgn0001089","FBgn0001090","FBgn0001091","FBgn0001092","FBgn0001098","FBgn0001104","FBgn0001105","FBgn0001108","FBgn0001114","FBgn0001122","FBgn0001123","FBgn0001124","FBgn0001125","FBgn0001128","FBgn0001133","FBgn0001134","FBgn0001137","FBgn0001139","FBgn0001142","FBgn0001145","FBgn0001149","FBgn0001150","FBgn0001168","FBgn0001169","FBgn0001179","FBgn0001185","FBgn0001186","FBgn0001187","FBgn0001189","FBgn0001197","FBgn0001202","FBgn0001203","FBgn0001205","FBgn0001206","FBgn0001215","FBgn0001218","FBgn0001219","FBgn0001220","FBgn0001222","FBgn0001224","FBgn0001225","FBgn0001226","FBgn0001227","FBgn0001230","FBgn0001233","FBgn0001234","FBgn0001247","FBgn0001248","FBgn0001250","FBgn0001255","FBgn0001257","FBgn0001258","FBgn0001259","FBgn0001263","FBgn0001276","FBgn0001280","FBgn0001291","FBgn0001296","FBgn0001297","FBgn0001301","FBgn0001308","FBgn0001311","FBgn0001316","FBgn0001321","FBgn0001324","FBgn0001330","FBgn0001332","FBgn0001337","FBgn0001341","FBgn0001404","FBgn0001491","FBgn0001565","FBgn0001612","FBgn0001624","FBgn0001941","FBgn0001942","FBgn0001961","FBgn0001965","FBgn0001967","FBgn0001970","FBgn0001974","FBgn0001977","FBgn0001978","FBgn0001986","FBgn0001987","FBgn0001989","FBgn0001990","FBgn0001992","FBgn0001994","FBgn0001995","FBgn0002021","FBgn0002022","FBgn0002031","FBgn0002036","FBgn0002044","FBgn0002069","FBgn0002121","FBgn0002174","FBgn0002183","FBgn0002283","FBgn0002284","FBgn0002306","FBgn0002354","FBgn0002413","FBgn0002431","FBgn0002440","FBgn0002441","FBgn0002466","FBgn0002521","FBgn0002524","FBgn0002525","FBgn0002526","FBgn0002527","FBgn0002528","FBgn0002531","FBgn0002542","FBgn0002543","FBgn0002552","FBgn0002564","FBgn0002566","FBgn0002567","FBgn0002576","FBgn0002577","FBgn0002579","FBgn0002590","FBgn0002592","FBgn0002593","FBgn0002607","FBgn0002609","FBgn0002622","FBgn0002626","FBgn0002638","FBgn0002641","FBgn0002643","FBgn0002645","FBgn0002652","FBgn0002673","FBgn0002707","FBgn0002715","FBgn0002716","FBgn0002719","FBgn0002723","FBgn0002733","FBgn0002734","FBgn0002736","FBgn0002773","FBgn0002774","FBgn0002775","FBgn0002778","FBgn0002780","FBgn0002781","FBgn0002783","FBgn0002787","FBgn0002791","FBgn0002868","FBgn0002872","FBgn0002873","FBgn0002878","FBgn0002891","FBgn0002899","FBgn0002901","FBgn0002905","FBgn0002906","FBgn0002914","FBgn0002917","FBgn0002921","FBgn0002922","FBgn0002924","FBgn0002926","FBgn0002931","FBgn0002932","FBgn0002939","FBgn0002945","FBgn0002948","FBgn0002962","FBgn0002968","FBgn0002973","FBgn0002989","FBgn0003008","FBgn0003011","FBgn0003013","FBgn0003015","FBgn0003016","FBgn0003022","FBgn0003023","FBgn0003031","FBgn0003041","FBgn0003042","FBgn0003044","FBgn0003048","FBgn0003053","FBgn0003062","FBgn0003067","FBgn0003068","FBgn0003071","FBgn0003074","FBgn0003076","FBgn0003079","FBgn0003082","FBgn0003087","FBgn0003089","FBgn0003093","FBgn0003116","FBgn0003117","FBgn0003118","FBgn0003124","FBgn0003134","FBgn0003137","FBgn0003138","FBgn0003139","FBgn0003141","FBgn0003149","FBgn0003159","FBgn0003162","FBgn0003165","FBgn0003169","FBgn0003175","FBgn0003177","FBgn0003178","FBgn0003187","FBgn0003189","FBgn0003200","FBgn0003204","FBgn0003205","FBgn0003206","FBgn0003209","FBgn0003210","FBgn0003218","FBgn0003227","FBgn0003231","FBgn0003255","FBgn0003257","FBgn0003261","FBgn0003268","FBgn0003274","FBgn0003275","FBgn0003276","FBgn0003277","FBgn0003278","FBgn0003279","FBgn0003285","FBgn0003301","FBgn0003302","FBgn0003308","FBgn0003310","FBgn0003317","FBgn0003319","FBgn0003321","FBgn0003328","FBgn0003330","FBgn0003334","FBgn0003345","FBgn0003346","FBgn0003353","FBgn0003366","FBgn0003371","FBgn0003386","FBgn0003388","FBgn0003390","FBgn0003391","FBgn0003392","FBgn0003396","FBgn0003401","FBgn0003410","FBgn0003415","FBgn0003416","FBgn0003423","FBgn0003425","FBgn0003435","FBgn0003444","FBgn0003447","FBgn0003449","FBgn0003459","FBgn0003462","FBgn0003463","FBgn0003464","FBgn0003475","FBgn0003479","FBgn0003480","FBgn0003482","FBgn0003483","FBgn0003495","FBgn0003499","FBgn0003501","FBgn0003502","FBgn0003507","FBgn0003510","FBgn0003511","FBgn0003512","FBgn0003514","FBgn0003517","FBgn0003520","FBgn0003525","FBgn0003527","FBgn0003545","FBgn0003557","FBgn0003567","FBgn0003575","FBgn0003598","FBgn0003600","FBgn0003607","FBgn0003612","FBgn0003638","FBgn0003654","FBgn0003655","FBgn0003656","FBgn0003659","FBgn0003660","FBgn0003676","FBgn0003687","FBgn0003701","FBgn0003710","FBgn0003714","FBgn0003716","FBgn0003717","FBgn0003721","FBgn0003731","FBgn0003732","FBgn0003733","FBgn0003741","FBgn0003742","FBgn0003744","FBgn0003747","FBgn0003748","FBgn0003751","FBgn0003862","FBgn0003866","FBgn0003870","FBgn0003882","FBgn0003884","FBgn0003885","FBgn0003886","FBgn0003887","FBgn0003888","FBgn0003890","FBgn0003891","FBgn0003892","FBgn0003926","FBgn0003930","FBgn0003941","FBgn0003942","FBgn0003943","FBgn0003950","FBgn0003963","FBgn0003964","FBgn0003969","FBgn0003977","FBgn0003978","FBgn0003984","FBgn0003996","FBgn0003997","FBgn0004028","FBgn0004034","FBgn0004047","FBgn0004049","FBgn0004050","FBgn0004055","FBgn0004057","FBgn0004066","FBgn0004087","FBgn0004101","FBgn0004103","FBgn0004106","FBgn0004107","FBgn0004108","FBgn0004118","FBgn0004132","FBgn0004133","FBgn0004145","FBgn0004167","FBgn0004169","FBgn0004177","FBgn0004179","FBgn0004188","FBgn0004197","FBgn0004198","FBgn0004227","FBgn0004237","FBgn0004359","FBgn0004360","FBgn0004362","FBgn0004363","FBgn0004364","FBgn0004367","FBgn0004368","FBgn0004369","FBgn0004370","FBgn0004373","FBgn0004374","FBgn0004378","FBgn0004379","FBgn0004380","FBgn0004381","FBgn0004387","FBgn0004390","FBgn0004391","FBgn0004395","FBgn0004396","FBgn0004397","FBgn0004399","FBgn0004400","FBgn0004401","FBgn0004403","FBgn0004404","FBgn0004406","FBgn0004413","FBgn0004419","FBgn0004431","FBgn0004432","FBgn0004435","FBgn0004436","FBgn0004449","FBgn0004456","FBgn0004462","FBgn0004463","FBgn0004465","FBgn0004507","FBgn0004509","FBgn0004510","FBgn0004511","FBgn0004512","FBgn0004551","FBgn0004556","FBgn0004569","FBgn0004574","FBgn0004581","FBgn0004583","FBgn0004584","FBgn0004587","FBgn0004595","FBgn0004597","FBgn0004598","FBgn0004603","FBgn0004606","FBgn0004607","FBgn0004611","FBgn0004624","FBgn0004625","FBgn0004629","FBgn0004635","FBgn0004636","FBgn0004638","FBgn0004643","FBgn0004646","FBgn0004647","FBgn0004648","FBgn0004650","FBgn0004652","FBgn0004654","FBgn0004655","FBgn0004656","FBgn0004657","FBgn0004687","FBgn0004698","FBgn0004797","FBgn0004811","FBgn0004828","FBgn0004832","FBgn0004834","FBgn0004837","FBgn0004838","FBgn0004839","FBgn0004852","FBgn0004855","FBgn0004856","FBgn0004858","FBgn0004859","FBgn0004860","FBgn0004861","FBgn0004864","FBgn0004865","FBgn0004867","FBgn0004868","FBgn0004870","FBgn0004872","FBgn0004873","FBgn0004875","FBgn0004876","FBgn0004882","FBgn0004885","FBgn0004888","FBgn0004889","FBgn0004893","FBgn0004896","FBgn0004901","FBgn0004903","FBgn0004907","FBgn0004908","FBgn0004913","FBgn0004914","FBgn0004915","FBgn0004919","FBgn0004921","FBgn0004922","FBgn0004924","FBgn0004925","FBgn0004926","FBgn0004957","FBgn0005198","FBgn0005278","FBgn0005322","FBgn0005355","FBgn0005386","FBgn0005410","FBgn0005411","FBgn0005427","FBgn0005533","FBgn0005536","FBgn0005558","FBgn0005561","FBgn0005563","FBgn0005585","FBgn0005592","FBgn0005593","FBgn0005596","FBgn0005612","FBgn0005613","FBgn0005614","FBgn0005616","FBgn0005617","FBgn0005619","FBgn0005624","FBgn0005629","FBgn0005630","FBgn0005631","FBgn0005632","FBgn0005634","FBgn0005636","FBgn0005640","FBgn0005642","FBgn0005648","FBgn0005649","FBgn0005655","FBgn0005660","FBgn0005666","FBgn0005670","FBgn0005671","FBgn0005672","FBgn0005674","FBgn0005683","FBgn0005694","FBgn0005695","FBgn0005696","FBgn0005771","FBgn0005777","FBgn0008635","FBgn0008649","FBgn0008654","FBgn0010015","FBgn0010038","FBgn0010039","FBgn0010040","FBgn0010041","FBgn0010043","FBgn0010051","FBgn0010052","FBgn0010053","FBgn0010078","FBgn0010083","FBgn0010097","FBgn0010100","FBgn0010110","FBgn0010113","FBgn0010173","FBgn0010194","FBgn0010198","FBgn0010213","FBgn0010215","FBgn0010217","FBgn0010220","FBgn0010222","FBgn0010223","FBgn0010225","FBgn0010226","FBgn0010228","FBgn0010235","FBgn0010238","FBgn0010240","FBgn0010241","FBgn0010246","FBgn0010256","FBgn0010258","FBgn0010263","FBgn0010265","FBgn0010269","FBgn0010278","FBgn0010280","FBgn0010282","FBgn0010287","FBgn0010288","FBgn0010292","FBgn0010300","FBgn0010303","FBgn0010309","FBgn0010313","FBgn0010314","FBgn0010315","FBgn0010316","FBgn0010328","FBgn0010329","FBgn0010333","FBgn0010339","FBgn0010340","FBgn0010341","FBgn0010342","FBgn0010348","FBgn0010349","FBgn0010350","FBgn0010352","FBgn0010355","FBgn0010356","FBgn0010379","FBgn0010380","FBgn0010381","FBgn0010382","FBgn0010383","FBgn0010388","FBgn0010389","FBgn0010391","FBgn0010395","FBgn0010397","FBgn0010398","FBgn0010406","FBgn0010407","FBgn0010408","FBgn0010409","FBgn0010410","FBgn0010411","FBgn0010412","FBgn0010415","FBgn0010416","FBgn0010417","FBgn0010421","FBgn0010422","FBgn0010426","FBgn0010434","FBgn0010435","FBgn0010438","FBgn0010441","FBgn0010470","FBgn0010473","FBgn0010482","FBgn0010488","FBgn0010497","FBgn0010501","FBgn0010504","FBgn0010516","FBgn0010520","FBgn0010531","FBgn0010548","FBgn0010549","FBgn0010551","FBgn0010575","FBgn0010583","FBgn0010590","FBgn0010591","FBgn0010602","FBgn0010607","FBgn0010609","FBgn0010611","FBgn0010612","FBgn0010620","FBgn0010621","FBgn0010622","FBgn0010638","FBgn0010651","FBgn0010660","FBgn0010704","FBgn0010709","FBgn0010741","FBgn0010747","FBgn0010750","FBgn0010762","FBgn0010768","FBgn0010770","FBgn0010772","FBgn0010774","FBgn0010786","FBgn0010803","FBgn0010808","FBgn0010812","FBgn0010825","FBgn0010830","FBgn0010877","FBgn0010894","FBgn0010905","FBgn0010909","FBgn0010926","FBgn0011016","FBgn0011020","FBgn0011202","FBgn0011204","FBgn0011205","FBgn0011206","FBgn0011207","FBgn0011211","FBgn0011217","FBgn0011219","FBgn0011224","FBgn0011225","FBgn0011227","FBgn0011230","FBgn0011232","FBgn0011236","FBgn0011241","FBgn0011259","FBgn0011260","FBgn0011272","FBgn0011274","FBgn0011276","FBgn0011280","FBgn0011284","FBgn0011285","FBgn0011286","FBgn0011288","FBgn0011289","FBgn0011290","FBgn0011291","FBgn0011297","FBgn0011300","FBgn0011305","FBgn0011327","FBgn0011335","FBgn0011336","FBgn0011361","FBgn0011455","FBgn0011474","FBgn0011476","FBgn0011481","FBgn0011509","FBgn0011566","FBgn0011570","FBgn0011571","FBgn0011573","FBgn0011576","FBgn0011577","FBgn0011584","FBgn0011586","FBgn0011589","FBgn0011591","FBgn0011592","FBgn0011596","FBgn0011604","FBgn0011606","FBgn0011638","FBgn0011640","FBgn0011642","FBgn0011648","FBgn0011653","FBgn0011655","FBgn0011656","FBgn0011659","FBgn0011660","FBgn0011661","FBgn0011666","FBgn0011672","FBgn0011674","FBgn0011676","FBgn0011692","FBgn0011695","FBgn0011703","FBgn0011704","FBgn0011705","FBgn0011706","FBgn0011708","FBgn0011710","FBgn0011715","FBgn0011722","FBgn0011725","FBgn0011726","FBgn0011737","FBgn0011739","FBgn0011740","FBgn0011741","FBgn0011742","FBgn0011744","FBgn0011745","FBgn0011746","FBgn0011747","FBgn0011754","FBgn0011760","FBgn0011762","FBgn0011763","FBgn0011764","FBgn0011766","FBgn0011768","FBgn0011769","FBgn0011770","FBgn0011771","FBgn0011774","FBgn0011785","FBgn0011787","FBgn0011802","FBgn0011817","FBgn0011818","FBgn0011823","FBgn0011824","FBgn0011826","FBgn0011828","FBgn0011829","FBgn0011836","FBgn0011837","FBgn0012034","FBgn0012036","FBgn0012037","FBgn0012042","FBgn0012049","FBgn0012051","FBgn0012058","FBgn0012066","FBgn0013263","FBgn0013269","FBgn0013272","FBgn0013303","FBgn0013305","FBgn0013325","FBgn0013334","FBgn0013343","FBgn0013347","FBgn0013432","FBgn0013435","FBgn0013442","FBgn0013469","FBgn0013531","FBgn0013548","FBgn0013563","FBgn0013576","FBgn0013591","FBgn0013717","FBgn0013718","FBgn0013720","FBgn0013726","FBgn0013732","FBgn0013733","FBgn0013746","FBgn0013749","FBgn0013750","FBgn0013753","FBgn0013756","FBgn0013759","FBgn0013762","FBgn0013763","FBgn0013764","FBgn0013765","FBgn0013770","FBgn0013771","FBgn0013772","FBgn0013773","FBgn0013799","FBgn0013809","FBgn0013810","FBgn0013811","FBgn0013812","FBgn0013813","FBgn0013948","FBgn0013953","FBgn0013954","FBgn0013955","FBgn0013972","FBgn0013981","FBgn0013983","FBgn0013984","FBgn0013987","FBgn0013988","FBgn0013995","FBgn0013997","FBgn0013998","FBgn0014001","FBgn0014002","FBgn0014006","FBgn0014007","FBgn0014009","FBgn0014010","FBgn0014011","FBgn0014018","FBgn0014020","FBgn0014022","FBgn0014023","FBgn0014024","FBgn0014026","FBgn0014028","FBgn0014029","FBgn0014031","FBgn0014032","FBgn0014033","FBgn0014037","FBgn0014075","FBgn0014092","FBgn0014127","FBgn0014133","FBgn0014135","FBgn0014141","FBgn0014163","FBgn0014184","FBgn0014189","FBgn0014269","FBgn0014340","FBgn0014342","FBgn0014343","FBgn0014362","FBgn0014366","FBgn0014380","FBgn0014388","FBgn0014391","FBgn0014395","FBgn0014411","FBgn0014417","FBgn0014427","FBgn0014455","FBgn0014467","FBgn0014469","FBgn0014857","FBgn0014859","FBgn0014861","FBgn0014863","FBgn0014868","FBgn0014869","FBgn0014870","FBgn0014877","FBgn0014879","FBgn0014906","FBgn0014930","FBgn0014931","FBgn0014963","FBgn0015000","FBgn0015014","FBgn0015019","FBgn0015024","FBgn0015025","FBgn0015031","FBgn0015034","FBgn0015036","FBgn0015037","FBgn0015040","FBgn0015075","FBgn0015218","FBgn0015221","FBgn0015222","FBgn0015229","FBgn0015239","FBgn0015240","FBgn0015245","FBgn0015247","FBgn0015268","FBgn0015269","FBgn0015270","FBgn0015271","FBgn0015277","FBgn0015278","FBgn0015279","FBgn0015282","FBgn0015283","FBgn0015286","FBgn0015288","FBgn0015295","FBgn0015296","FBgn0015298","FBgn0015299","FBgn0015316","FBgn0015320","FBgn0015321","FBgn0015324","FBgn0015331","FBgn0015336","FBgn0015338","FBgn0015359","FBgn0015360","FBgn0015371","FBgn0015372","FBgn0015374","FBgn0015376","FBgn0015379","FBgn0015391","FBgn0015393","FBgn0015396","FBgn0015399","FBgn0015402","FBgn0015477","FBgn0015509","FBgn0015513","FBgn0015520","FBgn0015521","FBgn0015522","FBgn0015527","FBgn0015541","FBgn0015542","FBgn0015543","FBgn0015544","FBgn0015546","FBgn0015553","FBgn0015558","FBgn0015562","FBgn0015565","FBgn0015567","FBgn0015568","FBgn0015569","FBgn0015571","FBgn0015572","FBgn0015575","FBgn0015576","FBgn0015577","FBgn0015582","FBgn0015589","FBgn0015600","FBgn0015602","FBgn0015609","FBgn0015610","FBgn0015614","FBgn0015615","FBgn0015617","FBgn0015618","FBgn0015621","FBgn0015622","FBgn0015623","FBgn0015624","FBgn0015625","FBgn0015657","FBgn0015663","FBgn0015664","FBgn0015714","FBgn0015737","FBgn0015754","FBgn0015756","FBgn0015765","FBgn0015766","FBgn0015772","FBgn0015773","FBgn0015774","FBgn0015776","FBgn0015777","FBgn0015778","FBgn0015781","FBgn0015789","FBgn0015790","FBgn0015791","FBgn0015793","FBgn0015794","FBgn0015795","FBgn0015796","FBgn0015797","FBgn0015799","FBgn0015801","FBgn0015803","FBgn0015805","FBgn0015806","FBgn0015808","FBgn0015816","FBgn0015818","FBgn0015828","FBgn0015829","FBgn0015834","FBgn0015838","FBgn0015844","FBgn0015872","FBgn0015903","FBgn0015905","FBgn0015907","FBgn0015924","FBgn0015925","FBgn0015926","FBgn0015929","FBgn0015949","FBgn0016031","FBgn0016032","FBgn0016034","FBgn0016038","FBgn0016041","FBgn0016054","FBgn0016059","FBgn0016070","FBgn0016075","FBgn0016076","FBgn0016078","FBgn0016080","FBgn0016081","FBgn0016119","FBgn0016120","FBgn0016122","FBgn0016123","FBgn0016126","FBgn0016131","FBgn0016641","FBgn0016672","FBgn0016684","FBgn0016685","FBgn0016687","FBgn0016691","FBgn0016693","FBgn0016694","FBgn0016696","FBgn0016697","FBgn0016698","FBgn0016700","FBgn0016701","FBgn0016715","FBgn0016726","FBgn0016754","FBgn0016756","FBgn0016762","FBgn0016792","FBgn0016794","FBgn0016797","FBgn0016917","FBgn0016919","FBgn0016926","FBgn0016930","FBgn0016940","FBgn0016970","FBgn0016974","FBgn0016977","FBgn0016978","FBgn0016983","FBgn0016984","FBgn0017397","FBgn0017414","FBgn0017418","FBgn0017429","FBgn0017430","FBgn0017453","FBgn0017456","FBgn0017457","FBgn0017482","FBgn0017545","FBgn0017549","FBgn0017550","FBgn0017551","FBgn0017558","FBgn0017566","FBgn0017567","FBgn0017572","FBgn0017577","FBgn0017578","FBgn0017579","FBgn0017581","FBgn0019624","FBgn0019637","FBgn0019643","FBgn0019644","FBgn0019650","FBgn0019660","FBgn0019661","FBgn0019662","FBgn0019686","FBgn0019830","FBgn0019886","FBgn0019890","FBgn0019925","FBgn0019932","FBgn0019936","FBgn0019938","FBgn0019947","FBgn0019948","FBgn0019949","FBgn0019952","FBgn0019957","FBgn0019960","FBgn0019968","FBgn0019972","FBgn0019982","FBgn0019985","FBgn0019990","FBgn0020018","FBgn0020224","FBgn0020235","FBgn0020236","FBgn0020238","FBgn0020240","FBgn0020245","FBgn0020248","FBgn0020249","FBgn0020251","FBgn0020255","FBgn0020257","FBgn0020261","FBgn0020269","FBgn0020270","FBgn0020272","FBgn0020278","FBgn0020279","FBgn0020294","FBgn0020303","FBgn0020304","FBgn0020305","FBgn0020306","FBgn0020307","FBgn0020309","FBgn0020312","FBgn0020367","FBgn0020368","FBgn0020369","FBgn0020370","FBgn0020372","FBgn0020376","FBgn0020379","FBgn0020381","FBgn0020385","FBgn0020386","FBgn0020388","FBgn0020389","FBgn0020391","FBgn0020392","FBgn0020407","FBgn0020412","FBgn0020414","FBgn0020415","FBgn0020416","FBgn0020440","FBgn0020443","FBgn0020445","FBgn0020493","FBgn0020496","FBgn0020497","FBgn0020503","FBgn0020510","FBgn0020513","FBgn0020521","FBgn0020545","FBgn0020611","FBgn0020616","FBgn0020618","FBgn0020620","FBgn0020621","FBgn0020622","FBgn0020623","FBgn0020626","FBgn0020633","FBgn0020647","FBgn0020653","FBgn0020655","FBgn0020756","FBgn0020762","FBgn0020764","FBgn0020766","FBgn0020767","FBgn0020887","FBgn0020909","FBgn0020910","FBgn0020930","FBgn0021750","FBgn0021760","FBgn0021761","FBgn0021764","FBgn0021765","FBgn0021767","FBgn0021768","FBgn0021776","FBgn0021795","FBgn0021796","FBgn0021800","FBgn0021814","FBgn0021818","FBgn0021825","FBgn0021847","FBgn0021856","FBgn0021872","FBgn0021873","FBgn0021874","FBgn0021875","FBgn0021895","FBgn0021906","FBgn0021944","FBgn0021953","FBgn0021967","FBgn0021979","FBgn0021995","FBgn0022023","FBgn0022027","FBgn0022029","FBgn0022063","FBgn0022069","FBgn0022070","FBgn0022085","FBgn0022097","FBgn0022131","FBgn0022153","FBgn0022160","FBgn0022201","FBgn0022213","FBgn0022224","FBgn0022238","FBgn0022246","FBgn0022268","FBgn0022288","FBgn0022338","FBgn0022343","FBgn0022344","FBgn0022349","FBgn0022355","FBgn0022359","FBgn0022361","FBgn0022382","FBgn0022699","FBgn0022702","FBgn0022708","FBgn0022709","FBgn0022710","FBgn0022720","FBgn0022724","FBgn0022764","FBgn0022768","FBgn0022770","FBgn0022772","FBgn0022774","FBgn0022786","FBgn0022787","FBgn0022800","FBgn0022893","FBgn0022935","FBgn0022936","FBgn0022942","FBgn0022943","FBgn0022959","FBgn0022960","FBgn0022984","FBgn0022985","FBgn0022986","FBgn0022987","FBgn0023000","FBgn0023023","FBgn0023081","FBgn0023083","FBgn0023090","FBgn0023091","FBgn0023094","FBgn0023095","FBgn0023096","FBgn0023097","FBgn0023129","FBgn0023130","FBgn0023143","FBgn0023167","FBgn0023169","FBgn0023170","FBgn0023171","FBgn0023172","FBgn0023174","FBgn0023175","FBgn0023177","FBgn0023180","FBgn0023181","FBgn0023211","FBgn0023212","FBgn0023213","FBgn0023214","FBgn0023215","FBgn0023216","FBgn0023388","FBgn0023395","FBgn0023407","FBgn0023416","FBgn0023423","FBgn0023441","FBgn0023444","FBgn0023458","FBgn0023477","FBgn0023479","FBgn0023506","FBgn0023507","FBgn0023508","FBgn0023509","FBgn0023510","FBgn0023511","FBgn0023512","FBgn0023513","FBgn0023514","FBgn0023515","FBgn0023516","FBgn0023517","FBgn0023518","FBgn0023519","FBgn0023520","FBgn0023521","FBgn0023522","FBgn0023523","FBgn0023524","FBgn0023525","FBgn0023526","FBgn0023527","FBgn0023528","FBgn0023529","FBgn0023530","FBgn0023531","FBgn0023535","FBgn0023536","FBgn0023537","FBgn0023540","FBgn0023541","FBgn0023542","FBgn0023545","FBgn0023549","FBgn0024150","FBgn0024177","FBgn0024183","FBgn0024187","FBgn0024188","FBgn0024191","FBgn0024194","FBgn0024196","FBgn0024222","FBgn0024227","FBgn0024230","FBgn0024234","FBgn0024236","FBgn0024238","FBgn0024245","FBgn0024248","FBgn0024250","FBgn0024252","FBgn0024273","FBgn0024277","FBgn0024285","FBgn0024288","FBgn0024291","FBgn0024308","FBgn0024314","FBgn0024315","FBgn0024320","FBgn0024321","FBgn0024326","FBgn0024329","FBgn0024330","FBgn0024332","FBgn0024352","FBgn0024360","FBgn0024361","FBgn0024362","FBgn0024364","FBgn0024365","FBgn0024366","FBgn0024371","FBgn0024432","FBgn0024491","FBgn0024509","FBgn0024510","FBgn0024542","FBgn0024555","FBgn0024556","FBgn0024558","FBgn0024689","FBgn0024698","FBgn0024728","FBgn0024732","FBgn0024733","FBgn0024734","FBgn0024753","FBgn0024754","FBgn0024806","FBgn0024807","FBgn0024811","FBgn0024814","FBgn0024832","FBgn0024833","FBgn0024836","FBgn0024841","FBgn0024846","FBgn0024887","FBgn0024889","FBgn0024891","FBgn0024897","FBgn0024909","FBgn0024912","FBgn0024920","FBgn0024921","FBgn0024939","FBgn0024943","FBgn0024945","FBgn0024947","FBgn0024956","FBgn0024957","FBgn0024958","FBgn0024973","FBgn0024975","FBgn0024977","FBgn0024980","FBgn0024983","FBgn0024984","FBgn0024985","FBgn0024986","FBgn0024987","FBgn0024991","FBgn0024992","FBgn0024993","FBgn0024994","FBgn0024996","FBgn0024997","FBgn0024998","FBgn0025109","FBgn0025117","FBgn0025140","FBgn0025185","FBgn0025186","FBgn0025286","FBgn0025335","FBgn0025336","FBgn0025352","FBgn0025355","FBgn0025366","FBgn0025373","FBgn0025381","FBgn0025382","FBgn0025383","FBgn0025387","FBgn0025388","FBgn0025390","FBgn0025394","FBgn0025455","FBgn0025456","FBgn0025457","FBgn0025458","FBgn0025463","FBgn0025469","FBgn0025519","FBgn0025525","FBgn0025549","FBgn0025558","FBgn0025571","FBgn0025574","FBgn0025582","FBgn0025592","FBgn0025593","FBgn0025608","FBgn0025615","FBgn0025616","FBgn0025621","FBgn0025624","FBgn0025625","FBgn0025626","FBgn0025627","FBgn0025628","FBgn0025629","FBgn0025630","FBgn0025631","FBgn0025632","FBgn0025633","FBgn0025634","FBgn0025635","FBgn0025637","FBgn0025638","FBgn0025639","FBgn0025640","FBgn0025641","FBgn0025642","FBgn0025643","FBgn0025674","FBgn0025676","FBgn0025678","FBgn0025681","FBgn0025682","FBgn0025683","FBgn0025684","FBgn0025687","FBgn0025692","FBgn0025693","FBgn0025697","FBgn0025698","FBgn0025700","FBgn0025701","FBgn0025702","FBgn0025712","FBgn0025716","FBgn0025720","FBgn0025724","FBgn0025725","FBgn0025726","FBgn0025739","FBgn0025740","FBgn0025741","FBgn0025742","FBgn0025743","FBgn0025777","FBgn0025781","FBgn0025790","FBgn0025800","FBgn0025802","FBgn0025803","FBgn0025806","FBgn0025807","FBgn0025808","FBgn0025809","FBgn0025814","FBgn0025815","FBgn0025820","FBgn0025825","FBgn0025830","FBgn0025832","FBgn0025836","FBgn0025838","FBgn0025839","FBgn0025864","FBgn0025865","FBgn0025866","FBgn0025874","FBgn0025879","FBgn0025885","FBgn0025936","FBgn0026015","FBgn0026056","FBgn0026059","FBgn0026060","FBgn0026076","FBgn0026079","FBgn0026080","FBgn0026083","FBgn0026084","FBgn0026085","FBgn0026086","FBgn0026088","FBgn0026089","FBgn0026090","FBgn0026143","FBgn0026144","FBgn0026147","FBgn0026148","FBgn0026149","FBgn0026150","FBgn0026160","FBgn0026170","FBgn0026174","FBgn0026175","FBgn0026176","FBgn0026179","FBgn0026181","FBgn0026189","FBgn0026192","FBgn0026196","FBgn0026197","FBgn0026199","FBgn0026206","FBgn0026207","FBgn0026208","FBgn0026238","FBgn0026239","FBgn0026250","FBgn0026252","FBgn0026255","FBgn0026257","FBgn0026259","FBgn0026261","FBgn0026262","FBgn0026263","FBgn0026309","FBgn0026313","FBgn0026315","FBgn0026316","FBgn0026317","FBgn0026318","FBgn0026319","FBgn0026323","FBgn0026324","FBgn0026326","FBgn0026361","FBgn0026369","FBgn0026370","FBgn0026371","FBgn0026372","FBgn0026373","FBgn0026374","FBgn0026375","FBgn0026376","FBgn0026378","FBgn0026379","FBgn0026380","FBgn0026393","FBgn0026400","FBgn0026401","FBgn0026402","FBgn0026404","FBgn0026409","FBgn0026415","FBgn0026417","FBgn0026418","FBgn0026427","FBgn0026428","FBgn0026430","FBgn0026431","FBgn0026432","FBgn0026433","FBgn0026439","FBgn0026441","FBgn0026479","FBgn0026533","FBgn0026562","FBgn0026566","FBgn0026567","FBgn0026570","FBgn0026573","FBgn0026575","FBgn0026576","FBgn0026577","FBgn0026582","FBgn0026585","FBgn0026592","FBgn0026593","FBgn0026597","FBgn0026598","FBgn0026602","FBgn0026611","FBgn0026616","FBgn0026619","FBgn0026620","FBgn0026630","FBgn0026634","FBgn0026666","FBgn0026679","FBgn0026702","FBgn0026713","FBgn0026718","FBgn0026721","FBgn0026722","FBgn0026737","FBgn0026738","FBgn0026741","FBgn0026749","FBgn0026751","FBgn0026753","FBgn0026758","FBgn0026760","FBgn0026761","FBgn0026777","FBgn0026787","FBgn0026869","FBgn0026872","FBgn0026873","FBgn0026874","FBgn0026876","FBgn0026878","FBgn0026879","FBgn0027052","FBgn0027053","FBgn0027054","FBgn0027055","FBgn0027057","FBgn0027066","FBgn0027070","FBgn0027079","FBgn0027080","FBgn0027081","FBgn0027082","FBgn0027083","FBgn0027084","FBgn0027085","FBgn0027086","FBgn0027087","FBgn0027088","FBgn0027090","FBgn0027091","FBgn0027093","FBgn0027094","FBgn0027095","FBgn0027101","FBgn0027103","FBgn0027108","FBgn0027111","FBgn0027259","FBgn0027279","FBgn0027280","FBgn0027287","FBgn0027291","FBgn0027296","FBgn0027329","FBgn0027330","FBgn0027334","FBgn0027335","FBgn0027338","FBgn0027339","FBgn0027342","FBgn0027348","FBgn0027356","FBgn0027359","FBgn0027363","FBgn0027375","FBgn0027378","FBgn0027453","FBgn0027490","FBgn0027491","FBgn0027492","FBgn0027493","FBgn0027495","FBgn0027496","FBgn0027497","FBgn0027498","FBgn0027499","FBgn0027500","FBgn0027503","FBgn0027504","FBgn0027505","FBgn0027506","FBgn0027507","FBgn0027508","FBgn0027509","FBgn0027512","FBgn0027513","FBgn0027514","FBgn0027515","FBgn0027518","FBgn0027521","FBgn0027524","FBgn0027525","FBgn0027526","FBgn0027529","FBgn0027532","FBgn0027534","FBgn0027535","FBgn0027537","FBgn0027538","FBgn0027539","FBgn0027544","FBgn0027547","FBgn0027548","FBgn0027549","FBgn0027550","FBgn0027552","FBgn0027553","FBgn0027554","FBgn0027556","FBgn0027558","FBgn0027559","FBgn0027560","FBgn0027561","FBgn0027563","FBgn0027564","FBgn0027565","FBgn0027567","FBgn0027568","FBgn0027569","FBgn0027570","FBgn0027571","FBgn0027572","FBgn0027574","FBgn0027575","FBgn0027578","FBgn0027579","FBgn0027580","FBgn0027581","FBgn0027582","FBgn0027583","FBgn0027585","FBgn0027587","FBgn0027588","FBgn0027590","FBgn0027592","FBgn0027594","FBgn0027596","FBgn0027597","FBgn0027598","FBgn0027599","FBgn0027601","FBgn0027602","FBgn0027603","FBgn0027604","FBgn0027605","FBgn0027607","FBgn0027609","FBgn0027610","FBgn0027611","FBgn0027615","FBgn0027616","FBgn0027617","FBgn0027619","FBgn0027620","FBgn0027621","FBgn0027654","FBgn0027655","FBgn0027657","FBgn0027660","FBgn0027779","FBgn0027780","FBgn0027783","FBgn0027784","FBgn0027785","FBgn0027786","FBgn0027791","FBgn0027793","FBgn0027794","FBgn0027795","FBgn0027835","FBgn0027836","FBgn0027841","FBgn0027842","FBgn0027843","FBgn0027864","FBgn0027865","FBgn0027866","FBgn0027868","FBgn0027872","FBgn0027873","FBgn0027885","FBgn0027889","FBgn0027903","FBgn0027914","FBgn0027929","FBgn0027930","FBgn0027932","FBgn0027936","FBgn0027945","FBgn0027948","FBgn0027949","FBgn0027950","FBgn0027951","FBgn0028274","FBgn0028292","FBgn0028325","FBgn0028327","FBgn0028331","FBgn0028336","FBgn0028341","FBgn0028342","FBgn0028343","FBgn0028360","FBgn0028372","FBgn0028373","FBgn0028375","FBgn0028379","FBgn0028380","FBgn0028382","FBgn0028386","FBgn0028387","FBgn0028394","FBgn0028397","FBgn0028398","FBgn0028399","FBgn0028401","FBgn0028402","FBgn0028406","FBgn0028408","FBgn0028411","FBgn0028418","FBgn0028419","FBgn0028420","FBgn0028421","FBgn0028424","FBgn0028425","FBgn0028426","FBgn0028427","FBgn0028428","FBgn0028429","FBgn0028430","FBgn0028433","FBgn0028434","FBgn0028436","FBgn0028467","FBgn0028468","FBgn0028470","FBgn0028471","FBgn0028473","FBgn0028474","FBgn0028475","FBgn0028476","FBgn0028479","FBgn0028480","FBgn0028482","FBgn0028484","FBgn0028485","FBgn0028487","FBgn0028490","FBgn0028491","FBgn0028494","FBgn0028497","FBgn0028499","FBgn0028500","FBgn0028504","FBgn0028506","FBgn0028507","FBgn0028509","FBgn0028513","FBgn0028514","FBgn0028515","FBgn0028519","FBgn0028523","FBgn0028530","FBgn0028535","FBgn0028538","FBgn0028539","FBgn0028540","FBgn0028541","FBgn0028542","FBgn0028543","FBgn0028546","FBgn0028550","FBgn0028552","FBgn0028554","FBgn0028562","FBgn0028563","FBgn0028572","FBgn0028573","FBgn0028577","FBgn0028579","FBgn0028582","FBgn0028622","FBgn0028646","FBgn0028647","FBgn0028648","FBgn0028658","FBgn0028662","FBgn0028663","FBgn0028664","FBgn0028665","FBgn0028670","FBgn0028671","FBgn0028675","FBgn0028683","FBgn0028684","FBgn0028685","FBgn0028686","FBgn0028687","FBgn0028688","FBgn0028689","FBgn0028690","FBgn0028691","FBgn0028692","FBgn0028693","FBgn0028694","FBgn0028695","FBgn0028696","FBgn0028700","FBgn0028703","FBgn0028704","FBgn0028707","FBgn0028708","FBgn0028717","FBgn0028734","FBgn0028737","FBgn0028738","FBgn0028740","FBgn0028743","FBgn0028744","FBgn0028746","FBgn0028836","FBgn0028837","FBgn0028859","FBgn0028863","FBgn0028886","FBgn0028894","FBgn0028895","FBgn0028897","FBgn0028899","FBgn0028903","FBgn0028905","FBgn0028906","FBgn0028916","FBgn0028919","FBgn0028925","FBgn0028926","FBgn0028931","FBgn0028932","FBgn0028936","FBgn0028939","FBgn0028940","FBgn0028941","FBgn0028952","FBgn0028953","FBgn0028954","FBgn0028955","FBgn0028956","FBgn0028962","FBgn0028965","FBgn0028968","FBgn0028969","FBgn0028970","FBgn0028974","FBgn0028978","FBgn0028979","FBgn0028980","FBgn0028982","FBgn0028983","FBgn0028984","FBgn0028985","FBgn0028988","FBgn0028990","FBgn0028991","FBgn0028992","FBgn0028996","FBgn0028997","FBgn0029002","FBgn0029006","FBgn0029067","FBgn0029079","FBgn0029088","FBgn0029090","FBgn0029092","FBgn0029093","FBgn0029094","FBgn0029095","FBgn0029113","FBgn0029114","FBgn0029117","FBgn0029118","FBgn0029121","FBgn0029128","FBgn0029131","FBgn0029133","FBgn0029134","FBgn0029137","FBgn0029147","FBgn0029148","FBgn0029152","FBgn0029155","FBgn0029157","FBgn0029158","FBgn0029173","FBgn0029174","FBgn0029176","FBgn0029502","FBgn0029503","FBgn0029504","FBgn0029506","FBgn0029507","FBgn0029508","FBgn0029512","FBgn0029514","FBgn0029521","FBgn0029522","FBgn0029523","FBgn0029524","FBgn0029525","FBgn0029526","FBgn0029529","FBgn0029538","FBgn0029568","FBgn0029573","FBgn0029580","FBgn0029587","FBgn0029588","FBgn0029594","FBgn0029596","FBgn0029608","FBgn0029629","FBgn0029643","FBgn0029648","FBgn0029658","FBgn0029659","FBgn0029660","FBgn0029661","FBgn0029662","FBgn0029663","FBgn0029664","FBgn0029665","FBgn0029666","FBgn0029667","FBgn0029669","FBgn0029672","FBgn0029676","FBgn0029685","FBgn0029686","FBgn0029687","FBgn0029688","FBgn0029689","FBgn0029693","FBgn0029697","FBgn0029704","FBgn0029706","FBgn0029709","FBgn0029710","FBgn0029711","FBgn0029712","FBgn0029713","FBgn0029714","FBgn0029715","FBgn0029718","FBgn0029719","FBgn0029722","FBgn0029736","FBgn0029737","FBgn0029738","FBgn0029747","FBgn0029750","FBgn0029755","FBgn0029756","FBgn0029761","FBgn0029762","FBgn0029763","FBgn0029764","FBgn0029765","FBgn0029766","FBgn0029771","FBgn0029778","FBgn0029783","FBgn0029785","FBgn0029789","FBgn0029791","FBgn0029795","FBgn0029798","FBgn0029799","FBgn0029800","FBgn0029801","FBgn0029803","FBgn0029804","FBgn0029807","FBgn0029808","FBgn0029813","FBgn0029818","FBgn0029819","FBgn0029820","FBgn0029821","FBgn0029822","FBgn0029823","FBgn0029824","FBgn0029825","FBgn0029826","FBgn0029830","FBgn0029833","FBgn0029834","FBgn0029840","FBgn0029846","FBgn0029848","FBgn0029849","FBgn0029850","FBgn0029851","FBgn0029853","FBgn0029854","FBgn0029856","FBgn0029857","FBgn0029858","FBgn0029861","FBgn0029866","FBgn0029867","FBgn0029868","FBgn0029870","FBgn0029873","FBgn0029874","FBgn0029877","FBgn0029878","FBgn0029879","FBgn0029880","FBgn0029881","FBgn0029882","FBgn0029885","FBgn0029887","FBgn0029888","FBgn0029890","FBgn0029891","FBgn0029892","FBgn0029893","FBgn0029894","FBgn0029895","FBgn0029896","FBgn0029897","FBgn0029898","FBgn0029899","FBgn0029903","FBgn0029905","FBgn0029909","FBgn0029911","FBgn0029912","FBgn0029913","FBgn0029914","FBgn0029915","FBgn0029924","FBgn0029925","FBgn0029928","FBgn0029929","FBgn0029935","FBgn0029936","FBgn0029937","FBgn0029939","FBgn0029941","FBgn0029942","FBgn0029943","FBgn0029944","FBgn0029945","FBgn0029950","FBgn0029951","FBgn0029957","FBgn0029958","FBgn0029959","FBgn0029968","FBgn0029969","FBgn0029970","FBgn0029971","FBgn0029974","FBgn0029975","FBgn0029976","FBgn0029977","FBgn0029979","FBgn0029980","FBgn0029986","FBgn0029989","FBgn0029990","FBgn0029992","FBgn0029994","FBgn0029996","FBgn0029997","FBgn0029999","FBgn0030000","FBgn0030001","FBgn0030003","FBgn0030004","FBgn0030006","FBgn0030007","FBgn0030008","FBgn0030010","FBgn0030011","FBgn0030012","FBgn0030013","FBgn0030017","FBgn0030018","FBgn0030025","FBgn0030026","FBgn0030028","FBgn0030029","FBgn0030030","FBgn0030034","FBgn0030035","FBgn0030037","FBgn0030038","FBgn0030039","FBgn0030040","FBgn0030041","FBgn0030048","FBgn0030049","FBgn0030051","FBgn0030052","FBgn0030053","FBgn0030054","FBgn0030055","FBgn0030056","FBgn0030057","FBgn0030060","FBgn0030061","FBgn0030063","FBgn0030065","FBgn0030066","FBgn0030067","FBgn0030071","FBgn0030073","FBgn0030077","FBgn0030078","FBgn0030079","FBgn0030081","FBgn0030082","FBgn0030086","FBgn0030087","FBgn0030088","FBgn0030089","FBgn0030090","FBgn0030091","FBgn0030092","FBgn0030093","FBgn0030096","FBgn0030099","FBgn0030100","FBgn0030101","FBgn0030109","FBgn0030114","FBgn0030120","FBgn0030121","FBgn0030122","FBgn0030136","FBgn0030137","FBgn0030141","FBgn0030142","FBgn0030144","FBgn0030145","FBgn0030151","FBgn0030160","FBgn0030170","FBgn0030171","FBgn0030177","FBgn0030178","FBgn0030183","FBgn0030189","FBgn0030195","FBgn0030196","FBgn0030205","FBgn0030206","FBgn0030208","FBgn0030217","FBgn0030218","FBgn0030228","FBgn0030230","FBgn0030234","FBgn0030235","FBgn0030237","FBgn0030239","FBgn0030240","FBgn0030241","FBgn0030242","FBgn0030243","FBgn0030244","FBgn0030245","FBgn0030246","FBgn0030249","FBgn0030252","FBgn0030262","FBgn0030263","FBgn0030266","FBgn0030268","FBgn0030269","FBgn0030270","FBgn0030271","FBgn0030272","FBgn0030274","FBgn0030276","FBgn0030286","FBgn0030289","FBgn0030291","FBgn0030292","FBgn0030293","FBgn0030294","FBgn0030299","FBgn0030300","FBgn0030301","FBgn0030305","FBgn0030306","FBgn0030309","FBgn0030310","FBgn0030311","FBgn0030314","FBgn0030316","FBgn0030317","FBgn0030318","FBgn0030319","FBgn0030320","FBgn0030321","FBgn0030322","FBgn0030323","FBgn0030326","FBgn0030327","FBgn0030328","FBgn0030329","FBgn0030330","FBgn0030331","FBgn0030332","FBgn0030334","FBgn0030336","FBgn0030340","FBgn0030341","FBgn0030342","FBgn0030343","FBgn0030344","FBgn0030345","FBgn0030346","FBgn0030347","FBgn0030348","FBgn0030349","FBgn0030350","FBgn0030351","FBgn0030352","FBgn0030354","FBgn0030357","FBgn0030358","FBgn0030360","FBgn0030361","FBgn0030362","FBgn0030364","FBgn0030365","FBgn0030366","FBgn0030367","FBgn0030369","FBgn0030373","FBgn0030377","FBgn0030391","FBgn0030394","FBgn0030396","FBgn0030399","FBgn0030400","FBgn0030403","FBgn0030406","FBgn0030407","FBgn0030410","FBgn0030411","FBgn0030412","FBgn0030417","FBgn0030418","FBgn0030420","FBgn0030421","FBgn0030425","FBgn0030431","FBgn0030432","FBgn0030433","FBgn0030434","FBgn0030435","FBgn0030447","FBgn0030448","FBgn0030451","FBgn0030452","FBgn0030456","FBgn0030457","FBgn0030459","FBgn0030460","FBgn0030465","FBgn0030466","FBgn0030467","FBgn0030468","FBgn0030469","FBgn0030474","FBgn0030478","FBgn0030479","FBgn0030480","FBgn0030481","FBgn0030482","FBgn0030484","FBgn0030485","FBgn0030486","FBgn0030499","FBgn0030500","FBgn0030501","FBgn0030502","FBgn0030503","FBgn0030504","FBgn0030505","FBgn0030506","FBgn0030507","FBgn0030508","FBgn0030510","FBgn0030511","FBgn0030512","FBgn0030514","FBgn0030518","FBgn0030519","FBgn0030520","FBgn0030521","FBgn0030522","FBgn0030524","FBgn0030528","FBgn0030529","FBgn0030530","FBgn0030532","FBgn0030545","FBgn0030552","FBgn0030554","FBgn0030555","FBgn0030556","FBgn0030562","FBgn0030571","FBgn0030572","FBgn0030573","FBgn0030574","FBgn0030575","FBgn0030576","FBgn0030581","FBgn0030582","FBgn0030583","FBgn0030584","FBgn0030587","FBgn0030588","FBgn0030590","FBgn0030592","FBgn0030593","FBgn0030594","FBgn0030596","FBgn0030597","FBgn0030598","FBgn0030600","FBgn0030603","FBgn0030605","FBgn0030606","FBgn0030607","FBgn0030610","FBgn0030611","FBgn0030612","FBgn0030613","FBgn0030615","FBgn0030616","FBgn0030625","FBgn0030626","FBgn0030627","FBgn0030628","FBgn0030629","FBgn0030630","FBgn0030631","FBgn0030634","FBgn0030636","FBgn0030638","FBgn0030642","FBgn0030643","FBgn0030645","FBgn0030646","FBgn0030647","FBgn0030648","FBgn0030653","FBgn0030654","FBgn0030655","FBgn0030657","FBgn0030658","FBgn0030659","FBgn0030660","FBgn0030661","FBgn0030662","FBgn0030664","FBgn0030665","FBgn0030666","FBgn0030667","FBgn0030668","FBgn0030669","FBgn0030670","FBgn0030671","FBgn0030672","FBgn0030673","FBgn0030674","FBgn0030675","FBgn0030676","FBgn0030678","FBgn0030679","FBgn0030680","FBgn0030683","FBgn0030685","FBgn0030686","FBgn0030687","FBgn0030691","FBgn0030692","FBgn0030693","FBgn0030694","FBgn0030695","FBgn0030696","FBgn0030699","FBgn0030700","FBgn0030701","FBgn0030703","FBgn0030704","FBgn0030706","FBgn0030710","FBgn0030711","FBgn0030716","FBgn0030717","FBgn0030718","FBgn0030719","FBgn0030720","FBgn0030721","FBgn0030722","FBgn0030723","FBgn0030724","FBgn0030725","FBgn0030731","FBgn0030734","FBgn0030735","FBgn0030737","FBgn0030738","FBgn0030739","FBgn0030740","FBgn0030742","FBgn0030743","FBgn0030744","FBgn0030745","FBgn0030746","FBgn0030747","FBgn0030748","FBgn0030749","FBgn0030752","FBgn0030753","FBgn0030755","FBgn0030757","FBgn0030758","FBgn0030759","FBgn0030761","FBgn0030764","FBgn0030765","FBgn0030766","FBgn0030768","FBgn0030786","FBgn0030787","FBgn0030788","FBgn0030789","FBgn0030790","FBgn0030791","FBgn0030792","FBgn0030793","FBgn0030794","FBgn0030795","FBgn0030796","FBgn0030797","FBgn0030799","FBgn0030800","FBgn0030801","FBgn0030802","FBgn0030803","FBgn0030805","FBgn0030806","FBgn0030807","FBgn0030808","FBgn0030809","FBgn0030812","FBgn0030813","FBgn0030816","FBgn0030817","FBgn0030828","FBgn0030833","FBgn0030834","FBgn0030838","FBgn0030839","FBgn0030842","FBgn0030847","FBgn0030850","FBgn0030851","FBgn0030852","FBgn0030853","FBgn0030854","FBgn0030855","FBgn0030858","FBgn0030859","FBgn0030863","FBgn0030864","FBgn0030869","FBgn0030871","FBgn0030872","FBgn0030873","FBgn0030874","FBgn0030876","FBgn0030877","FBgn0030878","FBgn0030883","FBgn0030884","FBgn0030890","FBgn0030891","FBgn0030892","FBgn0030893","FBgn0030894","FBgn0030904","FBgn0030915","FBgn0030918","FBgn0030925","FBgn0030927","FBgn0030928","FBgn0030929","FBgn0030930","FBgn0030931","FBgn0030932","FBgn0030933","FBgn0030938","FBgn0030941","FBgn0030943","FBgn0030944","FBgn0030945","FBgn0030946","FBgn0030954","FBgn0030955","FBgn0030956","FBgn0030958","FBgn0030959","FBgn0030960","FBgn0030961","FBgn0030963","FBgn0030966","FBgn0030968","FBgn0030969","FBgn0030970","FBgn0030973","FBgn0030974","FBgn0030985","FBgn0030989","FBgn0030990","FBgn0030991","FBgn0030992","FBgn0030993","FBgn0030994","FBgn0030995","FBgn0030996","FBgn0030997","FBgn0031002","FBgn0031003","FBgn0031006","FBgn0031008","FBgn0031010","FBgn0031011","FBgn0031012","FBgn0031016","FBgn0031018","FBgn0031020","FBgn0031021","FBgn0031022","FBgn0031023","FBgn0031030","FBgn0031031","FBgn0031032","FBgn0031033","FBgn0031034","FBgn0031035","FBgn0031036","FBgn0031037","FBgn0031039","FBgn0031040","FBgn0031042","FBgn0031043","FBgn0031044","FBgn0031045","FBgn0031047","FBgn0031048","FBgn0031049","FBgn0031050","FBgn0031051","FBgn0031052","FBgn0031053","FBgn0031054","FBgn0031055","FBgn0031057","FBgn0031058","FBgn0031059","FBgn0031060","FBgn0031061","FBgn0031062","FBgn0031066","FBgn0031068","FBgn0031069","FBgn0031070","FBgn0031074","FBgn0031077","FBgn0031078","FBgn0031079","FBgn0031080","FBgn0031081","FBgn0031088","FBgn0031089","FBgn0031090","FBgn0031091","FBgn0031092","FBgn0031093","FBgn0031094","FBgn0031097","FBgn0031098","FBgn0031099","FBgn0031106","FBgn0031107","FBgn0031114","FBgn0031115","FBgn0031116","FBgn0031117","FBgn0031118","FBgn0031119","FBgn0031126","FBgn0031127","FBgn0031128","FBgn0031142","FBgn0031143","FBgn0031144","FBgn0031145","FBgn0031146","FBgn0031148","FBgn0031149","FBgn0031150","FBgn0031155","FBgn0031157","FBgn0031161","FBgn0031168","FBgn0031169","FBgn0031170","FBgn0031174","FBgn0031182","FBgn0031183","FBgn0031184","FBgn0031186","FBgn0031187","FBgn0031188","FBgn0031189","FBgn0031190","FBgn0031191","FBgn0031194","FBgn0031195","FBgn0031196","FBgn0031201","FBgn0031213","FBgn0031214","FBgn0031216","FBgn0031217","FBgn0031219","FBgn0031220","FBgn0031224","FBgn0031227","FBgn0031228","FBgn0031229","FBgn0031231","FBgn0031233","FBgn0031238","FBgn0031239","FBgn0031240","FBgn0031244","FBgn0031245","FBgn0031247","FBgn0031250","FBgn0031252","FBgn0031253","FBgn0031254","FBgn0031255","FBgn0031256","FBgn0031257","FBgn0031258","FBgn0031260","FBgn0031261","FBgn0031263","FBgn0031264","FBgn0031265","FBgn0031266","FBgn0031267","FBgn0031268","FBgn0031270","FBgn0031281","FBgn0031282","FBgn0031283","FBgn0031284","FBgn0031285","FBgn0031286","FBgn0031287","FBgn0031292","FBgn0031296","FBgn0031298","FBgn0031299","FBgn0031300","FBgn0031302","FBgn0031304","FBgn0031307","FBgn0031308","FBgn0031309","FBgn0031310","FBgn0031312","FBgn0031313","FBgn0031314","FBgn0031315","FBgn0031317","FBgn0031318","FBgn0031319","FBgn0031320","FBgn0031321","FBgn0031322","FBgn0031327","FBgn0031344","FBgn0031351","FBgn0031356","FBgn0031357","FBgn0031359","FBgn0031360","FBgn0031361","FBgn0031362","FBgn0031374","FBgn0031376","FBgn0031377","FBgn0031378","FBgn0031379","FBgn0031381","FBgn0031384","FBgn0031389","FBgn0031390","FBgn0031391","FBgn0031392","FBgn0031395","FBgn0031397","FBgn0031398","FBgn0031399","FBgn0031401","FBgn0031403","FBgn0031405","FBgn0031408","FBgn0031413","FBgn0031414","FBgn0031418","FBgn0031419","FBgn0031420","FBgn0031421","FBgn0031422","FBgn0031428","FBgn0031429","FBgn0031434","FBgn0031435","FBgn0031436","FBgn0031437","FBgn0031449","FBgn0031450","FBgn0031451","FBgn0031452","FBgn0031453","FBgn0031456","FBgn0031457","FBgn0031458","FBgn0031459","FBgn0031460","FBgn0031461","FBgn0031464","FBgn0031470","FBgn0031471","FBgn0031474","FBgn0031476","FBgn0031478","FBgn0031479","FBgn0031483","FBgn0031484","FBgn0031485","FBgn0031488","FBgn0031489","FBgn0031490","FBgn0031491","FBgn0031492","FBgn0031493","FBgn0031494","FBgn0031495","FBgn0031496","FBgn0031497","FBgn0031498","FBgn0031500","FBgn0031505","FBgn0031513","FBgn0031515","FBgn0031516","FBgn0031517","FBgn0031518","FBgn0031519","FBgn0031529","FBgn0031530","FBgn0031534","FBgn0031535","FBgn0031536","FBgn0031537","FBgn0031538","FBgn0031540","FBgn0031544","FBgn0031547","FBgn0031548","FBgn0031549","FBgn0031566","FBgn0031568","FBgn0031571","FBgn0031573","FBgn0031574","FBgn0031575","FBgn0031580","FBgn0031581","FBgn0031589","FBgn0031590","FBgn0031597","FBgn0031598","FBgn0031599","FBgn0031600","FBgn0031601","FBgn0031603","FBgn0031604","FBgn0031606","FBgn0031607","FBgn0031608","FBgn0031609","FBgn0031610","FBgn0031611","FBgn0031622","FBgn0031628","FBgn0031630","FBgn0031631","FBgn0031633","FBgn0031634","FBgn0031635","FBgn0031637","FBgn0031638","FBgn0031639","FBgn0031640","FBgn0031643","FBgn0031644","FBgn0031645","FBgn0031646","FBgn0031651","FBgn0031652","FBgn0031655","FBgn0031656","FBgn0031657","FBgn0031659","FBgn0031660","FBgn0031661","FBgn0031662","FBgn0031663","FBgn0031664","FBgn0031670","FBgn0031673","FBgn0031675","FBgn0031676","FBgn0031677","FBgn0031678","FBgn0031681","FBgn0031682","FBgn0031683","FBgn0031684","FBgn0031688","FBgn0031689","FBgn0031690","FBgn0031693","FBgn0031694","FBgn0031695","FBgn0031696","FBgn0031698","FBgn0031703","FBgn0031707","FBgn0031708","FBgn0031710","FBgn0031711","FBgn0031713","FBgn0031717","FBgn0031735","FBgn0031736","FBgn0031737","FBgn0031738","FBgn0031739","FBgn0031740","FBgn0031747","FBgn0031752","FBgn0031756","FBgn0031757","FBgn0031758","FBgn0031759","FBgn0031760","FBgn0031762","FBgn0031764","FBgn0031768","FBgn0031769","FBgn0031771","FBgn0031772","FBgn0031773","FBgn0031774","FBgn0031775","FBgn0031776","FBgn0031777","FBgn0031779","FBgn0031781","FBgn0031782","FBgn0031799","FBgn0031805","FBgn0031811","FBgn0031812","FBgn0031813","FBgn0031814","FBgn0031815","FBgn0031816","FBgn0031817","FBgn0031818","FBgn0031822","FBgn0031824","FBgn0031829","FBgn0031830","FBgn0031831","FBgn0031832","FBgn0031834","FBgn0031836","FBgn0031842","FBgn0031844","FBgn0031845","FBgn0031848","FBgn0031850","FBgn0031851","FBgn0031857","FBgn0031865","FBgn0031868","FBgn0031869","FBgn0031871","FBgn0031872","FBgn0031873","FBgn0031874","FBgn0031875","FBgn0031876","FBgn0031877","FBgn0031878","FBgn0031880","FBgn0031881","FBgn0031882","FBgn0031883","FBgn0031885","FBgn0031886","FBgn0031887","FBgn0031888","FBgn0031893","FBgn0031894","FBgn0031895","FBgn0031896","FBgn0031897","FBgn0031904","FBgn0031907","FBgn0031909","FBgn0031912","FBgn0031913","FBgn0031914","FBgn0031920","FBgn0031936","FBgn0031937","FBgn0031947","FBgn0031948","FBgn0031950","FBgn0031951","FBgn0031952","FBgn0031955","FBgn0031957","FBgn0031961","FBgn0031962","FBgn0031968","FBgn0031969","FBgn0031970","FBgn0031971","FBgn0031972","FBgn0031973","FBgn0031974","FBgn0031975","FBgn0031976","FBgn0031977","FBgn0031979","FBgn0031980","FBgn0031981","FBgn0031985","FBgn0031987","FBgn0031988","FBgn0031990","FBgn0031992","FBgn0031993","FBgn0031995","FBgn0031996","FBgn0031997","FBgn0031998","FBgn0031999","FBgn0032000","FBgn0032001","FBgn0032002","FBgn0032003","FBgn0032004","FBgn0032005","FBgn0032006","FBgn0032010","FBgn0032013","FBgn0032014","FBgn0032015","FBgn0032016","FBgn0032017","FBgn0032018","FBgn0032019","FBgn0032020","FBgn0032024","FBgn0032025","FBgn0032026","FBgn0032029","FBgn0032030","FBgn0032031","FBgn0032032","FBgn0032033","FBgn0032034","FBgn0032035","FBgn0032036","FBgn0032040","FBgn0032042","FBgn0032047","FBgn0032050","FBgn0032051","FBgn0032052","FBgn0032053","FBgn0032054","FBgn0032059","FBgn0032061","FBgn0032078","FBgn0032079","FBgn0032080","FBgn0032082","FBgn0032083","FBgn0032084","FBgn0032085","FBgn0032101","FBgn0032105","FBgn0032117","FBgn0032120","FBgn0032123","FBgn0032129","FBgn0032130","FBgn0032135","FBgn0032136","FBgn0032138","FBgn0032139","FBgn0032140","FBgn0032142","FBgn0032147","FBgn0032149","FBgn0032150","FBgn0032153","FBgn0032154","FBgn0032156","FBgn0032157","FBgn0032160","FBgn0032161","FBgn0032162","FBgn0032163","FBgn0032166","FBgn0032167","FBgn0032168","FBgn0032169","FBgn0032170","FBgn0032171","FBgn0032172","FBgn0032178","FBgn0032189","FBgn0032191","FBgn0032192","FBgn0032193","FBgn0032194","FBgn0032195","FBgn0032196","FBgn0032197","FBgn0032198","FBgn0032200","FBgn0032202","FBgn0032204","FBgn0032205","FBgn0032208","FBgn0032210","FBgn0032213","FBgn0032214","FBgn0032215","FBgn0032216","FBgn0032217","FBgn0032218","FBgn0032219","FBgn0032221","FBgn0032222","FBgn0032223","FBgn0032224","FBgn0032228","FBgn0032229","FBgn0032230","FBgn0032231","FBgn0032233","FBgn0032234","FBgn0032235","FBgn0032236","FBgn0032237","FBgn0032242","FBgn0032243","FBgn0032244","FBgn0032246","FBgn0032247","FBgn0032248","FBgn0032249","FBgn0032250","FBgn0032251","FBgn0032252","FBgn0032253","FBgn0032256","FBgn0032258","FBgn0032259","FBgn0032261","FBgn0032262","FBgn0032264","FBgn0032290","FBgn0032292","FBgn0032293","FBgn0032295","FBgn0032296","FBgn0032297","FBgn0032298","FBgn0032305","FBgn0032310","FBgn0032321","FBgn0032322","FBgn0032329","FBgn0032330","FBgn0032339","FBgn0032340","FBgn0032341","FBgn0032343","FBgn0032345","FBgn0032346","FBgn0032348","FBgn0032350","FBgn0032354","FBgn0032358","FBgn0032361","FBgn0032363","FBgn0032373","FBgn0032377","FBgn0032378","FBgn0032382","FBgn0032387","FBgn0032388","FBgn0032390","FBgn0032391","FBgn0032393","FBgn0032394","FBgn0032395","FBgn0032397","FBgn0032398","FBgn0032399","FBgn0032400","FBgn0032401","FBgn0032402","FBgn0032405","FBgn0032407","FBgn0032408","FBgn0032409","FBgn0032420","FBgn0032421","FBgn0032422","FBgn0032428","FBgn0032429","FBgn0032430","FBgn0032433","FBgn0032436","FBgn0032444","FBgn0032445","FBgn0032446","FBgn0032447","FBgn0032449","FBgn0032450","FBgn0032451","FBgn0032452","FBgn0032453","FBgn0032454","FBgn0032455","FBgn0032456","FBgn0032465","FBgn0032467","FBgn0032469","FBgn0032470","FBgn0032473","FBgn0032474","FBgn0032475","FBgn0032476","FBgn0032477","FBgn0032478","FBgn0032479","FBgn0032480","FBgn0032481","FBgn0032482","FBgn0032483","FBgn0032485","FBgn0032486","FBgn0032487","FBgn0032488","FBgn0032497","FBgn0032498","FBgn0032499","FBgn0032504","FBgn0032509","FBgn0032511","FBgn0032512","FBgn0032513","FBgn0032514","FBgn0032515","FBgn0032516","FBgn0032517","FBgn0032518","FBgn0032520","FBgn0032521","FBgn0032524","FBgn0032530","FBgn0032533","FBgn0032535","FBgn0032586","FBgn0032587","FBgn0032590","FBgn0032595","FBgn0032596","FBgn0032597","FBgn0032598","FBgn0032600","FBgn0032601","FBgn0032602","FBgn0032603","FBgn0032613","FBgn0032620","FBgn0032621","FBgn0032622","FBgn0032633","FBgn0032634","FBgn0032635","FBgn0032637","FBgn0032638","FBgn0032640","FBgn0032642","FBgn0032643","FBgn0032644","FBgn0032646","FBgn0032647","FBgn0032656","FBgn0032666","FBgn0032670","FBgn0032671","FBgn0032679","FBgn0032680","FBgn0032681","FBgn0032683","FBgn0032684","FBgn0032685","FBgn0032688","FBgn0032689","FBgn0032690","FBgn0032691","FBgn0032693","FBgn0032694","FBgn0032698","FBgn0032699","FBgn0032700","FBgn0032701","FBgn0032702","FBgn0032703","FBgn0032704","FBgn0032705","FBgn0032713","FBgn0032715","FBgn0032717","FBgn0032719","FBgn0032720","FBgn0032721","FBgn0032723","FBgn0032724","FBgn0032725","FBgn0032726","FBgn0032727","FBgn0032728","FBgn0032729","FBgn0032730","FBgn0032731","FBgn0032732","FBgn0032744","FBgn0032746","FBgn0032748","FBgn0032750","FBgn0032751","FBgn0032752","FBgn0032763","FBgn0032772","FBgn0032773","FBgn0032774","FBgn0032775","FBgn0032779","FBgn0032780","FBgn0032781","FBgn0032782","FBgn0032783","FBgn0032785","FBgn0032787","FBgn0032790","FBgn0032791","FBgn0032793","FBgn0032796","FBgn0032797","FBgn0032798","FBgn0032799","FBgn0032800","FBgn0032801","FBgn0032805","FBgn0032808","FBgn0032809","FBgn0032810","FBgn0032811","FBgn0032812","FBgn0032813","FBgn0032814","FBgn0032815","FBgn0032816","FBgn0032817","FBgn0032818","FBgn0032819","FBgn0032820","FBgn0032821","FBgn0032822","FBgn0032833","FBgn0032843","FBgn0032845","FBgn0032846","FBgn0032847","FBgn0032848","FBgn0032849","FBgn0032858","FBgn0032859","FBgn0032863","FBgn0032864","FBgn0032869","FBgn0032870","FBgn0032871","FBgn0032872","FBgn0032873","FBgn0032876","FBgn0032877","FBgn0032879","FBgn0032880","FBgn0032881","FBgn0032882","FBgn0032883","FBgn0032884","FBgn0032886","FBgn0032889","FBgn0032891","FBgn0032895","FBgn0032897","FBgn0032899","FBgn0032900","FBgn0032901","FBgn0032904","FBgn0032906","FBgn0032907","FBgn0032908","FBgn0032910","FBgn0032915","FBgn0032916","FBgn0032919","FBgn0032921","FBgn0032922","FBgn0032923","FBgn0032924","FBgn0032925","FBgn0032926","FBgn0032929","FBgn0032934","FBgn0032935","FBgn0032938","FBgn0032940","FBgn0032943","FBgn0032946","FBgn0032949","FBgn0032955","FBgn0032956","FBgn0032957","FBgn0032961","FBgn0032964","FBgn0032965","FBgn0032967","FBgn0032968","FBgn0032972","FBgn0032973","FBgn0032974","FBgn0032976","FBgn0032978","FBgn0032979","FBgn0032981","FBgn0032986","FBgn0032987","FBgn0032988","FBgn0032997","FBgn0033000","FBgn0033005","FBgn0033010","FBgn0033015","FBgn0033017","FBgn0033019","FBgn0033021","FBgn0033027","FBgn0033028","FBgn0033029","FBgn0033032","FBgn0033033","FBgn0033038","FBgn0033039","FBgn0033049","FBgn0033050","FBgn0033051","FBgn0033052","FBgn0033055","FBgn0033059","FBgn0033060","FBgn0033061","FBgn0033062","FBgn0033065","FBgn0033068","FBgn0033073","FBgn0033074","FBgn0033075","FBgn0033079","FBgn0033081","FBgn0033083","FBgn0033087","FBgn0033088","FBgn0033089","FBgn0033090","FBgn0033092","FBgn0033093","FBgn0033095","FBgn0033100","FBgn0033101","FBgn0033104","FBgn0033107","FBgn0033108","FBgn0033109","FBgn0033110","FBgn0033113","FBgn0033115","FBgn0033117","FBgn0033121","FBgn0033122","FBgn0033124","FBgn0033127","FBgn0033128","FBgn0033130","FBgn0033132","FBgn0033133","FBgn0033134","FBgn0033136","FBgn0033137","FBgn0033153","FBgn0033155","FBgn0033159","FBgn0033160","FBgn0033162","FBgn0033166","FBgn0033169","FBgn0033170","FBgn0033174","FBgn0033177","FBgn0033178","FBgn0033179","FBgn0033182","FBgn0033183","FBgn0033184","FBgn0033185","FBgn0033186","FBgn0033187","FBgn0033188","FBgn0033189","FBgn0033190","FBgn0033191","FBgn0033192","FBgn0033194","FBgn0033195","FBgn0033196","FBgn0033199","FBgn0033204","FBgn0033205","FBgn0033206","FBgn0033208","FBgn0033209","FBgn0033210","FBgn0033212","FBgn0033214","FBgn0033221","FBgn0033224","FBgn0033225","FBgn0033226","FBgn0033232","FBgn0033233","FBgn0033234","FBgn0033235","FBgn0033236","FBgn0033240","FBgn0033241","FBgn0033243","FBgn0033244","FBgn0033246","FBgn0033247","FBgn0033252","FBgn0033257","FBgn0033258","FBgn0033259","FBgn0033260","FBgn0033261","FBgn0033264","FBgn0033265","FBgn0033266","FBgn0033268","FBgn0033269","FBgn0033272","FBgn0033273","FBgn0033274","FBgn0033277","FBgn0033282","FBgn0033286","FBgn0033288","FBgn0033292","FBgn0033302","FBgn0033304","FBgn0033308","FBgn0033309","FBgn0033310","FBgn0033312","FBgn0033313","FBgn0033315","FBgn0033316","FBgn0033317","FBgn0033320","FBgn0033321","FBgn0033337","FBgn0033339","FBgn0033340","FBgn0033341","FBgn0033342","FBgn0033344","FBgn0033347","FBgn0033348","FBgn0033349","FBgn0033350","FBgn0033351","FBgn0033352","FBgn0033353","FBgn0033354","FBgn0033356","FBgn0033357","FBgn0033359","FBgn0033362","FBgn0033363","FBgn0033364","FBgn0033366","FBgn0033367","FBgn0033368","FBgn0033372","FBgn0033373","FBgn0033375","FBgn0033376","FBgn0033377","FBgn0033378","FBgn0033379","FBgn0033380","FBgn0033381","FBgn0033382","FBgn0033387","FBgn0033388","FBgn0033389","FBgn0033391","FBgn0033392","FBgn0033393","FBgn0033397","FBgn0033400","FBgn0033401","FBgn0033402","FBgn0033403","FBgn0033405","FBgn0033413","FBgn0033421","FBgn0033422","FBgn0033423","FBgn0033426","FBgn0033427","FBgn0033428","FBgn0033429","FBgn0033431","FBgn0033434","FBgn0033437","FBgn0033438","FBgn0033446","FBgn0033448","FBgn0033449","FBgn0033450","FBgn0033451","FBgn0033452","FBgn0033453","FBgn0033454","FBgn0033457","FBgn0033458","FBgn0033459","FBgn0033460","FBgn0033463","FBgn0033465","FBgn0033466","FBgn0033467","FBgn0033468","FBgn0033469","FBgn0033471","FBgn0033473","FBgn0033474","FBgn0033475","FBgn0033476","FBgn0033477","FBgn0033478","FBgn0033479","FBgn0033480","FBgn0033481","FBgn0033482","FBgn0033483","FBgn0033485","FBgn0033486","FBgn0033491","FBgn0033494","FBgn0033495","FBgn0033501","FBgn0033502","FBgn0033504","FBgn0033507","FBgn0033518","FBgn0033519","FBgn0033523","FBgn0033524","FBgn0033526","FBgn0033527","FBgn0033528","FBgn0033529","FBgn0033538","FBgn0033539","FBgn0033540","FBgn0033541","FBgn0033543","FBgn0033544","FBgn0033547","FBgn0033548","FBgn0033549","FBgn0033550","FBgn0033551","FBgn0033554","FBgn0033555","FBgn0033556","FBgn0033557","FBgn0033562","FBgn0033566","FBgn0033569","FBgn0033570","FBgn0033571","FBgn0033578","FBgn0033579","FBgn0033581","FBgn0033584","FBgn0033605","FBgn0033607","FBgn0033608","FBgn0033609","FBgn0033610","FBgn0033615","FBgn0033616","FBgn0033624","FBgn0033627","FBgn0033631","FBgn0033635","FBgn0033636","FBgn0033638","FBgn0033639","FBgn0033644","FBgn0033645","FBgn0033649","FBgn0033653","FBgn0033654","FBgn0033656","FBgn0033661","FBgn0033663","FBgn0033665","FBgn0033668","FBgn0033669","FBgn0033672","FBgn0033673","FBgn0033677","FBgn0033683","FBgn0033685","FBgn0033686","FBgn0033688","FBgn0033690","FBgn0033691","FBgn0033692","FBgn0033696","FBgn0033697","FBgn0033698","FBgn0033699","FBgn0033702","FBgn0033706","FBgn0033712","FBgn0033713","FBgn0033714","FBgn0033715","FBgn0033716","FBgn0033717","FBgn0033718","FBgn0033720","FBgn0033724","FBgn0033725","FBgn0033734","FBgn0033735","FBgn0033737","FBgn0033738","FBgn0033739","FBgn0033740","FBgn0033741","FBgn0033744","FBgn0033748","FBgn0033749","FBgn0033750","FBgn0033751","FBgn0033752","FBgn0033754","FBgn0033755","FBgn0033756","FBgn0033757","FBgn0033761","FBgn0033762","FBgn0033763","FBgn0033764","FBgn0033765","FBgn0033766","FBgn0033767","FBgn0033769","FBgn0033775","FBgn0033777","FBgn0033778","FBgn0033781","FBgn0033782","FBgn0033783","FBgn0033784","FBgn0033785","FBgn0033786","FBgn0033787","FBgn0033798","FBgn0033799","FBgn0033802","FBgn0033806","FBgn0033808","FBgn0033809","FBgn0033810","FBgn0033812","FBgn0033813","FBgn0033814","FBgn0033816","FBgn0033821","FBgn0033836","FBgn0033842","FBgn0033844","FBgn0033845","FBgn0033846","FBgn0033853","FBgn0033857","FBgn0033859","FBgn0033869","FBgn0033871","FBgn0033872","FBgn0033875","FBgn0033876","FBgn0033879","FBgn0033880","FBgn0033882","FBgn0033883","FBgn0033884","FBgn0033886","FBgn0033887","FBgn0033888","FBgn0033889","FBgn0033890","FBgn0033891","FBgn0033893","FBgn0033897","FBgn0033899","FBgn0033900","FBgn0033901","FBgn0033902","FBgn0033903","FBgn0033905","FBgn0033906","FBgn0033907","FBgn0033912","FBgn0033913","FBgn0033915","FBgn0033916","FBgn0033917","FBgn0033918","FBgn0033919","FBgn0033921","FBgn0033924","FBgn0033925","FBgn0033926","FBgn0033928","FBgn0033929","FBgn0033934","FBgn0033935","FBgn0033936","FBgn0033943","FBgn0033945","FBgn0033948","FBgn0033949","FBgn0033951","FBgn0033960","FBgn0033961","FBgn0033962","FBgn0033963","FBgn0033971","FBgn0033972","FBgn0033973","FBgn0033978","FBgn0033979","FBgn0033980","FBgn0033982","FBgn0033984","FBgn0033987","FBgn0033988","FBgn0033989","FBgn0033990","FBgn0033993","FBgn0033994","FBgn0033995","FBgn0033996","FBgn0033998","FBgn0034000","FBgn0034001","FBgn0034002","FBgn0034005","FBgn0034008","FBgn0034009","FBgn0034010","FBgn0034011","FBgn0034012","FBgn0034013","FBgn0034021","FBgn0034025","FBgn0034027","FBgn0034029","FBgn0034030","FBgn0034032","FBgn0034033","FBgn0034035","FBgn0034037","FBgn0034046","FBgn0034049","FBgn0034050","FBgn0034051","FBgn0034053","FBgn0034054","FBgn0034057","FBgn0034058","FBgn0034060","FBgn0034061","FBgn0034062","FBgn0034063","FBgn0034065","FBgn0034066","FBgn0034067","FBgn0034068","FBgn0034071","FBgn0034072","FBgn0034073","FBgn0034075","FBgn0034076","FBgn0034082","FBgn0034083","FBgn0034084","FBgn0034085","FBgn0034086","FBgn0034087","FBgn0034088","FBgn0034089","FBgn0034091","FBgn0034093","FBgn0034094","FBgn0034095","FBgn0034098","FBgn0034109","FBgn0034110","FBgn0034113","FBgn0034114","FBgn0034117","FBgn0034118","FBgn0034126","FBgn0034127","FBgn0034136","FBgn0034137","FBgn0034138","FBgn0034139","FBgn0034140","FBgn0034141","FBgn0034142","FBgn0034154","FBgn0034155","FBgn0034158","FBgn0034159","FBgn0034160","FBgn0034162","FBgn0034166","FBgn0034172","FBgn0034175","FBgn0034176","FBgn0034177","FBgn0034179","FBgn0034180","FBgn0034181","FBgn0034182","FBgn0034184","FBgn0034186","FBgn0034187","FBgn0034191","FBgn0034194","FBgn0034198","FBgn0034199","FBgn0034200","FBgn0034210","FBgn0034214","FBgn0034215","FBgn0034217","FBgn0034219","FBgn0034221","FBgn0034223","FBgn0034225","FBgn0034230","FBgn0034231","FBgn0034232","FBgn0034237","FBgn0034238","FBgn0034239","FBgn0034240","FBgn0034242","FBgn0034243","FBgn0034245","FBgn0034246","FBgn0034248","FBgn0034249","FBgn0034253","FBgn0034255","FBgn0034258","FBgn0034259","FBgn0034261","FBgn0034264","FBgn0034265","FBgn0034267","FBgn0034269","FBgn0034270","FBgn0034271","FBgn0034277","FBgn0034279","FBgn0034281","FBgn0034282","FBgn0034290","FBgn0034299","FBgn0034300","FBgn0034304","FBgn0034307","FBgn0034308","FBgn0034310","FBgn0034312","FBgn0034313","FBgn0034314","FBgn0034315","FBgn0034321","FBgn0034327","FBgn0034345","FBgn0034346","FBgn0034351","FBgn0034354","FBgn0034356","FBgn0034360","FBgn0034361","FBgn0034362","FBgn0034365","FBgn0034366","FBgn0034367","FBgn0034368","FBgn0034371","FBgn0034376","FBgn0034378","FBgn0034379","FBgn0034380","FBgn0034389","FBgn0034390","FBgn0034391","FBgn0034392","FBgn0034396","FBgn0034397","FBgn0034398","FBgn0034399","FBgn0034400","FBgn0034401","FBgn0034402","FBgn0034403","FBgn0034405","FBgn0034407","FBgn0034408","FBgn0034410","FBgn0034411","FBgn0034417","FBgn0034418","FBgn0034419","FBgn0034420","FBgn0034421","FBgn0034422","FBgn0034425","FBgn0034430","FBgn0034432","FBgn0034433","FBgn0034434","FBgn0034436","FBgn0034438","FBgn0034442","FBgn0034443","FBgn0034447","FBgn0034451","FBgn0034452","FBgn0034455","FBgn0034476","FBgn0034479","FBgn0034488","FBgn0034490","FBgn0034491","FBgn0034493","FBgn0034494","FBgn0034495","FBgn0034496","FBgn0034497","FBgn0034498","FBgn0034500","FBgn0034501","FBgn0034503","FBgn0034504","FBgn0034521","FBgn0034523","FBgn0034527","FBgn0034528","FBgn0034529","FBgn0034530","FBgn0034532","FBgn0034534","FBgn0034535","FBgn0034537","FBgn0034542","FBgn0034543","FBgn0034564","FBgn0034568","FBgn0034569","FBgn0034570","FBgn0034572","FBgn0034573","FBgn0034576","FBgn0034577","FBgn0034578","FBgn0034579","FBgn0034583","FBgn0034585","FBgn0034590","FBgn0034595","FBgn0034598","FBgn0034599","FBgn0034602","FBgn0034603","FBgn0034605","FBgn0034611","FBgn0034612","FBgn0034614","FBgn0034617","FBgn0034618","FBgn0034626","FBgn0034627","FBgn0034628","FBgn0034629","FBgn0034631","FBgn0034634","FBgn0034636","FBgn0034638","FBgn0034639","FBgn0034641","FBgn0034643","FBgn0034644","FBgn0034645","FBgn0034646","FBgn0034647","FBgn0034650","FBgn0034654","FBgn0034655","FBgn0034656","FBgn0034657","FBgn0034662","FBgn0034674","FBgn0034684","FBgn0034687","FBgn0034688","FBgn0034689","FBgn0034691","FBgn0034694","FBgn0034697","FBgn0034703","FBgn0034704","FBgn0034705","FBgn0034706","FBgn0034707","FBgn0034708","FBgn0034709","FBgn0034718","FBgn0034722","FBgn0034724","FBgn0034726","FBgn0034727","FBgn0034728","FBgn0034729","FBgn0034733","FBgn0034734","FBgn0034735","FBgn0034736","FBgn0034741","FBgn0034742","FBgn0034743","FBgn0034744","FBgn0034745","FBgn0034748","FBgn0034750","FBgn0034751","FBgn0034753","FBgn0034756","FBgn0034758","FBgn0034759","FBgn0034761","FBgn0034763","FBgn0034786","FBgn0034789","FBgn0034791","FBgn0034792","FBgn0034793","FBgn0034794","FBgn0034797","FBgn0034800","FBgn0034802","FBgn0034803","FBgn0034804","FBgn0034814","FBgn0034816","FBgn0034817","FBgn0034849","FBgn0034850","FBgn0034853","FBgn0034854","FBgn0034856","FBgn0034858","FBgn0034859","FBgn0034873","FBgn0034876","FBgn0034877","FBgn0034878","FBgn0034879","FBgn0034880","FBgn0034885","FBgn0034886","FBgn0034887","FBgn0034893","FBgn0034894","FBgn0034897","FBgn0034898","FBgn0034902","FBgn0034906","FBgn0034908","FBgn0034909","FBgn0034911","FBgn0034913","FBgn0034914","FBgn0034915","FBgn0034918","FBgn0034919","FBgn0034921","FBgn0034922","FBgn0034923","FBgn0034924","FBgn0034925","FBgn0034926","FBgn0034928","FBgn0034931","FBgn0034933","FBgn0034936","FBgn0034937","FBgn0034938","FBgn0034939","FBgn0034940","FBgn0034945","FBgn0034946","FBgn0034948","FBgn0034950","FBgn0034951","FBgn0034956","FBgn0034958","FBgn0034959","FBgn0034961","FBgn0034962","FBgn0034963","FBgn0034964","FBgn0034965","FBgn0034966","FBgn0034967","FBgn0034968","FBgn0034970","FBgn0034971","FBgn0034975","FBgn0034976","FBgn0034982","FBgn0034985","FBgn0034986","FBgn0034987","FBgn0034988","FBgn0034989","FBgn0034997","FBgn0034999","FBgn0035001","FBgn0035002","FBgn0035016","FBgn0035020","FBgn0035021","FBgn0035023","FBgn0035024","FBgn0035025","FBgn0035026","FBgn0035027","FBgn0035028","FBgn0035032","FBgn0035033","FBgn0035035","FBgn0035036","FBgn0035039","FBgn0035044","FBgn0035046","FBgn0035047","FBgn0035049","FBgn0035059","FBgn0035060","FBgn0035063","FBgn0035064","FBgn0035065","FBgn0035073","FBgn0035076","FBgn0035077","FBgn0035078","FBgn0035082","FBgn0035083","FBgn0035084","FBgn0035085","FBgn0035086","FBgn0035087","FBgn0035088","FBgn0035089","FBgn0035091","FBgn0035097","FBgn0035099","FBgn0035100","FBgn0035101","FBgn0035102","FBgn0035106","FBgn0035107","FBgn0035109","FBgn0035110","FBgn0035111","FBgn0035113","FBgn0035120","FBgn0035121","FBgn0035122","FBgn0035131","FBgn0035132","FBgn0035133","FBgn0035134","FBgn0035136","FBgn0035137","FBgn0035140","FBgn0035141","FBgn0035142","FBgn0035143","FBgn0035144","FBgn0035145","FBgn0035146","FBgn0035147","FBgn0035148","FBgn0035149","FBgn0035150","FBgn0035151","FBgn0035152","FBgn0035153","FBgn0035154","FBgn0035155","FBgn0035157","FBgn0035158","FBgn0035159","FBgn0035160","FBgn0035161","FBgn0035162","FBgn0035164","FBgn0035165","FBgn0035166","FBgn0035167","FBgn0035168","FBgn0035169","FBgn0035170","FBgn0035171","FBgn0035173","FBgn0035181","FBgn0035189","FBgn0035194","FBgn0035195","FBgn0035202","FBgn0035203","FBgn0035204","FBgn0035205","FBgn0035206","FBgn0035207","FBgn0035208","FBgn0035209","FBgn0035210","FBgn0035211","FBgn0035213","FBgn0035216","FBgn0035227","FBgn0035228","FBgn0035229","FBgn0035231","FBgn0035232","FBgn0035233","FBgn0035234","FBgn0035235","FBgn0035236","FBgn0035237","FBgn0035238","FBgn0035241","FBgn0035243","FBgn0035244","FBgn0035246","FBgn0035247","FBgn0035248","FBgn0035249","FBgn0035251","FBgn0035252","FBgn0035253","FBgn0035254","FBgn0035260","FBgn0035262","FBgn0035263","FBgn0035264","FBgn0035266","FBgn0035267","FBgn0035268","FBgn0035270","FBgn0035271","FBgn0035272","FBgn0035283","FBgn0035285","FBgn0035286","FBgn0035287","FBgn0035294","FBgn0035295","FBgn0035296","FBgn0035297","FBgn0035298","FBgn0035308","FBgn0035309","FBgn0035311","FBgn0035312","FBgn0035317","FBgn0035318","FBgn0035321","FBgn0035323","FBgn0035333","FBgn0035334","FBgn0035335","FBgn0035336","FBgn0035337","FBgn0035338","FBgn0035344","FBgn0035346","FBgn0035347","FBgn0035348","FBgn0035355","FBgn0035356","FBgn0035357","FBgn0035358","FBgn0035359","FBgn0035360","FBgn0035367","FBgn0035370","FBgn0035371","FBgn0035372","FBgn0035374","FBgn0035375","FBgn0035376","FBgn0035377","FBgn0035378","FBgn0035383","FBgn0035388","FBgn0035390","FBgn0035392","FBgn0035393","FBgn0035397","FBgn0035399","FBgn0035400","FBgn0035401","FBgn0035402","FBgn0035403","FBgn0035404","FBgn0035405","FBgn0035407","FBgn0035409","FBgn0035410","FBgn0035411","FBgn0035414","FBgn0035415","FBgn0035416","FBgn0035420","FBgn0035421","FBgn0035422","FBgn0035423","FBgn0035424","FBgn0035425","FBgn0035432","FBgn0035435","FBgn0035437","FBgn0035438","FBgn0035439","FBgn0035440","FBgn0035443","FBgn0035444","FBgn0035445","FBgn0035449","FBgn0035452","FBgn0035461","FBgn0035462","FBgn0035464","FBgn0035469","FBgn0035470","FBgn0035471","FBgn0035473","FBgn0035475","FBgn0035476","FBgn0035477","FBgn0035480","FBgn0035482","FBgn0035483","FBgn0035484","FBgn0035488","FBgn0035489","FBgn0035496","FBgn0035497","FBgn0035498","FBgn0035499","FBgn0035500","FBgn0035501","FBgn0035507","FBgn0035509","FBgn0035512","FBgn0035513","FBgn0035514","FBgn0035515","FBgn0035517","FBgn0035518","FBgn0035519","FBgn0035520","FBgn0035523","FBgn0035524","FBgn0035526","FBgn0035528","FBgn0035529","FBgn0035532","FBgn0035533","FBgn0035534","FBgn0035537","FBgn0035539","FBgn0035540","FBgn0035541","FBgn0035542","FBgn0035558","FBgn0035577","FBgn0035581","FBgn0035586","FBgn0035587","FBgn0035588","FBgn0035589","FBgn0035590","FBgn0035591","FBgn0035592","FBgn0035593","FBgn0035600","FBgn0035601","FBgn0035603","FBgn0035608","FBgn0035612","FBgn0035617","FBgn0035622","FBgn0035623","FBgn0035624","FBgn0035626","FBgn0035627","FBgn0035630","FBgn0035631","FBgn0035632","FBgn0035639","FBgn0035640","FBgn0035641","FBgn0035642","FBgn0035644","FBgn0035670","FBgn0035673","FBgn0035674","FBgn0035675","FBgn0035676","FBgn0035678","FBgn0035679","FBgn0035688","FBgn0035689","FBgn0035690","FBgn0035691","FBgn0035692","FBgn0035693","FBgn0035702","FBgn0035703","FBgn0035704","FBgn0035707","FBgn0035708","FBgn0035710","FBgn0035713","FBgn0035714","FBgn0035715","FBgn0035719","FBgn0035720","FBgn0035721","FBgn0035722","FBgn0035724","FBgn0035725","FBgn0035726","FBgn0035727","FBgn0035751","FBgn0035753","FBgn0035754","FBgn0035760","FBgn0035761","FBgn0035762","FBgn0035763","FBgn0035765","FBgn0035766","FBgn0035767","FBgn0035768","FBgn0035769","FBgn0035770","FBgn0035771","FBgn0035772","FBgn0035777","FBgn0035789","FBgn0035793","FBgn0035802","FBgn0035805","FBgn0035806","FBgn0035807","FBgn0035811","FBgn0035812","FBgn0035824","FBgn0035825","FBgn0035827","FBgn0035829","FBgn0035830","FBgn0035831","FBgn0035832","FBgn0035833","FBgn0035838","FBgn0035839","FBgn0035842","FBgn0035844","FBgn0035848","FBgn0035849","FBgn0035850","FBgn0035851","FBgn0035852","FBgn0035853","FBgn0035854","FBgn0035855","FBgn0035856","FBgn0035866","FBgn0035867","FBgn0035868","FBgn0035871","FBgn0035872","FBgn0035876","FBgn0035877","FBgn0035878","FBgn0035879","FBgn0035880","FBgn0035888","FBgn0035890","FBgn0035891","FBgn0035892","FBgn0035895","FBgn0035896","FBgn0035898","FBgn0035900","FBgn0035901","FBgn0035902","FBgn0035903","FBgn0035904","FBgn0035906","FBgn0035907","FBgn0035909","FBgn0035911","FBgn0035916","FBgn0035918","FBgn0035923","FBgn0035932","FBgn0035933","FBgn0035936","FBgn0035937","FBgn0035938","FBgn0035941","FBgn0035942","FBgn0035944","FBgn0035945","FBgn0035947","FBgn0035948","FBgn0035949","FBgn0035950","FBgn0035951","FBgn0035953","FBgn0035957","FBgn0035959","FBgn0035960","FBgn0035964","FBgn0035965","FBgn0035968","FBgn0035969","FBgn0035975","FBgn0035976","FBgn0035977","FBgn0035978","FBgn0035980","FBgn0035981","FBgn0035983","FBgn0035985","FBgn0035986","FBgn0035987","FBgn0035988","FBgn0035989","FBgn0035993","FBgn0035995","FBgn0035996","FBgn0035997","FBgn0035998","FBgn0035999","FBgn0036000","FBgn0036004","FBgn0036005","FBgn0036007","FBgn0036008","FBgn0036017","FBgn0036018","FBgn0036019","FBgn0036020","FBgn0036028","FBgn0036030","FBgn0036032","FBgn0036035","FBgn0036036","FBgn0036038","FBgn0036039","FBgn0036043","FBgn0036052","FBgn0036053","FBgn0036058","FBgn0036059","FBgn0036062","FBgn0036063","FBgn0036068","FBgn0036082","FBgn0036083","FBgn0036090","FBgn0036093","FBgn0036094","FBgn0036096","FBgn0036099","FBgn0036101","FBgn0036104","FBgn0036105","FBgn0036107","FBgn0036110","FBgn0036111","FBgn0036116","FBgn0036117","FBgn0036118","FBgn0036121","FBgn0036122","FBgn0036124","FBgn0036126","FBgn0036133","FBgn0036134","FBgn0036135","FBgn0036136","FBgn0036141","FBgn0036142","FBgn0036144","FBgn0036145","FBgn0036146","FBgn0036147","FBgn0036150","FBgn0036152","FBgn0036153","FBgn0036154","FBgn0036155","FBgn0036159","FBgn0036162","FBgn0036165","FBgn0036173","FBgn0036180","FBgn0036181","FBgn0036184","FBgn0036186","FBgn0036187","FBgn0036188","FBgn0036191","FBgn0036192","FBgn0036193","FBgn0036194","FBgn0036196","FBgn0036198","FBgn0036199","FBgn0036207","FBgn0036210","FBgn0036211","FBgn0036212","FBgn0036213","FBgn0036214","FBgn0036237","FBgn0036239","FBgn0036240","FBgn0036248","FBgn0036249","FBgn0036254","FBgn0036255","FBgn0036257","FBgn0036258","FBgn0036260","FBgn0036262","FBgn0036263","FBgn0036266","FBgn0036271","FBgn0036272","FBgn0036273","FBgn0036277","FBgn0036279","FBgn0036282","FBgn0036286","FBgn0036287","FBgn0036288","FBgn0036289","FBgn0036290","FBgn0036291","FBgn0036292","FBgn0036298","FBgn0036299","FBgn0036300","FBgn0036301","FBgn0036302","FBgn0036305","FBgn0036306","FBgn0036309","FBgn0036312","FBgn0036314","FBgn0036316","FBgn0036317","FBgn0036318","FBgn0036319","FBgn0036324","FBgn0036331","FBgn0036332","FBgn0036333","FBgn0036334","FBgn0036335","FBgn0036336","FBgn0036337","FBgn0036338","FBgn0036340","FBgn0036341","FBgn0036342","FBgn0036353","FBgn0036354","FBgn0036356","FBgn0036365","FBgn0036366","FBgn0036368","FBgn0036369","FBgn0036372","FBgn0036373","FBgn0036374","FBgn0036376","FBgn0036379","FBgn0036381","FBgn0036382","FBgn0036386","FBgn0036389","FBgn0036395","FBgn0036396","FBgn0036397","FBgn0036398","FBgn0036402","FBgn0036405","FBgn0036406","FBgn0036410","FBgn0036419","FBgn0036423","FBgn0036428","FBgn0036433","FBgn0036446","FBgn0036448","FBgn0036449","FBgn0036450","FBgn0036451","FBgn0036454","FBgn0036460","FBgn0036461","FBgn0036462","FBgn0036463","FBgn0036476","FBgn0036478","FBgn0036480","FBgn0036481","FBgn0036483","FBgn0036484","FBgn0036486","FBgn0036487","FBgn0036488","FBgn0036489","FBgn0036493","FBgn0036500","FBgn0036501","FBgn0036502","FBgn0036505","FBgn0036509","FBgn0036510","FBgn0036511","FBgn0036512","FBgn0036514","FBgn0036515","FBgn0036516","FBgn0036518","FBgn0036519","FBgn0036520","FBgn0036522","FBgn0036533","FBgn0036534","FBgn0036536","FBgn0036537","FBgn0036538","FBgn0036544","FBgn0036545","FBgn0036546","FBgn0036547","FBgn0036548","FBgn0036549","FBgn0036551","FBgn0036552","FBgn0036556","FBgn0036557","FBgn0036558","FBgn0036560","FBgn0036564","FBgn0036565","FBgn0036566","FBgn0036567","FBgn0036568","FBgn0036569","FBgn0036570","FBgn0036571","FBgn0036573","FBgn0036574","FBgn0036575","FBgn0036576","FBgn0036578","FBgn0036579","FBgn0036580","FBgn0036581","FBgn0036614","FBgn0036615","FBgn0036616","FBgn0036620","FBgn0036621","FBgn0036622","FBgn0036623","FBgn0036624","FBgn0036629","FBgn0036640","FBgn0036641","FBgn0036643","FBgn0036646","FBgn0036648","FBgn0036652","FBgn0036654","FBgn0036655","FBgn0036660","FBgn0036661","FBgn0036662","FBgn0036663","FBgn0036666","FBgn0036667","FBgn0036668","FBgn0036671","FBgn0036684","FBgn0036685","FBgn0036686","FBgn0036688","FBgn0036689","FBgn0036691","FBgn0036695","FBgn0036696","FBgn0036697","FBgn0036702","FBgn0036710","FBgn0036714","FBgn0036723","FBgn0036726","FBgn0036728","FBgn0036732","FBgn0036733","FBgn0036734","FBgn0036735","FBgn0036740","FBgn0036741","FBgn0036742","FBgn0036745","FBgn0036746","FBgn0036749","FBgn0036752","FBgn0036754","FBgn0036759","FBgn0036760","FBgn0036761","FBgn0036762","FBgn0036763","FBgn0036764","FBgn0036765","FBgn0036769","FBgn0036770","FBgn0036771","FBgn0036772","FBgn0036774","FBgn0036775","FBgn0036777","FBgn0036801","FBgn0036804","FBgn0036805","FBgn0036806","FBgn0036809","FBgn0036811","FBgn0036812","FBgn0036813","FBgn0036814","FBgn0036815","FBgn0036816","FBgn0036817","FBgn0036818","FBgn0036819","FBgn0036820","FBgn0036821","FBgn0036824","FBgn0036825","FBgn0036826","FBgn0036827","FBgn0036828","FBgn0036837","FBgn0036838","FBgn0036842","FBgn0036843","FBgn0036844","FBgn0036846","FBgn0036847","FBgn0036848","FBgn0036849","FBgn0036850","FBgn0036853","FBgn0036857","FBgn0036862","FBgn0036875","FBgn0036882","FBgn0036886","FBgn0036887","FBgn0036888","FBgn0036889","FBgn0036891","FBgn0036892","FBgn0036893","FBgn0036896","FBgn0036897","FBgn0036900","FBgn0036906","FBgn0036908","FBgn0036909","FBgn0036911","FBgn0036913","FBgn0036915","FBgn0036916","FBgn0036918","FBgn0036919","FBgn0036920","FBgn0036921","FBgn0036922","FBgn0036926","FBgn0036927","FBgn0036928","FBgn0036929","FBgn0036931","FBgn0036932","FBgn0036942","FBgn0036945","FBgn0036958","FBgn0036959","FBgn0036964","FBgn0036967","FBgn0036968","FBgn0036973","FBgn0036974","FBgn0036975","FBgn0036978","FBgn0036980","FBgn0036984","FBgn0036985","FBgn0036987","FBgn0036988","FBgn0036990","FBgn0036991","FBgn0036992","FBgn0036994","FBgn0036995","FBgn0036997","FBgn0036998","FBgn0036999","FBgn0037000","FBgn0037001","FBgn0037003","FBgn0037004","FBgn0037005","FBgn0037007","FBgn0037008","FBgn0037009","FBgn0037010","FBgn0037011","FBgn0037012","FBgn0037013","FBgn0037016","FBgn0037017","FBgn0037018","FBgn0037019","FBgn0037020","FBgn0037021","FBgn0037022","FBgn0037024","FBgn0037025","FBgn0037026","FBgn0037027","FBgn0037028","FBgn0037031","FBgn0037037","FBgn0037044","FBgn0037045","FBgn0037046","FBgn0037050","FBgn0037051","FBgn0037057","FBgn0037061","FBgn0037063","FBgn0037064","FBgn0037065","FBgn0037070","FBgn0037071","FBgn0037073","FBgn0037074","FBgn0037076","FBgn0037078","FBgn0037081","FBgn0037082","FBgn0037084","FBgn0037087","FBgn0037092","FBgn0037093","FBgn0037094","FBgn0037097","FBgn0037098","FBgn0037102","FBgn0037105","FBgn0037106","FBgn0037108","FBgn0037109","FBgn0037110","FBgn0037116","FBgn0037117","FBgn0037120","FBgn0037121","FBgn0037126","FBgn0037130","FBgn0037133","FBgn0037134","FBgn0037135","FBgn0037137","FBgn0037138","FBgn0037140","FBgn0037141","FBgn0037142","FBgn0037144","FBgn0037149","FBgn0037150","FBgn0037151","FBgn0037153","FBgn0037156","FBgn0037182","FBgn0037183","FBgn0037184","FBgn0037185","FBgn0037186","FBgn0037188","FBgn0037199","FBgn0037200","FBgn0037202","FBgn0037203","FBgn0037205","FBgn0037206","FBgn0037207","FBgn0037212","FBgn0037213","FBgn0037215","FBgn0037217","FBgn0037218","FBgn0037220","FBgn0037222","FBgn0037223","FBgn0037225","FBgn0037228","FBgn0037229","FBgn0037230","FBgn0037231","FBgn0037232","FBgn0037234","FBgn0037235","FBgn0037236","FBgn0037238","FBgn0037239","FBgn0037240","FBgn0037241","FBgn0037242","FBgn0037244","FBgn0037245","FBgn0037248","FBgn0037249","FBgn0037250","FBgn0037251","FBgn0037252","FBgn0037255","FBgn0037260","FBgn0037261","FBgn0037262","FBgn0037263","FBgn0037265","FBgn0037270","FBgn0037275","FBgn0037279","FBgn0037282","FBgn0037290","FBgn0037293","FBgn0037297","FBgn0037298","FBgn0037299","FBgn0037301","FBgn0037304","FBgn0037305","FBgn0037309","FBgn0037312","FBgn0037313","FBgn0037315","FBgn0037317","FBgn0037321","FBgn0037322","FBgn0037327","FBgn0037328","FBgn0037329","FBgn0037330","FBgn0037332","FBgn0037336","FBgn0037338","FBgn0037339","FBgn0037340","FBgn0037341","FBgn0037342","FBgn0037344","FBgn0037345","FBgn0037347","FBgn0037350","FBgn0037351","FBgn0037352","FBgn0037354","FBgn0037356","FBgn0037357","FBgn0037358","FBgn0037359","FBgn0037360","FBgn0037363","FBgn0037364","FBgn0037368","FBgn0037369","FBgn0037370","FBgn0037371","FBgn0037372","FBgn0037374","FBgn0037375","FBgn0037376","FBgn0037377","FBgn0037378","FBgn0037379","FBgn0037382","FBgn0037383","FBgn0037384","FBgn0037386","FBgn0037387","FBgn0037391","FBgn0037405","FBgn0037429","FBgn0037432","FBgn0037433","FBgn0037434","FBgn0037435","FBgn0037439","FBgn0037440","FBgn0037442","FBgn0037443","FBgn0037445","FBgn0037446","FBgn0037448","FBgn0037449","FBgn0037465","FBgn0037466","FBgn0037467","FBgn0037468","FBgn0037469","FBgn0037470","FBgn0037472","FBgn0037473","FBgn0037478","FBgn0037481","FBgn0037482","FBgn0037483","FBgn0037489","FBgn0037490","FBgn0037491","FBgn0037492","FBgn0037504","FBgn0037513","FBgn0037515","FBgn0037518","FBgn0037525","FBgn0037526","FBgn0037529","FBgn0037530","FBgn0037531","FBgn0037533","FBgn0037535","FBgn0037536","FBgn0037537","FBgn0037538","FBgn0037539","FBgn0037540","FBgn0037541","FBgn0037543","FBgn0037544","FBgn0037548","FBgn0037549","FBgn0037550","FBgn0037551","FBgn0037552","FBgn0037553","FBgn0037554","FBgn0037555","FBgn0037556","FBgn0037560","FBgn0037561","FBgn0037562","FBgn0037566","FBgn0037567","FBgn0037569","FBgn0037573","FBgn0037574","FBgn0037578","FBgn0037580","FBgn0037581","FBgn0037583","FBgn0037584","FBgn0037592","FBgn0037602","FBgn0037603","FBgn0037606","FBgn0037607","FBgn0037608","FBgn0037609","FBgn0037610","FBgn0037611","FBgn0037612","FBgn0037613","FBgn0037614","FBgn0037616","FBgn0037617","FBgn0037619","FBgn0037620","FBgn0037621","FBgn0037622","FBgn0037623","FBgn0037624","FBgn0037630","FBgn0037632","FBgn0037633","FBgn0037634","FBgn0037635","FBgn0037636","FBgn0037637","FBgn0037638","FBgn0037640","FBgn0037643","FBgn0037644","FBgn0037646","FBgn0037647","FBgn0037648","FBgn0037652","FBgn0037653","FBgn0037654","FBgn0037655","FBgn0037656","FBgn0037657","FBgn0037659","FBgn0037660","FBgn0037661","FBgn0037662","FBgn0037667","FBgn0037669","FBgn0037670","FBgn0037671","FBgn0037672","FBgn0037678","FBgn0037679","FBgn0037680","FBgn0037683","FBgn0037684","FBgn0037686","FBgn0037687","FBgn0037688","FBgn0037696","FBgn0037698","FBgn0037700","FBgn0037702","FBgn0037703","FBgn0037705","FBgn0037707","FBgn0037708","FBgn0037709","FBgn0037710","FBgn0037712","FBgn0037713","FBgn0037714","FBgn0037715","FBgn0037716","FBgn0037717","FBgn0037718","FBgn0037719","FBgn0037720","FBgn0037721","FBgn0037722","FBgn0037723","FBgn0037726","FBgn0037728","FBgn0037730","FBgn0037734","FBgn0037737","FBgn0037739","FBgn0037741","FBgn0037742","FBgn0037743","FBgn0037744","FBgn0037746","FBgn0037747","FBgn0037749","FBgn0037750","FBgn0037751","FBgn0037752","FBgn0037753","FBgn0037754","FBgn0037755","FBgn0037756","FBgn0037757","FBgn0037758","FBgn0037759","FBgn0037760","FBgn0037770","FBgn0037773","FBgn0037777","FBgn0037778","FBgn0037779","FBgn0037780","FBgn0037781","FBgn0037788","FBgn0037792","FBgn0037794","FBgn0037801","FBgn0037802","FBgn0037804","FBgn0037806","FBgn0037807","FBgn0037808","FBgn0037809","FBgn0037810","FBgn0037811","FBgn0037812","FBgn0037814","FBgn0037815","FBgn0037816","FBgn0037817","FBgn0037822","FBgn0037824","FBgn0037831","FBgn0037834","FBgn0037837","FBgn0037838","FBgn0037842","FBgn0037843","FBgn0037844","FBgn0037846","FBgn0037847","FBgn0037848","FBgn0037850","FBgn0037852","FBgn0037853","FBgn0037855","FBgn0037856","FBgn0037872","FBgn0037873","FBgn0037874","FBgn0037875","FBgn0037876","FBgn0037877","FBgn0037878","FBgn0037880","FBgn0037881","FBgn0037882","FBgn0037883","FBgn0037884","FBgn0037885","FBgn0037890","FBgn0037891","FBgn0037892","FBgn0037893","FBgn0037894","FBgn0037896","FBgn0037897","FBgn0037898","FBgn0037899","FBgn0037900","FBgn0037901","FBgn0037902","FBgn0037906","FBgn0037911","FBgn0037912","FBgn0037913","FBgn0037915","FBgn0037916","FBgn0037917","FBgn0037918","FBgn0037920","FBgn0037921","FBgn0037923","FBgn0037924","FBgn0037926","FBgn0037930","FBgn0037931","FBgn0037933","FBgn0037934","FBgn0037935","FBgn0037937","FBgn0037938","FBgn0037942","FBgn0037943","FBgn0037944","FBgn0037949","FBgn0037950","FBgn0037955","FBgn0037956","FBgn0037958","FBgn0037960","FBgn0037963","FBgn0037973","FBgn0037974","FBgn0037975","FBgn0037978","FBgn0037979","FBgn0037980","FBgn0037981","FBgn0037989","FBgn0037998","FBgn0037999","FBgn0038013","FBgn0038014","FBgn0038016","FBgn0038020","FBgn0038034","FBgn0038035","FBgn0038037","FBgn0038038","FBgn0038039","FBgn0038042","FBgn0038043","FBgn0038046","FBgn0038047","FBgn0038049","FBgn0038053","FBgn0038055","FBgn0038056","FBgn0038057","FBgn0038058","FBgn0038065","FBgn0038067","FBgn0038074","FBgn0038080","FBgn0038083","FBgn0038088","FBgn0038092","FBgn0038095","FBgn0038098","FBgn0038099","FBgn0038100","FBgn0038102","FBgn0038105","FBgn0038106","FBgn0038107","FBgn0038108","FBgn0038109","FBgn0038110","FBgn0038111","FBgn0038118","FBgn0038128","FBgn0038129","FBgn0038143","FBgn0038144","FBgn0038145","FBgn0038146","FBgn0038149","FBgn0038156","FBgn0038166","FBgn0038167","FBgn0038168","FBgn0038170","FBgn0038183","FBgn0038194","FBgn0038195","FBgn0038196","FBgn0038197","FBgn0038198","FBgn0038206","FBgn0038220","FBgn0038221","FBgn0038223","FBgn0038224","FBgn0038233","FBgn0038234","FBgn0038235","FBgn0038237","FBgn0038243","FBgn0038244","FBgn0038248","FBgn0038250","FBgn0038251","FBgn0038256","FBgn0038257","FBgn0038258","FBgn0038260","FBgn0038261","FBgn0038268","FBgn0038269","FBgn0038271","FBgn0038272","FBgn0038274","FBgn0038275","FBgn0038277","FBgn0038286","FBgn0038289","FBgn0038290","FBgn0038291","FBgn0038292","FBgn0038293","FBgn0038296","FBgn0038299","FBgn0038300","FBgn0038301","FBgn0038302","FBgn0038303","FBgn0038304","FBgn0038306","FBgn0038309","FBgn0038311","FBgn0038312","FBgn0038313","FBgn0038316","FBgn0038318","FBgn0038319","FBgn0038320","FBgn0038321","FBgn0038323","FBgn0038324","FBgn0038325","FBgn0038326","FBgn0038327","FBgn0038330","FBgn0038331","FBgn0038332","FBgn0038341","FBgn0038344","FBgn0038346","FBgn0038347","FBgn0038348","FBgn0038349","FBgn0038351","FBgn0038353","FBgn0038360","FBgn0038361","FBgn0038363","FBgn0038364","FBgn0038365","FBgn0038366","FBgn0038369","FBgn0038371","FBgn0038373","FBgn0038376","FBgn0038381","FBgn0038387","FBgn0038388","FBgn0038389","FBgn0038390","FBgn0038391","FBgn0038392","FBgn0038394","FBgn0038395","FBgn0038396","FBgn0038397","FBgn0038400","FBgn0038401","FBgn0038402","FBgn0038404","FBgn0038407","FBgn0038414","FBgn0038415","FBgn0038416","FBgn0038418","FBgn0038419","FBgn0038420","FBgn0038421","FBgn0038424","FBgn0038425","FBgn0038426","FBgn0038427","FBgn0038428","FBgn0038429","FBgn0038431","FBgn0038432","FBgn0038435","FBgn0038436","FBgn0038437","FBgn0038438","FBgn0038446","FBgn0038453","FBgn0038454","FBgn0038460","FBgn0038461","FBgn0038462","FBgn0038463","FBgn0038464","FBgn0038465","FBgn0038466","FBgn0038467","FBgn0038469","FBgn0038470","FBgn0038471","FBgn0038472","FBgn0038473","FBgn0038474","FBgn0038475","FBgn0038476","FBgn0038478","FBgn0038484","FBgn0038488","FBgn0038490","FBgn0038491","FBgn0038499","FBgn0038501","FBgn0038504","FBgn0038515","FBgn0038516","FBgn0038519","FBgn0038524","FBgn0038527","FBgn0038528","FBgn0038530","FBgn0038532","FBgn0038533","FBgn0038535","FBgn0038536","FBgn0038544","FBgn0038545","FBgn0038546","FBgn0038547","FBgn0038548","FBgn0038549","FBgn0038551","FBgn0038552","FBgn0038554","FBgn0038564","FBgn0038569","FBgn0038575","FBgn0038576","FBgn0038577","FBgn0038578","FBgn0038581","FBgn0038582","FBgn0038583","FBgn0038584","FBgn0038585","FBgn0038586","FBgn0038587","FBgn0038588","FBgn0038589","FBgn0038590","FBgn0038593","FBgn0038595","FBgn0038597","FBgn0038598","FBgn0038601","FBgn0038603","FBgn0038608","FBgn0038609","FBgn0038612","FBgn0038617","FBgn0038619","FBgn0038621","FBgn0038629","FBgn0038633","FBgn0038638","FBgn0038639","FBgn0038640","FBgn0038649","FBgn0038651","FBgn0038652","FBgn0038653","FBgn0038658","FBgn0038659","FBgn0038660","FBgn0038662","FBgn0038666","FBgn0038667","FBgn0038672","FBgn0038673","FBgn0038674","FBgn0038675","FBgn0038678","FBgn0038679","FBgn0038680","FBgn0038681","FBgn0038682","FBgn0038683","FBgn0038686","FBgn0038690","FBgn0038691","FBgn0038692","FBgn0038693","FBgn0038704","FBgn0038705","FBgn0038717","FBgn0038720","FBgn0038721","FBgn0038722","FBgn0038723","FBgn0038725","FBgn0038735","FBgn0038736","FBgn0038737","FBgn0038738","FBgn0038739","FBgn0038741","FBgn0038742","FBgn0038744","FBgn0038745","FBgn0038746","FBgn0038747","FBgn0038755","FBgn0038760","FBgn0038763","FBgn0038765","FBgn0038766","FBgn0038767","FBgn0038768","FBgn0038769","FBgn0038771","FBgn0038772","FBgn0038780","FBgn0038784","FBgn0038787","FBgn0038788","FBgn0038803","FBgn0038804","FBgn0038805","FBgn0038806","FBgn0038808","FBgn0038809","FBgn0038810","FBgn0038811","FBgn0038815","FBgn0038816","FBgn0038821","FBgn0038826","FBgn0038827","FBgn0038828","FBgn0038829","FBgn0038830","FBgn0038832","FBgn0038834","FBgn0038842","FBgn0038846","FBgn0038851","FBgn0038852","FBgn0038853","FBgn0038854","FBgn0038855","FBgn0038856","FBgn0038857","FBgn0038858","FBgn0038860","FBgn0038861","FBgn0038862","FBgn0038865","FBgn0038866","FBgn0038868","FBgn0038869","FBgn0038870","FBgn0038871","FBgn0038872","FBgn0038873","FBgn0038874","FBgn0038876","FBgn0038877","FBgn0038881","FBgn0038889","FBgn0038890","FBgn0038893","FBgn0038894","FBgn0038903","FBgn0038909","FBgn0038912","FBgn0038916","FBgn0038917","FBgn0038922","FBgn0038923","FBgn0038924","FBgn0038925","FBgn0038926","FBgn0038927","FBgn0038928","FBgn0038947","FBgn0038948","FBgn0038950","FBgn0038951","FBgn0038952","FBgn0038953","FBgn0038957","FBgn0038961","FBgn0038964","FBgn0038965","FBgn0038966","FBgn0038968","FBgn0038972","FBgn0038973","FBgn0038974","FBgn0038975","FBgn0038976","FBgn0038977","FBgn0038981","FBgn0038984","FBgn0038989","FBgn0038993","FBgn0038996","FBgn0039000","FBgn0039003","FBgn0039004","FBgn0039005","FBgn0039006","FBgn0039013","FBgn0039016","FBgn0039017","FBgn0039018","FBgn0039019","FBgn0039020","FBgn0039022","FBgn0039023","FBgn0039024","FBgn0039025","FBgn0039026","FBgn0039043","FBgn0039044","FBgn0039045","FBgn0039049","FBgn0039051","FBgn0039052","FBgn0039054","FBgn0039055","FBgn0039056","FBgn0039058","FBgn0039059","FBgn0039060","FBgn0039061","FBgn0039064","FBgn0039065","FBgn0039066","FBgn0039067","FBgn0039068","FBgn0039069","FBgn0039071","FBgn0039073","FBgn0039094","FBgn0039098","FBgn0039099","FBgn0039101","FBgn0039102","FBgn0039108","FBgn0039109","FBgn0039110","FBgn0039111","FBgn0039112","FBgn0039113","FBgn0039114","FBgn0039115","FBgn0039116","FBgn0039117","FBgn0039118","FBgn0039120","FBgn0039125","FBgn0039126","FBgn0039127","FBgn0039128","FBgn0039129","FBgn0039130","FBgn0039132","FBgn0039135","FBgn0039136","FBgn0039137","FBgn0039139","FBgn0039140","FBgn0039141","FBgn0039145","FBgn0039149","FBgn0039150","FBgn0039152","FBgn0039153","FBgn0039156","FBgn0039157","FBgn0039158","FBgn0039159","FBgn0039160","FBgn0039163","FBgn0039164","FBgn0039165","FBgn0039167","FBgn0039169","FBgn0039170","FBgn0039172","FBgn0039175","FBgn0039178","FBgn0039179","FBgn0039180","FBgn0039182","FBgn0039183","FBgn0039186","FBgn0039187","FBgn0039188","FBgn0039189","FBgn0039201","FBgn0039204","FBgn0039205","FBgn0039206","FBgn0039207","FBgn0039208","FBgn0039209","FBgn0039210","FBgn0039212","FBgn0039213","FBgn0039214","FBgn0039215","FBgn0039218","FBgn0039219","FBgn0039223","FBgn0039226","FBgn0039227","FBgn0039229","FBgn0039233","FBgn0039234","FBgn0039240","FBgn0039241","FBgn0039249","FBgn0039250","FBgn0039251","FBgn0039252","FBgn0039254","FBgn0039255","FBgn0039257","FBgn0039258","FBgn0039259","FBgn0039260","FBgn0039261","FBgn0039265","FBgn0039266","FBgn0039269","FBgn0039270","FBgn0039271","FBgn0039273","FBgn0039274","FBgn0039290","FBgn0039291","FBgn0039293","FBgn0039294","FBgn0039296","FBgn0039297","FBgn0039300","FBgn0039301","FBgn0039302","FBgn0039303","FBgn0039304","FBgn0039305","FBgn0039306","FBgn0039321","FBgn0039323","FBgn0039328","FBgn0039329","FBgn0039335","FBgn0039336","FBgn0039337","FBgn0039338","FBgn0039339","FBgn0039341","FBgn0039347","FBgn0039348","FBgn0039349","FBgn0039350","FBgn0039352","FBgn0039354","FBgn0039355","FBgn0039356","FBgn0039357","FBgn0039358","FBgn0039359","FBgn0039360","FBgn0039371","FBgn0039379","FBgn0039381","FBgn0039385","FBgn0039386","FBgn0039387","FBgn0039402","FBgn0039404","FBgn0039406","FBgn0039407","FBgn0039415","FBgn0039417","FBgn0039419","FBgn0039420","FBgn0039427","FBgn0039429","FBgn0039430","FBgn0039431","FBgn0039449","FBgn0039450","FBgn0039451","FBgn0039454","FBgn0039459","FBgn0039461","FBgn0039462","FBgn0039464","FBgn0039465","FBgn0039466","FBgn0039467","FBgn0039478","FBgn0039479","FBgn0039485","FBgn0039487","FBgn0039488","FBgn0039489","FBgn0039490","FBgn0039492","FBgn0039494","FBgn0039498","FBgn0039500","FBgn0039501","FBgn0039503","FBgn0039504","FBgn0039505","FBgn0039507","FBgn0039508","FBgn0039509","FBgn0039510","FBgn0039525","FBgn0039527","FBgn0039528","FBgn0039530","FBgn0039531","FBgn0039532","FBgn0039536","FBgn0039537","FBgn0039538","FBgn0039540","FBgn0039543","FBgn0039544","FBgn0039554","FBgn0039555","FBgn0039557","FBgn0039558","FBgn0039559","FBgn0039560","FBgn0039561","FBgn0039562","FBgn0039563","FBgn0039564","FBgn0039565","FBgn0039566","FBgn0039580","FBgn0039581","FBgn0039588","FBgn0039589","FBgn0039590","FBgn0039593","FBgn0039594","FBgn0039600","FBgn0039601","FBgn0039602","FBgn0039611","FBgn0039612","FBgn0039613","FBgn0039620","FBgn0039621","FBgn0039622","FBgn0039623","FBgn0039625","FBgn0039626","FBgn0039627","FBgn0039630","FBgn0039631","FBgn0039632","FBgn0039633","FBgn0039634","FBgn0039635","FBgn0039636","FBgn0039637","FBgn0039638","FBgn0039639","FBgn0039640","FBgn0039641","FBgn0039642","FBgn0039644","FBgn0039647","FBgn0039650","FBgn0039651","FBgn0039654","FBgn0039655","FBgn0039663","FBgn0039664","FBgn0039665","FBgn0039667","FBgn0039668","FBgn0039673","FBgn0039674","FBgn0039679","FBgn0039680","FBgn0039682","FBgn0039687","FBgn0039688","FBgn0039689","FBgn0039690","FBgn0039691","FBgn0039694","FBgn0039695","FBgn0039696","FBgn0039697","FBgn0039698","FBgn0039702","FBgn0039704","FBgn0039705","FBgn0039710","FBgn0039711","FBgn0039712","FBgn0039713","FBgn0039714","FBgn0039726","FBgn0039727","FBgn0039730","FBgn0039731","FBgn0039732","FBgn0039733","FBgn0039735","FBgn0039736","FBgn0039737","FBgn0039738","FBgn0039740","FBgn0039741","FBgn0039742","FBgn0039743","FBgn0039747","FBgn0039751","FBgn0039755","FBgn0039756","FBgn0039757","FBgn0039759","FBgn0039764","FBgn0039765","FBgn0039766","FBgn0039767","FBgn0039773","FBgn0039774","FBgn0039776","FBgn0039788","FBgn0039789","FBgn0039790","FBgn0039797","FBgn0039798","FBgn0039800","FBgn0039801","FBgn0039802","FBgn0039804","FBgn0039808","FBgn0039809","FBgn0039827","FBgn0039828","FBgn0039829","FBgn0039830","FBgn0039831","FBgn0039833","FBgn0039835","FBgn0039836","FBgn0039844","FBgn0039846","FBgn0039848","FBgn0039849","FBgn0039854","FBgn0039855","FBgn0039856","FBgn0039857","FBgn0039858","FBgn0039859","FBgn0039860","FBgn0039861","FBgn0039862","FBgn0039863","FBgn0039867","FBgn0039868","FBgn0039869","FBgn0039870","FBgn0039874","FBgn0039875","FBgn0039876","FBgn0039877","FBgn0039881","FBgn0039882","FBgn0039883","FBgn0039886","FBgn0039889","FBgn0039890","FBgn0039896","FBgn0039897","FBgn0039902","FBgn0039904","FBgn0039905","FBgn0039907","FBgn0039908","FBgn0039909","FBgn0039911","FBgn0039914","FBgn0039915","FBgn0039916","FBgn0039920","FBgn0039923","FBgn0039924","FBgn0039925","FBgn0039927","FBgn0039928","FBgn0039929","FBgn0039930","FBgn0039932","FBgn0039936","FBgn0039938","FBgn0039965","FBgn0039966","FBgn0039969","FBgn0039970","FBgn0039972","FBgn0039977","FBgn0039979","FBgn0039994","FBgn0039997","FBgn0040002","FBgn0040005","FBgn0040007","FBgn0040009","FBgn0040010","FBgn0040011","FBgn0040045","FBgn0040056","FBgn0040064","FBgn0040066","FBgn0040068","FBgn0040070","FBgn0040071","FBgn0040075","FBgn0040078","FBgn0040079","FBgn0040080","FBgn0040087","FBgn0040089","FBgn0040091","FBgn0040099","FBgn0040104","FBgn0040153","FBgn0040206","FBgn0040207","FBgn0040208","FBgn0040212","FBgn0040227","FBgn0040228","FBgn0040230","FBgn0040232","FBgn0040234","FBgn0040235","FBgn0040236","FBgn0040237","FBgn0040238","FBgn0040239","FBgn0040250","FBgn0040251","FBgn0040252","FBgn0040256","FBgn0040257","FBgn0040259","FBgn0040260","FBgn0040262","FBgn0040268","FBgn0040271","FBgn0040273","FBgn0040281","FBgn0040282","FBgn0040283","FBgn0040284","FBgn0040285","FBgn0040286","FBgn0040290","FBgn0040294","FBgn0040297","FBgn0040298","FBgn0040299","FBgn0040305","FBgn0040308","FBgn0040309","FBgn0040319","FBgn0040321","FBgn0040322","FBgn0040323","FBgn0040324","FBgn0040333","FBgn0040334","FBgn0040335","FBgn0040336","FBgn0040337","FBgn0040339","FBgn0040340","FBgn0040342","FBgn0040344","FBgn0040346","FBgn0040347","FBgn0040348","FBgn0040351","FBgn0040364","FBgn0040366","FBgn0040367","FBgn0040372","FBgn0040373","FBgn0040375","FBgn0040376","FBgn0040377","FBgn0040382","FBgn0040383","FBgn0040384","FBgn0040388","FBgn0040389","FBgn0040391","FBgn0040392","FBgn0040394","FBgn0040395","FBgn0040396","FBgn0040397","FBgn0040398","FBgn0040465","FBgn0040466","FBgn0040467","FBgn0040475","FBgn0040477","FBgn0040491","FBgn0040493","FBgn0040503","FBgn0040505","FBgn0040512","FBgn0040528","FBgn0040529","FBgn0040532","FBgn0040534","FBgn0040551","FBgn0040554","FBgn0040571","FBgn0040575","FBgn0040588","FBgn0040600","FBgn0040602","FBgn0040609","FBgn0040636","FBgn0040650","FBgn0040658","FBgn0040660","FBgn0040666","FBgn0040670","FBgn0040687","FBgn0040751","FBgn0040752","FBgn0040754","FBgn0040763","FBgn0040765","FBgn0040766","FBgn0040773","FBgn0040777","FBgn0040778","FBgn0040780","FBgn0040784","FBgn0040786","FBgn0040793","FBgn0040813","FBgn0040827","FBgn0040837","FBgn0040850","FBgn0040862","FBgn0040890","FBgn0040899","FBgn0040906","FBgn0040907","FBgn0040918","FBgn0040928","FBgn0040929","FBgn0040931","FBgn0040942","FBgn0040954","FBgn0040964","FBgn0040985","FBgn0041004","FBgn0041005","FBgn0041087","FBgn0041092","FBgn0041094","FBgn0041096","FBgn0041100","FBgn0041111","FBgn0041147","FBgn0041150","FBgn0041161","FBgn0041164","FBgn0041171","FBgn0041174","FBgn0041180","FBgn0041182","FBgn0041183","FBgn0041184","FBgn0041186","FBgn0041188","FBgn0041191","FBgn0041203","FBgn0041205","FBgn0041210","FBgn0041225","FBgn0041229","FBgn0041241","FBgn0041243","FBgn0041244","FBgn0041342","FBgn0041581","FBgn0041582","FBgn0041585","FBgn0041588","FBgn0041604","FBgn0041605","FBgn0041607","FBgn0041621","FBgn0041627","FBgn0041629","FBgn0041630","FBgn0041702","FBgn0041710","FBgn0041712","FBgn0041721","FBgn0041723","FBgn0041775","FBgn0041781","FBgn0041789","FBgn0042083","FBgn0042085","FBgn0042092","FBgn0042094","FBgn0042102","FBgn0042104","FBgn0042106","FBgn0042111","FBgn0042112","FBgn0042125","FBgn0042126","FBgn0042127","FBgn0042132","FBgn0042133","FBgn0042134","FBgn0042135","FBgn0042138","FBgn0042174","FBgn0042175","FBgn0042177","FBgn0042178","FBgn0042180","FBgn0042185","FBgn0042205","FBgn0042206","FBgn0042207","FBgn0042213","FBgn0042627","FBgn0042650","FBgn0042693","FBgn0042696","FBgn0042701","FBgn0042712","FBgn0043001","FBgn0043010","FBgn0043012","FBgn0043025","FBgn0043070","FBgn0043362","FBgn0043364","FBgn0043455","FBgn0043456","FBgn0043457","FBgn0043458","FBgn0043575","FBgn0043783","FBgn0043791","FBgn0043796","FBgn0043799","FBgn0043806","FBgn0043841","FBgn0043853","FBgn0043854","FBgn0043884","FBgn0043900","FBgn0043903","FBgn0043904","FBgn0044020","FBgn0044028","FBgn0044030","FBgn0044047","FBgn0044323","FBgn0044324","FBgn0044452","FBgn0044510","FBgn0044511","FBgn0044823","FBgn0044826","FBgn0044871","FBgn0044872","FBgn0045035","FBgn0045063","FBgn0045064","FBgn0045073","FBgn0045468","FBgn0045469","FBgn0045470","FBgn0045474","FBgn0045495","FBgn0045761","FBgn0045770","FBgn0045800","FBgn0045823","FBgn0045842","FBgn0045852","FBgn0045862","FBgn0045866","FBgn0045980","FBgn0046114","FBgn0046214","FBgn0046222","FBgn0046225","FBgn0046247","FBgn0046258","FBgn0046272","FBgn0046296","FBgn0046301","FBgn0046322","FBgn0046685","FBgn0046687","FBgn0046692","FBgn0046696","FBgn0046704","FBgn0046763","FBgn0046776","FBgn0047038","FBgn0047095","FBgn0047114","FBgn0047135","FBgn0047178","FBgn0047205","FBgn0050000","FBgn0050001","FBgn0050005","FBgn0050007","FBgn0050008","FBgn0050010","FBgn0050011","FBgn0050015","FBgn0050020","FBgn0050022","FBgn0050033","FBgn0050035","FBgn0050043","FBgn0050044","FBgn0050046","FBgn0050047","FBgn0050051","FBgn0050052","FBgn0050055","FBgn0050062","FBgn0050069","FBgn0050077","FBgn0050081","FBgn0050085","FBgn0050087","FBgn0050088","FBgn0050089","FBgn0050090","FBgn0050094","FBgn0050095","FBgn0050096","FBgn0050104","FBgn0050105","FBgn0050109","FBgn0050115","FBgn0050118","FBgn0050122","FBgn0050147","FBgn0050148","FBgn0050156","FBgn0050157","FBgn0050159","FBgn0050169","FBgn0050178","FBgn0050183","FBgn0050184","FBgn0050185","FBgn0050187","FBgn0050196","FBgn0050217","FBgn0050269","FBgn0050273","FBgn0050281","FBgn0050284","FBgn0050285","FBgn0050291","FBgn0050295","FBgn0050323","FBgn0050338","FBgn0050339","FBgn0050340","FBgn0050342","FBgn0050344","FBgn0050345","FBgn0050349","FBgn0050356","FBgn0050357","FBgn0050359","FBgn0050372","FBgn0050373","FBgn0050377","FBgn0050381","FBgn0050383","FBgn0050389","FBgn0050390","FBgn0050392","FBgn0050394","FBgn0050398","FBgn0050403","FBgn0050404","FBgn0050410","FBgn0050414","FBgn0050420","FBgn0050421","FBgn0050423","FBgn0050424","FBgn0050428","FBgn0050438","FBgn0050440","FBgn0050443","FBgn0050446","FBgn0050456","FBgn0050460","FBgn0050463","FBgn0050466","FBgn0050467","FBgn0050476","FBgn0050482","FBgn0050484","FBgn0050485","FBgn0050489","FBgn0050490","FBgn0050491","FBgn0050492","FBgn0050493","FBgn0050496","FBgn0050497","FBgn0050499","FBgn0050502","FBgn0051004","FBgn0051005","FBgn0051019","FBgn0051025","FBgn0051029","FBgn0051032","FBgn0051038","FBgn0051040","FBgn0051044","FBgn0051048","FBgn0051051","FBgn0051053","FBgn0051063","FBgn0051064","FBgn0051065","FBgn0051068","FBgn0051072","FBgn0051075","FBgn0051082","FBgn0051088","FBgn0051092","FBgn0051098","FBgn0051108","FBgn0051109","FBgn0051111","FBgn0051115","FBgn0051116","FBgn0051118","FBgn0051119","FBgn0051120","FBgn0051122","FBgn0051125","FBgn0051126","FBgn0051133","FBgn0051140","FBgn0051145","FBgn0051150","FBgn0051151","FBgn0051155","FBgn0051156","FBgn0051158","FBgn0051159","FBgn0051161","FBgn0051163","FBgn0051169","FBgn0051174","FBgn0051184","FBgn0051195","FBgn0051211","FBgn0051213","FBgn0051216","FBgn0051217","FBgn0051219","FBgn0051223","FBgn0051224","FBgn0051229","FBgn0051230","FBgn0051231","FBgn0051232","FBgn0051249","FBgn0051251","FBgn0051262","FBgn0051278","FBgn0051296","FBgn0051301","FBgn0051302","FBgn0051304","FBgn0051313","FBgn0051314","FBgn0051320","FBgn0051324","FBgn0051326","FBgn0051337","FBgn0051342","FBgn0051344","FBgn0051347","FBgn0051352","FBgn0051357","FBgn0051360","FBgn0051361","FBgn0051363","FBgn0051365","FBgn0051367","FBgn0051368","FBgn0051370","FBgn0051374","FBgn0051388","FBgn0051389","FBgn0051390","FBgn0051392","FBgn0051415","FBgn0051431","FBgn0051436","FBgn0051439","FBgn0051441","FBgn0051445","FBgn0051450","FBgn0051453","FBgn0051457","FBgn0051460","FBgn0051462","FBgn0051467","FBgn0051472","FBgn0051477","FBgn0051495","FBgn0051510","FBgn0051516","FBgn0051522","FBgn0051523","FBgn0051525","FBgn0051526","FBgn0051531","FBgn0051534","FBgn0051536","FBgn0051538","FBgn0051546","FBgn0051547","FBgn0051548","FBgn0051549","FBgn0051550","FBgn0051551","FBgn0051555","FBgn0051600","FBgn0051601","FBgn0051607","FBgn0051612","FBgn0051614","FBgn0051619","FBgn0051624","FBgn0051627","FBgn0051630","FBgn0051633","FBgn0051634","FBgn0051635","FBgn0051637","FBgn0051638","FBgn0051641","FBgn0051642","FBgn0051648","FBgn0051658","FBgn0051663","FBgn0051668","FBgn0051673","FBgn0051674","FBgn0051675","FBgn0051678","FBgn0051687","FBgn0051688","FBgn0051694","FBgn0051697","FBgn0051710","FBgn0051712","FBgn0051713","FBgn0051715","FBgn0051716","FBgn0051717","FBgn0051719","FBgn0051720","FBgn0051729","FBgn0051739","FBgn0051755","FBgn0051759","FBgn0051763","FBgn0051769","FBgn0051774","FBgn0051777","FBgn0051778","FBgn0051782","FBgn0051789","FBgn0051790","FBgn0051793","FBgn0051798","FBgn0051800","FBgn0051803","FBgn0051812","FBgn0051815","FBgn0051816","FBgn0051849","FBgn0051852","FBgn0051855","FBgn0051856","FBgn0051858","FBgn0051864","FBgn0051865","FBgn0051866","FBgn0051869","FBgn0051874","FBgn0051875","FBgn0051897","FBgn0051898","FBgn0051900","FBgn0051908","FBgn0051915","FBgn0051922","FBgn0051935","FBgn0051950","FBgn0051955","FBgn0051957","FBgn0051961","FBgn0051973","FBgn0051974","FBgn0051989","FBgn0051992","FBgn0051997","FBgn0051998","FBgn0051999","FBgn0052000","FBgn0052011","FBgn0052016","FBgn0052017","FBgn0052021","FBgn0052022","FBgn0052024","FBgn0052026","FBgn0052027","FBgn0052036","FBgn0052037","FBgn0052038","FBgn0052039","FBgn0052040","FBgn0052043","FBgn0052056","FBgn0052058","FBgn0052060","FBgn0052062","FBgn0052065","FBgn0052066","FBgn0052068","FBgn0052069","FBgn0052075","FBgn0052076","FBgn0052082","FBgn0052088","FBgn0052091","FBgn0052095","FBgn0052099","FBgn0052100","FBgn0052103","FBgn0052104","FBgn0052109","FBgn0052112","FBgn0052113","FBgn0052115","FBgn0052132","FBgn0052133","FBgn0052135","FBgn0052138","FBgn0052152","FBgn0052155","FBgn0052159","FBgn0052163","FBgn0052170","FBgn0052176","FBgn0052177","FBgn0052179","FBgn0052181","FBgn0052182","FBgn0052185","FBgn0052190","FBgn0052193","FBgn0052195","FBgn0052196","FBgn0052202","FBgn0052206","FBgn0052207","FBgn0052210","FBgn0052221","FBgn0052226","FBgn0052227","FBgn0052230","FBgn0052243","FBgn0052250","FBgn0052251","FBgn0052262","FBgn0052264","FBgn0052267","FBgn0052268","FBgn0052271","FBgn0052278","FBgn0052280","FBgn0052281","FBgn0052296","FBgn0052302","FBgn0052311","FBgn0052313","FBgn0052318","FBgn0052343","FBgn0052344","FBgn0052350","FBgn0052352","FBgn0052353","FBgn0052354","FBgn0052355","FBgn0052364","FBgn0052365","FBgn0052368","FBgn0052369","FBgn0052372","FBgn0052380","FBgn0052393","FBgn0052394","FBgn0052397","FBgn0052407","FBgn0052409","FBgn0052412","FBgn0052418","FBgn0052423","FBgn0052425","FBgn0052428","FBgn0052436","FBgn0052438","FBgn0052441","FBgn0052442","FBgn0052445","FBgn0052446","FBgn0052448","FBgn0052451","FBgn0052452","FBgn0052457","FBgn0052458","FBgn0052473","FBgn0052476","FBgn0052477","FBgn0052479","FBgn0052480","FBgn0052483","FBgn0052484","FBgn0052485","FBgn0052486","FBgn0052495","FBgn0052499","FBgn0052512","FBgn0052521","FBgn0052528","FBgn0052529","FBgn0052532","FBgn0052533","FBgn0052536","FBgn0052537","FBgn0052549","FBgn0052554","FBgn0052556","FBgn0052557","FBgn0052572","FBgn0052576","FBgn0052579","FBgn0052580","FBgn0052581","FBgn0052582","FBgn0052590","FBgn0052594","FBgn0052595","FBgn0052606","FBgn0052613","FBgn0052625","FBgn0052626","FBgn0052627","FBgn0052628","FBgn0052638","FBgn0052639","FBgn0052640","FBgn0052641","FBgn0052649","FBgn0052650","FBgn0052654","FBgn0052655","FBgn0052662","FBgn0052663","FBgn0052666","FBgn0052668","FBgn0052669","FBgn0052672","FBgn0052675","FBgn0052676","FBgn0052677","FBgn0052681","FBgn0052685","FBgn0052686","FBgn0052687","FBgn0052699","FBgn0052700","FBgn0052702","FBgn0052703","FBgn0052707","FBgn0052708","FBgn0052711","FBgn0052719","FBgn0052720","FBgn0052732","FBgn0052743","FBgn0052745","FBgn0052756","FBgn0052758","FBgn0052767","FBgn0052772","FBgn0052783","FBgn0052791","FBgn0052801","FBgn0052803","FBgn0052808","FBgn0052813","FBgn0052815","FBgn0052816","FBgn0052823","FBgn0052829","FBgn0052845","FBgn0052850","FBgn0052856","FBgn0052865","FBgn0052939","FBgn0052944","FBgn0052982","FBgn0052984","FBgn0052985","FBgn0053002","FBgn0053007","FBgn0053017","FBgn0053051","FBgn0053054","FBgn0053056","FBgn0053057","FBgn0053080","FBgn0053082","FBgn0053087","FBgn0053094","FBgn0053095","FBgn0053096","FBgn0053097","FBgn0053099","FBgn0053100","FBgn0053105","FBgn0053107","FBgn0053108","FBgn0053111","FBgn0053113","FBgn0053116","FBgn0053120","FBgn0053123","FBgn0053124","FBgn0053127","FBgn0053129","FBgn0053137","FBgn0053138","FBgn0053139","FBgn0053144","FBgn0053145","FBgn0053156","FBgn0053158","FBgn0053159","FBgn0053169","FBgn0053170","FBgn0053172","FBgn0053173","FBgn0053177","FBgn0053178","FBgn0053180","FBgn0053181","FBgn0053182","FBgn0053193","FBgn0053196","FBgn0053197","FBgn0053198","FBgn0053203","FBgn0053207","FBgn0053208","FBgn0053213","FBgn0053214","FBgn0053217","FBgn0053225","FBgn0053229","FBgn0053230","FBgn0053260","FBgn0053267","FBgn0053276","FBgn0053278","FBgn0053288","FBgn0053294","FBgn0053300","FBgn0053303","FBgn0053307","FBgn0053317","FBgn0053324","FBgn0053328","FBgn0053329","FBgn0053330","FBgn0053346","FBgn0053460","FBgn0053461","FBgn0053464","FBgn0053465","FBgn0053468","FBgn0053481","FBgn0053493","FBgn0053494","FBgn0053498","FBgn0053503","FBgn0053505","FBgn0053506","FBgn0053508","FBgn0053509","FBgn0053510","FBgn0053511","FBgn0053513","FBgn0053516","FBgn0053519","FBgn0053520","FBgn0053522","FBgn0053523","FBgn0053526","FBgn0053527","FBgn0053542","FBgn0053548","FBgn0053554","FBgn0053555","FBgn0053556","FBgn0053558","FBgn0053635","FBgn0053640","FBgn0053641","FBgn0053642","FBgn0053643","FBgn0053644","FBgn0053645","FBgn0053653","FBgn0053696","FBgn0053757","FBgn0053774","FBgn0053867","FBgn0053868","FBgn0053909","FBgn0053914","FBgn0053926","FBgn0053936","FBgn0053937","FBgn0053963","FBgn0053967","FBgn0053969","FBgn0053970","FBgn0053971","FBgn0053977","FBgn0053978","FBgn0053983","FBgn0053987","FBgn0053988","FBgn0053995","FBgn0054001","FBgn0054003","FBgn0054008","FBgn0054015","FBgn0054039","FBgn0054054","FBgn0054056","FBgn0054057","FBgn0058005","FBgn0058006","FBgn0058045","FBgn0058127","FBgn0058298","FBgn0058439","FBgn0058470","FBgn0060296","FBgn0061196","FBgn0061198","FBgn0061200","FBgn0061469","FBgn0061476","FBgn0061492","FBgn0061515","FBgn0062413","FBgn0062440","FBgn0062442","FBgn0062449","FBgn0062463","FBgn0062517","FBgn0062565","FBgn0062928","FBgn0062978","FBgn0063449","FBgn0063485","FBgn0063491","FBgn0063492","FBgn0063493","FBgn0063494","FBgn0063495","FBgn0063497","FBgn0063498","FBgn0063649","FBgn0063667","FBgn0064225","FBgn0064766","FBgn0064912","FBgn0065032","FBgn0065035","FBgn0065053","FBgn0065064","FBgn0065088","FBgn0065098","FBgn0065099","FBgn0065104","FBgn0066084","FBgn0066101","FBgn0066114","FBgn0066292","FBgn0067102","FBgn0067317","FBgn0067628","FBgn0067629","FBgn0067779","FBgn0067783","FBgn0067861","FBgn0067864","FBgn0067895","FBgn0069242","FBgn0069354","FBgn0069913","FBgn0069921","FBgn0069973","FBgn0082582","FBgn0082585","FBgn0082598","FBgn0082831","FBgn0083047","FBgn0083048","FBgn0083050","FBgn0083068","FBgn0083077","FBgn0083120","FBgn0083121","FBgn0083123","FBgn0083124","FBgn0083167","FBgn0083919","FBgn0083940","FBgn0083953","FBgn0083959","FBgn0083960","FBgn0083961","FBgn0083962","FBgn0083968","FBgn0083969","FBgn0083970","FBgn0083972","FBgn0083973","FBgn0083974","FBgn0083977","FBgn0083983","FBgn0083985","FBgn0083986","FBgn0083990","FBgn0084001","FBgn0084015","FBgn0084049","FBgn0085188","FBgn0085192","FBgn0085203","FBgn0085212","FBgn0085215","FBgn0085216","FBgn0085220","FBgn0085223","FBgn0085224","FBgn0085226","FBgn0085229","FBgn0085242","FBgn0085249","FBgn0085257","FBgn0085260","FBgn0085261","FBgn0085271","FBgn0085275","FBgn0085279","FBgn0085281","FBgn0085284","FBgn0085290","FBgn0085292","FBgn0085298","FBgn0085317","FBgn0085322","FBgn0085334","FBgn0085335","FBgn0085339","FBgn0085348","FBgn0085350","FBgn0085351","FBgn0085352","FBgn0085353","FBgn0085354","FBgn0085357","FBgn0085359","FBgn0085360","FBgn0085363","FBgn0085370","FBgn0085375","FBgn0085376","FBgn0085377","FBgn0085378","FBgn0085379","FBgn0085384","FBgn0085386","FBgn0085388","FBgn0085390","FBgn0085395","FBgn0085399","FBgn0085402","FBgn0085403","FBgn0085405","FBgn0085407","FBgn0085408","FBgn0085411","FBgn0085412","FBgn0085413","FBgn0085417","FBgn0085419","FBgn0085421","FBgn0085423","FBgn0085425","FBgn0085427","FBgn0085428","FBgn0085429","FBgn0085430","FBgn0085432","FBgn0085433","FBgn0085434","FBgn0085435","FBgn0085436","FBgn0085437","FBgn0085438","FBgn0085440","FBgn0085442","FBgn0085443","FBgn0085444","FBgn0085445","FBgn0085446","FBgn0085450","FBgn0085451","FBgn0085452","FBgn0085453","FBgn0085459","FBgn0085460","FBgn0085465","FBgn0085466","FBgn0085468","FBgn0085478","FBgn0085483","FBgn0085484","FBgn0085488","FBgn0085489","FBgn0085787","FBgn0085789","FBgn0085792","FBgn0086031","FBgn0086038","FBgn0086129","FBgn0086130","FBgn0086133","FBgn0086134","FBgn0086200","FBgn0086251","FBgn0086253","FBgn0086254","FBgn0086346","FBgn0086347","FBgn0086348","FBgn0086350","FBgn0086355","FBgn0086356","FBgn0086357","FBgn0086358","FBgn0086359","FBgn0086361","FBgn0086362","FBgn0086364","FBgn0086365","FBgn0086367","FBgn0086370","FBgn0086371","FBgn0086372","FBgn0086377","FBgn0086384","FBgn0086441","FBgn0086442","FBgn0086443","FBgn0086444","FBgn0086445","FBgn0086446","FBgn0086447","FBgn0086448","FBgn0086450","FBgn0086451","FBgn0086472","FBgn0086475","FBgn0086532","FBgn0086558","FBgn0086605","FBgn0086608","FBgn0086613","FBgn0086655","FBgn0086656","FBgn0086657","FBgn0086661","FBgn0086664","FBgn0086666","FBgn0086674","FBgn0086676","FBgn0086679","FBgn0086681","FBgn0086683","FBgn0086685","FBgn0086686","FBgn0086687","FBgn0086689","FBgn0086690","FBgn0086691","FBgn0086693","FBgn0086694","FBgn0086695","FBgn0086697","FBgn0086698","FBgn0086699","FBgn0086704","FBgn0086706","FBgn0086707","FBgn0086708","FBgn0086710","FBgn0086712","FBgn0086736","FBgn0086757","FBgn0086758","FBgn0086768","FBgn0086778","FBgn0086779","FBgn0086782","FBgn0086783","FBgn0086784","FBgn0086785","FBgn0086855","FBgn0086856","FBgn0086895","FBgn0086897","FBgn0086898","FBgn0086899","FBgn0086901","FBgn0086902","FBgn0086904","FBgn0086906","FBgn0086908","FBgn0086909","FBgn0086910","FBgn0086911","FBgn0086912","FBgn0087002","FBgn0087007","FBgn0087008","FBgn0087013","FBgn0087021","FBgn0087035","FBgn0087039","FBgn0243505","FBgn0243511","FBgn0243512","FBgn0243513","FBgn0243514","FBgn0243516","FBgn0243517","FBgn0250732","FBgn0250746","FBgn0250747","FBgn0250753","FBgn0250754","FBgn0250755","FBgn0250757","FBgn0250785","FBgn0250786","FBgn0250788","FBgn0250789","FBgn0250791","FBgn0250814","FBgn0250816","FBgn0250818","FBgn0250819","FBgn0250820","FBgn0250821","FBgn0250823","FBgn0250824","FBgn0250829","FBgn0250830","FBgn0250837","FBgn0250838","FBgn0250843","FBgn0250848","FBgn0250850","FBgn0250861","FBgn0250867","FBgn0250868","FBgn0250869","FBgn0250870","FBgn0250871","FBgn0250874","FBgn0250876","FBgn0250904","FBgn0250906","FBgn0250907","FBgn0259100","FBgn0259101","FBgn0259108","FBgn0259109","FBgn0259110","FBgn0259111","FBgn0259112","FBgn0259113","FBgn0259139","FBgn0259140","FBgn0259142","FBgn0259143","FBgn0259144","FBgn0259150","FBgn0259151","FBgn0259152","FBgn0259163","FBgn0259164","FBgn0259166","FBgn0259167","FBgn0259168","FBgn0259170","FBgn0259171","FBgn0259173","FBgn0259174","FBgn0259175","FBgn0259176","FBgn0259178","FBgn0259183","FBgn0259184","FBgn0259188","FBgn0259209","FBgn0259211","FBgn0259212","FBgn0259214","FBgn0259215","FBgn0259216","FBgn0259217","FBgn0259220","FBgn0259221","FBgn0259224","FBgn0259225","FBgn0259226","FBgn0259227","FBgn0259228","FBgn0259233","FBgn0259234","FBgn0259236","FBgn0259238","FBgn0259240","FBgn0259242","FBgn0259243","FBgn0259246","FBgn0259247","FBgn0259481","FBgn0259482","FBgn0259483","FBgn0259676","FBgn0259677","FBgn0259678","FBgn0259682","FBgn0259683","FBgn0259685","FBgn0259700","FBgn0259704","FBgn0259705","FBgn0259707","FBgn0259710","FBgn0259711","FBgn0259712","FBgn0259714","FBgn0259715","FBgn0259716","FBgn0259728","FBgn0259734","FBgn0259735","FBgn0259736","FBgn0259739","FBgn0259740","FBgn0259741","FBgn0259742","FBgn0259743","FBgn0259744","FBgn0259745","FBgn0259749","FBgn0259784","FBgn0259785","FBgn0259789","FBgn0259791","FBgn0259818","FBgn0259823","FBgn0259824","FBgn0259832","FBgn0259834","FBgn0259876","FBgn0259878","FBgn0259896","FBgn0259916","FBgn0259918","FBgn0259923","FBgn0259926","FBgn0259935","FBgn0259936","FBgn0259937","FBgn0259938","FBgn0259977","FBgn0259978","FBgn0259979","FBgn0259982","FBgn0259984","FBgn0259985","FBgn0259990","FBgn0259991","FBgn0259993","FBgn0259994","FBgn0260003","FBgn0260004","FBgn0260005","FBgn0260006","FBgn0260010","FBgn0260011","FBgn0260012","FBgn0260026","FBgn0260027","FBgn0260049","FBgn0260243","FBgn0260386","FBgn0260388","FBgn0260397","FBgn0260399","FBgn0260400","FBgn0260407","FBgn0260429","FBgn0260431","FBgn0260439","FBgn0260440","FBgn0260441","FBgn0260442","FBgn0260444","FBgn0260450","FBgn0260456","FBgn0260457","FBgn0260460","FBgn0260462","FBgn0260463","FBgn0260465","FBgn0260475","FBgn0260477","FBgn0260478","FBgn0260481","FBgn0260484","FBgn0260486","FBgn0260632","FBgn0260634","FBgn0260635","FBgn0260639","FBgn0260645","FBgn0260648","FBgn0260653","FBgn0260655","FBgn0260657","FBgn0260658","FBgn0260659","FBgn0260660","FBgn0260720","FBgn0260722","FBgn0260723","FBgn0260724","FBgn0260741","FBgn0260742","FBgn0260743","FBgn0260744","FBgn0260745","FBgn0260746","FBgn0260747","FBgn0260748","FBgn0260749","FBgn0260750","FBgn0260753","FBgn0260755","FBgn0260756","FBgn0260757","FBgn0260758","FBgn0260766","FBgn0260767","FBgn0260768","FBgn0260771","FBgn0260779","FBgn0260780","FBgn0260789","FBgn0260793","FBgn0260794","FBgn0260795","FBgn0260798","FBgn0260799","FBgn0260817","FBgn0260855","FBgn0260856","FBgn0260857","FBgn0260858","FBgn0260859","FBgn0260860","FBgn0260861","FBgn0260862","FBgn0260866","FBgn0260874","FBgn0260932","FBgn0260933","FBgn0260934","FBgn0260935","FBgn0260936","FBgn0260937","FBgn0260938","FBgn0260939","FBgn0260940","FBgn0260941","FBgn0260944","FBgn0260945","FBgn0260946","FBgn0260952","FBgn0260959","FBgn0260960","FBgn0260962","FBgn0260963","FBgn0260965","FBgn0260966","FBgn0260970","FBgn0260972","FBgn0260985","FBgn0260986","FBgn0260990","FBgn0260991","FBgn0260992","FBgn0260993","FBgn0261004","FBgn0261014","FBgn0261015","FBgn0261016","FBgn0261020","FBgn0261041","FBgn0261049","FBgn0261053","FBgn0261064","FBgn0261065","FBgn0261067","FBgn0261068","FBgn0261085","FBgn0261086","FBgn0261089","FBgn0261090","FBgn0261108","FBgn0261109","FBgn0261111","FBgn0261112","FBgn0261113","FBgn0261119","FBgn0261238","FBgn0261239","FBgn0261241","FBgn0261243","FBgn0261244","FBgn0261245","FBgn0261258","FBgn0261259","FBgn0261260","FBgn0261261","FBgn0261263","FBgn0261266","FBgn0261268","FBgn0261269","FBgn0261270","FBgn0261274","FBgn0261276","FBgn0261277","FBgn0261278","FBgn0261279","FBgn0261283","FBgn0261284","FBgn0261285","FBgn0261286","FBgn0261287","FBgn0261349","FBgn0261359","FBgn0261362","FBgn0261363","FBgn0261373","FBgn0261380","FBgn0261383","FBgn0261385","FBgn0261387","FBgn0261388","FBgn0261394","FBgn0261396","FBgn0261397","FBgn0261403","FBgn0261429","FBgn0261430","FBgn0261436","FBgn0261437","FBgn0261439","FBgn0261444","FBgn0261445","FBgn0261446","FBgn0261451","FBgn0261456","FBgn0261458","FBgn0261461","FBgn0261477","FBgn0261479","FBgn0261509","FBgn0261524","FBgn0261530","FBgn0261532","FBgn0261535","FBgn0261545","FBgn0261546","FBgn0261547","FBgn0261548","FBgn0261549","FBgn0261550","FBgn0261551","FBgn0261552","FBgn0261553","FBgn0261554","FBgn0261555","FBgn0261556","FBgn0261560","FBgn0261561","FBgn0261562","FBgn0261563","FBgn0261564","FBgn0261565","FBgn0261570","FBgn0261572","FBgn0261573","FBgn0261574","FBgn0261575"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":["rgba(62,85,137,1)","rgba(59,175,125,1)","rgba(42,122,142,1)","rgba(68,3,85,1)","rgba(43,141,138,1)","rgba(70,34,104,1)","rgba(202,223,60,1)","rgba(140,212,78,1)","rgba(69,52,120,1)","rgba(68,4,85,1)","rgba(69,49,117,1)","rgba(69,20,94,1)","rgba(38,161,134,1)","rgba(39,159,134,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(42,121,142,1)","rgba(86,186,111,1)","rgba(45,117,142,1)","rgba(53,106,140,1)","rgba(68,54,122,1)","rgba(57,98,139,1)","rgba(70,179,120,1)","rgba(41,154,135,1)","rgba(70,30,101,1)","rgba(69,45,114,1)","rgba(34,168,132,1)","rgba(124,209,81,1)","rgba(65,69,135,1)","rgba(59,93,139,1)","rgba(43,129,140,1)","rgba(44,132,140,1)","rgba(97,192,104,1)","rgba(44,131,140,1)","rgba(69,19,93,1)","rgba(44,132,140,1)","rgba(43,140,138,1)","rgba(40,157,135,1)","rgba(41,152,136,1)","rgba(42,121,142,1)","rgba(91,189,108,1)","rgba(43,126,141,1)","rgba(69,50,118,1)","rgba(44,139,138,1)","rgba(69,14,90,1)","rgba(80,183,115,1)","rgba(41,152,136,1)","rgba(68,53,120,1)","rgba(104,197,98,1)","rgba(36,164,133,1)","rgba(41,152,136,1)","rgba(39,169,131,1)","rgba(54,105,140,1)","rgba(44,136,139,1)","rgba(43,139,138,1)","rgba(37,163,133,1)","rgba(69,15,91,1)","rgba(57,98,139,1)","rgba(43,126,141,1)","rgba(56,100,140,1)","rgba(36,164,133,1)","rgba(38,160,134,1)","rgba(95,191,105,1)","rgba(39,160,134,1)","rgba(44,135,139,1)","rgba(43,143,138,1)","rgba(39,158,134,1)","rgba(40,157,135,1)","rgba(39,159,134,1)","rgba(64,77,136,1)","rgba(68,57,124,1)","rgba(66,66,133,1)","rgba(62,86,138,1)","rgba(45,170,129,1)","rgba(43,125,141,1)","rgba(43,170,130,1)","rgba(173,217,69,1)","rgba(41,153,135,1)","rgba(111,201,92,1)","rgba(64,73,136,1)","rgba(42,150,136,1)","rgba(43,139,138,1)","rgba(106,198,97,1)","rgba(50,111,141,1)","rgba(53,107,140,1)","rgba(69,51,118,1)","rgba(40,156,135,1)","rgba(43,128,140,1)","rgba(62,85,137,1)","rgba(67,59,126,1)","rgba(44,137,139,1)","rgba(98,193,103,1)","rgba(43,147,137,1)","rgba(44,139,138,1)","rgba(44,134,139,1)","rgba(43,130,140,1)","rgba(43,143,138,1)","rgba(44,131,140,1)","rgba(43,126,141,1)","rgba(37,162,133,1)","rgba(38,161,134,1)","rgba(50,111,141,1)","rgba(57,174,126,1)","rgba(36,166,133,1)","rgba(36,165,133,1)","rgba(38,161,134,1)","rgba(154,214,74,1)","rgba(68,5,86,1)","rgba(44,133,140,1)","rgba(43,131,140,1)","rgba(69,12,89,1)","rgba(36,165,133,1)","rgba(43,127,141,1)","rgba(41,152,136,1)","rgba(36,164,133,1)","rgba(70,38,107,1)","rgba(44,139,138,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(54,104,140,1)","rgba(43,141,138,1)","rgba(68,8,87,1)","rgba(43,130,140,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(68,5,86,1)","rgba(56,101,140,1)","rgba(44,138,139,1)","rgba(65,177,122,1)","rgba(57,98,139,1)","rgba(44,134,139,1)","rgba(43,140,138,1)","rgba(45,117,142,1)","rgba(253,231,37,1)","rgba(66,65,132,1)","rgba(218,225,54,1)","rgba(42,150,136,1)","rgba(43,130,140,1)","rgba(44,136,139,1)","rgba(50,110,141,1)","rgba(47,171,129,1)","rgba(42,150,136,1)","rgba(70,179,120,1)","rgba(90,188,109,1)","rgba(43,170,130,1)","rgba(45,117,142,1)","rgba(69,46,114,1)","rgba(42,149,136,1)","rgba(42,120,142,1)","rgba(44,137,139,1)","rgba(58,97,139,1)","rgba(43,144,137,1)","rgba(69,45,113,1)","rgba(69,50,118,1)","rgba(44,118,142,1)","rgba(58,97,139,1)","rgba(65,69,135,1)","rgba(43,145,137,1)","rgba(42,151,136,1)","rgba(38,161,134,1)","rgba(43,143,138,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(39,158,134,1)","rgba(59,93,139,1)","rgba(48,113,141,1)","rgba(60,92,138,1)","rgba(67,58,126,1)","rgba(44,133,140,1)","rgba(38,162,133,1)","rgba(43,146,137,1)","rgba(66,65,133,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(55,102,140,1)","rgba(64,77,136,1)","rgba(69,22,95,1)","rgba(43,145,137,1)","rgba(41,154,135,1)","rgba(65,70,135,1)","rgba(66,64,131,1)","rgba(58,174,125,1)","rgba(91,189,108,1)","rgba(43,125,141,1)","rgba(80,183,115,1)","rgba(56,174,126,1)","rgba(38,161,134,1)","rgba(155,214,74,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(43,147,137,1)","rgba(37,162,133,1)","rgba(50,111,141,1)","rgba(43,125,141,1)","rgba(69,16,91,1)","rgba(43,125,141,1)","rgba(54,173,127,1)","rgba(43,119,142,1)","rgba(42,122,142,1)","rgba(39,158,134,1)","rgba(66,66,133,1)","rgba(35,166,133,1)","rgba(42,150,136,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(37,163,133,1)","rgba(69,20,94,1)","rgba(42,148,136,1)","rgba(39,169,131,1)","rgba(44,134,139,1)","rgba(42,120,142,1)","rgba(42,122,142,1)","rgba(43,130,140,1)","rgba(76,181,117,1)","rgba(138,212,78,1)","rgba(205,223,60,1)","rgba(75,181,117,1)","rgba(43,143,138,1)","rgba(43,130,140,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(68,56,124,1)","rgba(55,102,140,1)","rgba(165,216,72,1)","rgba(43,142,138,1)","rgba(43,147,137,1)","rgba(35,167,132,1)","rgba(66,63,130,1)","rgba(69,43,111,1)","rgba(87,187,111,1)","rgba(42,151,136,1)","rgba(70,39,108,1)","rgba(66,63,131,1)","rgba(57,98,139,1)","rgba(43,127,141,1)","rgba(43,146,137,1)","rgba(64,77,136,1)","rgba(98,193,103,1)","rgba(43,144,137,1)","rgba(54,173,126,1)","rgba(70,33,103,1)","rgba(102,195,100,1)","rgba(69,23,96,1)","rgba(37,163,133,1)","rgba(43,123,142,1)","rgba(43,145,137,1)","rgba(44,133,140,1)","rgba(45,117,142,1)","rgba(42,148,137,1)","rgba(50,111,141,1)","rgba(38,160,134,1)","rgba(44,136,139,1)","rgba(74,181,118,1)","rgba(35,166,132,1)","rgba(179,219,68,1)","rgba(79,183,115,1)","rgba(40,155,135,1)","rgba(70,34,104,1)","rgba(43,127,141,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(73,180,118,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(44,137,139,1)","rgba(42,122,142,1)","rgba(68,4,86,1)","rgba(35,166,133,1)","rgba(43,126,141,1)","rgba(50,111,141,1)","rgba(42,148,137,1)","rgba(41,169,130,1)","rgba(64,77,136,1)","rgba(36,165,133,1)","rgba(53,173,127,1)","rgba(35,168,132,1)","rgba(43,144,137,1)","rgba(83,185,113,1)","rgba(45,117,142,1)","rgba(41,153,135,1)","rgba(69,12,89,1)","rgba(58,97,139,1)","rgba(35,167,132,1)","rgba(74,180,118,1)","rgba(67,62,129,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(42,121,142,1)","rgba(68,178,121,1)","rgba(64,177,123,1)","rgba(203,223,60,1)","rgba(153,214,74,1)","rgba(105,197,97,1)","rgba(68,4,86,1)","rgba(42,147,137,1)","rgba(43,139,138,1)","rgba(43,144,137,1)","rgba(70,41,110,1)","rgba(41,154,135,1)","rgba(65,68,135,1)","rgba(50,111,141,1)","rgba(58,97,139,1)","rgba(98,193,103,1)","rgba(142,212,77,1)","rgba(69,21,94,1)","rgba(121,208,82,1)","rgba(144,213,76,1)","rgba(66,65,132,1)","rgba(164,216,72,1)","rgba(151,214,75,1)","rgba(36,165,133,1)","rgba(42,121,142,1)","rgba(34,168,132,1)","rgba(79,183,116,1)","rgba(45,117,142,1)","rgba(62,83,137,1)","rgba(48,114,141,1)","rgba(56,100,139,1)","rgba(65,68,135,1)","rgba(86,186,111,1)","rgba(43,126,141,1)","rgba(47,115,141,1)","rgba(70,26,98,1)","rgba(43,128,140,1)","rgba(66,63,131,1)","rgba(43,140,138,1)","rgba(42,121,142,1)","rgba(43,146,137,1)","rgba(49,172,128,1)","rgba(59,175,125,1)","rgba(51,172,128,1)","rgba(41,154,135,1)","rgba(42,121,142,1)","rgba(62,86,138,1)","rgba(44,132,140,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(42,121,142,1)","rgba(55,103,140,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(39,159,134,1)","rgba(64,75,136,1)","rgba(105,197,98,1)","rgba(70,40,109,1)","rgba(64,177,123,1)","rgba(61,87,138,1)","rgba(63,82,137,1)","rgba(44,132,140,1)","rgba(68,7,87,1)","rgba(65,71,135,1)","rgba(49,113,141,1)","rgba(70,28,99,1)","rgba(49,171,128,1)","rgba(56,101,140,1)","rgba(45,117,142,1)","rgba(58,95,139,1)","rgba(69,18,92,1)","rgba(99,194,102,1)","rgba(68,54,122,1)","rgba(48,113,141,1)","rgba(42,148,136,1)","rgba(69,43,111,1)","rgba(154,214,74,1)","rgba(41,153,136,1)","rgba(55,103,140,1)","rgba(40,155,135,1)","rgba(62,176,123,1)","rgba(60,91,138,1)","rgba(38,162,133,1)","rgba(43,146,137,1)","rgba(56,100,140,1)","rgba(42,150,136,1)","rgba(39,160,134,1)","rgba(42,151,136,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(62,84,137,1)","rgba(69,23,95,1)","rgba(42,151,136,1)","rgba(58,96,139,1)","rgba(72,180,119,1)","rgba(79,183,115,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(61,175,124,1)","rgba(38,161,134,1)","rgba(44,132,140,1)","rgba(43,119,142,1)","rgba(69,25,97,1)","rgba(44,136,139,1)","rgba(63,82,137,1)","rgba(66,177,122,1)","rgba(42,150,136,1)","rgba(39,158,134,1)","rgba(50,172,128,1)","rgba(76,181,117,1)","rgba(70,27,99,1)","rgba(56,174,126,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(38,161,134,1)","rgba(44,136,139,1)","rgba(43,141,138,1)","rgba(42,150,136,1)","rgba(44,136,139,1)","rgba(52,107,140,1)","rgba(35,167,132,1)","rgba(70,39,108,1)","rgba(44,133,140,1)","rgba(108,199,95,1)","rgba(38,162,134,1)","rgba(126,210,80,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(97,192,104,1)","rgba(44,132,140,1)","rgba(186,220,65,1)","rgba(70,39,108,1)","rgba(43,144,137,1)","rgba(63,80,137,1)","rgba(52,108,141,1)","rgba(43,147,137,1)","rgba(42,149,136,1)","rgba(70,26,98,1)","rgba(42,148,137,1)","rgba(44,137,139,1)","rgba(44,131,140,1)","rgba(42,151,136,1)","rgba(37,162,133,1)","rgba(42,148,137,1)","rgba(60,91,138,1)","rgba(70,41,109,1)","rgba(36,165,133,1)","rgba(70,32,102,1)","rgba(68,6,87,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(36,165,133,1)","rgba(49,172,128,1)","rgba(53,107,140,1)","rgba(44,138,139,1)","rgba(38,160,134,1)","rgba(43,145,137,1)","rgba(43,128,141,1)","rgba(54,173,127,1)","rgba(58,95,139,1)","rgba(43,126,141,1)","rgba(67,60,127,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(41,153,136,1)","rgba(42,148,137,1)","rgba(55,173,126,1)","rgba(35,166,133,1)","rgba(43,123,141,1)","rgba(65,72,136,1)","rgba(67,59,127,1)","rgba(44,133,140,1)","rgba(63,176,123,1)","rgba(67,59,126,1)","rgba(49,112,141,1)","rgba(43,146,137,1)","rgba(71,179,119,1)","rgba(62,84,137,1)","rgba(45,117,142,1)","rgba(43,141,138,1)","rgba(58,174,125,1)","rgba(208,224,58,1)","rgba(39,159,134,1)","rgba(42,149,136,1)","rgba(68,55,123,1)","rgba(44,131,140,1)","rgba(36,164,133,1)","rgba(43,139,138,1)","rgba(43,140,138,1)","rgba(44,139,138,1)","rgba(40,169,131,1)","rgba(38,162,133,1)","rgba(40,155,135,1)","rgba(43,143,138,1)","rgba(43,128,141,1)","rgba(58,97,139,1)","rgba(42,151,136,1)","rgba(52,108,140,1)","rgba(36,165,133,1)","rgba(61,175,124,1)","rgba(46,116,142,1)","rgba(44,131,140,1)","rgba(68,5,86,1)","rgba(50,111,141,1)","rgba(44,131,140,1)","rgba(55,173,126,1)","rgba(51,172,128,1)","rgba(59,94,139,1)","rgba(75,181,118,1)","rgba(69,13,90,1)","rgba(59,94,139,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(70,29,100,1)","rgba(43,144,137,1)","rgba(69,47,115,1)","rgba(50,172,128,1)","rgba(38,162,133,1)","rgba(42,151,136,1)","rgba(43,141,138,1)","rgba(158,215,73,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(121,209,82,1)","rgba(80,183,115,1)","rgba(41,153,135,1)","rgba(103,196,99,1)","rgba(44,137,139,1)","rgba(69,19,93,1)","rgba(69,15,91,1)","rgba(120,207,84,1)","rgba(118,206,85,1)","rgba(79,183,116,1)","rgba(52,108,140,1)","rgba(40,169,131,1)","rgba(42,149,136,1)","rgba(39,159,134,1)","rgba(40,156,135,1)","rgba(43,127,141,1)","rgba(65,67,134,1)","rgba(63,80,137,1)","rgba(60,91,138,1)","rgba(69,11,89,1)","rgba(51,109,141,1)","rgba(65,67,134,1)","rgba(43,141,138,1)","rgba(44,136,139,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(35,166,133,1)","rgba(52,107,140,1)","rgba(43,119,142,1)","rgba(65,177,122,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(44,132,140,1)","rgba(69,50,118,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(111,201,92,1)","rgba(168,217,71,1)","rgba(69,52,119,1)","rgba(67,178,121,1)","rgba(43,145,137,1)","rgba(70,29,100,1)","rgba(42,147,137,1)","rgba(55,173,126,1)","rgba(43,144,137,1)","rgba(65,177,122,1)","rgba(63,82,137,1)","rgba(68,7,87,1)","rgba(42,170,130,1)","rgba(76,182,117,1)","rgba(62,83,137,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(39,158,134,1)","rgba(45,170,129,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(44,133,140,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(39,160,134,1)","rgba(36,164,133,1)","rgba(42,150,136,1)","rgba(44,135,139,1)","rgba(37,162,133,1)","rgba(37,169,131,1)","rgba(43,170,130,1)","rgba(51,110,141,1)","rgba(82,184,114,1)","rgba(106,198,97,1)","rgba(56,174,126,1)","rgba(44,134,139,1)","rgba(157,215,74,1)","rgba(42,150,136,1)","rgba(68,6,86,1)","rgba(85,186,112,1)","rgba(43,140,138,1)","rgba(41,153,136,1)","rgba(38,162,133,1)","rgba(49,172,128,1)","rgba(43,123,141,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(42,151,136,1)","rgba(43,130,140,1)","rgba(70,34,104,1)","rgba(43,126,141,1)","rgba(87,187,111,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(40,169,131,1)","rgba(70,36,105,1)","rgba(34,168,132,1)","rgba(40,156,135,1)","rgba(56,174,126,1)","rgba(65,68,135,1)","rgba(51,109,141,1)","rgba(48,171,128,1)","rgba(40,155,135,1)","rgba(104,197,98,1)","rgba(70,26,98,1)","rgba(42,121,142,1)","rgba(44,136,139,1)","rgba(42,147,137,1)","rgba(43,147,137,1)","rgba(44,133,140,1)","rgba(71,179,120,1)","rgba(42,150,136,1)","rgba(42,147,137,1)","rgba(65,68,135,1)","rgba(63,176,123,1)","rgba(42,147,137,1)","rgba(59,94,139,1)","rgba(40,157,135,1)","rgba(39,159,134,1)","rgba(39,158,134,1)","rgba(87,187,111,1)","rgba(80,183,115,1)","rgba(37,162,133,1)","rgba(36,164,133,1)","rgba(56,101,140,1)","rgba(44,134,139,1)","rgba(81,184,114,1)","rgba(50,110,141,1)","rgba(42,151,136,1)","rgba(44,118,142,1)","rgba(94,191,106,1)","rgba(69,18,93,1)","rgba(59,93,138,1)","rgba(42,120,142,1)","rgba(42,148,137,1)","rgba(67,62,129,1)","rgba(54,105,140,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(65,70,135,1)","rgba(157,215,74,1)","rgba(40,169,131,1)","rgba(62,85,137,1)","rgba(43,141,138,1)","rgba(52,172,127,1)","rgba(45,171,129,1)","rgba(43,145,137,1)","rgba(65,68,135,1)","rgba(69,8,87,1)","rgba(37,162,133,1)","rgba(41,155,135,1)","rgba(43,125,141,1)","rgba(68,7,87,1)","rgba(44,136,139,1)","rgba(44,135,139,1)","rgba(83,185,113,1)","rgba(58,95,139,1)","rgba(41,153,135,1)","rgba(43,145,137,1)","rgba(43,128,141,1)","rgba(62,86,137,1)","rgba(39,158,134,1)","rgba(170,217,70,1)","rgba(43,170,130,1)","rgba(52,172,127,1)","rgba(51,172,127,1)","rgba(48,114,141,1)","rgba(43,139,138,1)","rgba(37,162,133,1)","rgba(43,140,138,1)","rgba(44,134,139,1)","rgba(44,137,139,1)","rgba(40,156,135,1)","rgba(42,147,137,1)","rgba(42,121,142,1)","rgba(109,200,94,1)","rgba(53,173,127,1)","rgba(70,40,109,1)","rgba(68,4,86,1)","rgba(43,125,141,1)","rgba(118,206,86,1)","rgba(66,66,133,1)","rgba(145,213,76,1)","rgba(43,142,138,1)","rgba(44,135,139,1)","rgba(69,10,89,1)","rgba(64,75,136,1)","rgba(43,140,138,1)","rgba(42,148,136,1)","rgba(70,39,108,1)","rgba(43,142,138,1)","rgba(61,88,138,1)","rgba(99,194,102,1)","rgba(40,155,135,1)","rgba(88,187,110,1)","rgba(117,205,87,1)","rgba(42,121,142,1)","rgba(41,152,136,1)","rgba(43,141,138,1)","rgba(42,147,137,1)","rgba(37,163,133,1)","rgba(37,163,133,1)","rgba(58,96,139,1)","rgba(46,116,141,1)","rgba(61,88,138,1)","rgba(60,175,124,1)","rgba(61,175,124,1)","rgba(63,176,123,1)","rgba(47,114,141,1)","rgba(44,138,139,1)","rgba(51,109,141,1)","rgba(44,134,139,1)","rgba(70,40,109,1)","rgba(42,149,136,1)","rgba(75,181,117,1)","rgba(48,113,141,1)","rgba(43,146,137,1)","rgba(67,61,128,1)","rgba(65,72,136,1)","rgba(44,135,139,1)","rgba(69,17,92,1)","rgba(62,84,137,1)","rgba(61,87,138,1)","rgba(37,164,133,1)","rgba(69,45,113,1)","rgba(56,101,140,1)","rgba(128,210,80,1)","rgba(44,137,139,1)","rgba(61,88,138,1)","rgba(71,179,120,1)","rgba(52,172,127,1)","rgba(60,175,124,1)","rgba(34,168,132,1)","rgba(43,145,137,1)","rgba(158,215,73,1)","rgba(43,144,137,1)","rgba(41,153,135,1)","rgba(81,184,114,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(64,75,136,1)","rgba(103,196,100,1)","rgba(57,99,139,1)","rgba(42,148,137,1)","rgba(37,163,133,1)","rgba(39,158,134,1)","rgba(61,87,138,1)","rgba(69,8,88,1)","rgba(81,184,114,1)","rgba(43,170,130,1)","rgba(69,15,91,1)","rgba(70,32,102,1)","rgba(132,211,79,1)","rgba(45,117,142,1)","rgba(54,173,127,1)","rgba(50,172,128,1)","rgba(42,148,136,1)","rgba(56,101,140,1)","rgba(42,148,136,1)","rgba(43,129,140,1)","rgba(65,177,122,1)","rgba(67,178,121,1)","rgba(43,119,142,1)","rgba(65,68,135,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(43,124,141,1)","rgba(80,183,115,1)","rgba(66,66,133,1)","rgba(34,168,132,1)","rgba(44,135,139,1)","rgba(60,90,138,1)","rgba(72,179,119,1)","rgba(43,139,138,1)","rgba(51,172,128,1)","rgba(88,187,110,1)","rgba(44,132,140,1)","rgba(45,170,129,1)","rgba(39,158,134,1)","rgba(43,124,141,1)","rgba(39,158,134,1)","rgba(46,171,129,1)","rgba(69,24,96,1)","rgba(43,145,137,1)","rgba(68,53,120,1)","rgba(65,72,136,1)","rgba(69,18,93,1)","rgba(37,162,133,1)","rgba(43,142,138,1)","rgba(43,130,140,1)","rgba(60,92,138,1)","rgba(43,141,138,1)","rgba(66,66,133,1)","rgba(118,206,86,1)","rgba(126,210,80,1)","rgba(139,212,78,1)","rgba(124,209,81,1)","rgba(164,216,72,1)","rgba(41,155,135,1)","rgba(42,148,137,1)","rgba(43,125,141,1)","rgba(50,111,141,1)","rgba(43,140,138,1)","rgba(43,125,141,1)","rgba(112,202,91,1)","rgba(64,74,136,1)","rgba(44,135,139,1)","rgba(44,136,139,1)","rgba(64,176,123,1)","rgba(68,53,121,1)","rgba(69,20,94,1)","rgba(103,196,100,1)","rgba(58,96,139,1)","rgba(54,105,140,1)","rgba(44,131,140,1)","rgba(40,157,135,1)","rgba(59,94,139,1)","rgba(56,101,140,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(37,169,131,1)","rgba(48,171,128,1)","rgba(41,152,136,1)","rgba(43,145,137,1)","rgba(41,153,135,1)","rgba(40,155,135,1)","rgba(55,103,140,1)","rgba(58,174,125,1)","rgba(38,160,134,1)","rgba(42,148,137,1)","rgba(69,47,115,1)","rgba(93,190,107,1)","rgba(53,106,140,1)","rgba(42,149,136,1)","rgba(44,134,139,1)","rgba(38,161,134,1)","rgba(40,157,135,1)","rgba(54,105,140,1)","rgba(43,143,138,1)","rgba(40,155,135,1)","rgba(43,139,138,1)","rgba(36,164,133,1)","rgba(45,117,142,1)","rgba(43,126,141,1)","rgba(43,143,138,1)","rgba(41,153,136,1)","rgba(46,171,129,1)","rgba(42,151,136,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(39,158,134,1)","rgba(43,125,141,1)","rgba(35,166,132,1)","rgba(56,101,140,1)","rgba(39,160,134,1)","rgba(104,197,98,1)","rgba(44,131,140,1)","rgba(41,154,135,1)","rgba(43,119,142,1)","rgba(41,152,136,1)","rgba(42,151,136,1)","rgba(48,113,141,1)","rgba(50,111,141,1)","rgba(44,136,139,1)","rgba(101,194,101,1)","rgba(68,178,121,1)","rgba(84,185,112,1)","rgba(43,145,137,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(112,202,92,1)","rgba(43,130,140,1)","rgba(39,158,134,1)","rgba(60,92,138,1)","rgba(50,110,141,1)","rgba(54,105,140,1)","rgba(142,212,77,1)","rgba(44,136,139,1)","rgba(70,27,99,1)","rgba(69,18,92,1)","rgba(162,216,72,1)","rgba(43,130,140,1)","rgba(69,42,111,1)","rgba(69,50,118,1)","rgba(40,157,135,1)","rgba(43,130,140,1)","rgba(54,104,140,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(43,125,141,1)","rgba(47,114,141,1)","rgba(50,172,128,1)","rgba(41,153,135,1)","rgba(43,142,138,1)","rgba(43,143,138,1)","rgba(65,69,135,1)","rgba(43,144,137,1)","rgba(44,134,139,1)","rgba(44,118,142,1)","rgba(38,161,134,1)","rgba(43,170,130,1)","rgba(39,158,134,1)","rgba(62,85,137,1)","rgba(36,165,133,1)","rgba(40,156,135,1)","rgba(36,165,133,1)","rgba(70,30,101,1)","rgba(64,77,136,1)","rgba(43,126,141,1)","rgba(64,76,136,1)","rgba(37,169,131,1)","rgba(40,155,135,1)","rgba(48,171,128,1)","rgba(40,156,135,1)","rgba(42,122,142,1)","rgba(43,144,137,1)","rgba(69,9,88,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(123,209,81,1)","rgba(42,149,136,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(57,99,139,1)","rgba(40,157,135,1)","rgba(69,21,95,1)","rgba(36,164,133,1)","rgba(37,164,133,1)","rgba(41,153,135,1)","rgba(68,57,124,1)","rgba(42,151,136,1)","rgba(37,164,133,1)","rgba(43,143,138,1)","rgba(104,196,99,1)","rgba(44,133,140,1)","rgba(88,187,110,1)","rgba(44,132,140,1)","rgba(38,160,134,1)","rgba(44,134,139,1)","rgba(44,138,139,1)","rgba(39,160,134,1)","rgba(35,168,132,1)","rgba(41,153,136,1)","rgba(69,17,92,1)","rgba(61,175,124,1)","rgba(44,132,140,1)","rgba(70,179,120,1)","rgba(54,104,140,1)","rgba(39,158,134,1)","rgba(41,152,136,1)","rgba(71,179,119,1)","rgba(40,157,135,1)","rgba(59,95,139,1)","rgba(49,111,141,1)","rgba(41,153,135,1)","rgba(43,130,140,1)","rgba(37,163,133,1)","rgba(44,135,139,1)","rgba(48,113,141,1)","rgba(43,125,141,1)","rgba(43,131,140,1)","rgba(72,180,119,1)","rgba(43,143,138,1)","rgba(43,146,137,1)","rgba(168,217,71,1)","rgba(61,87,138,1)","rgba(43,131,140,1)","rgba(36,165,133,1)","rgba(40,155,135,1)","rgba(70,179,120,1)","rgba(44,137,139,1)","rgba(70,39,108,1)","rgba(35,167,132,1)","rgba(41,153,136,1)","rgba(43,145,137,1)","rgba(43,130,140,1)","rgba(62,84,137,1)","rgba(36,164,133,1)","rgba(106,198,97,1)","rgba(60,91,138,1)","rgba(38,169,131,1)","rgba(122,209,81,1)","rgba(43,145,137,1)","rgba(37,164,133,1)","rgba(43,130,140,1)","rgba(42,120,142,1)","rgba(61,89,138,1)","rgba(69,25,97,1)","rgba(69,22,95,1)","rgba(49,112,141,1)","rgba(44,137,139,1)","rgba(51,109,141,1)","rgba(37,163,133,1)","rgba(70,179,120,1)","rgba(44,118,142,1)","rgba(48,113,141,1)","rgba(43,119,142,1)","rgba(38,162,134,1)","rgba(59,94,139,1)","rgba(135,211,79,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(40,157,134,1)","rgba(43,129,140,1)","rgba(68,178,121,1)","rgba(34,168,132,1)","rgba(46,116,142,1)","rgba(132,211,79,1)","rgba(43,142,138,1)","rgba(36,165,133,1)","rgba(138,212,78,1)","rgba(63,78,136,1)","rgba(69,10,88,1)","rgba(58,96,139,1)","rgba(44,137,139,1)","rgba(70,37,107,1)","rgba(43,125,141,1)","rgba(44,118,142,1)","rgba(68,5,86,1)","rgba(69,45,114,1)","rgba(42,122,142,1)","rgba(66,63,130,1)","rgba(63,176,123,1)","rgba(39,158,134,1)","rgba(69,23,96,1)","rgba(51,172,127,1)","rgba(43,129,140,1)","rgba(63,176,123,1)","rgba(36,164,133,1)","rgba(36,164,133,1)","rgba(69,11,89,1)","rgba(40,157,135,1)","rgba(43,146,137,1)","rgba(39,159,134,1)","rgba(111,201,92,1)","rgba(42,151,136,1)","rgba(42,151,136,1)","rgba(40,156,135,1)","rgba(36,165,133,1)","rgba(43,143,138,1)","rgba(36,165,133,1)","rgba(97,192,104,1)","rgba(50,110,141,1)","rgba(43,128,141,1)","rgba(42,149,136,1)","rgba(159,215,73,1)","rgba(38,161,134,1)","rgba(43,170,130,1)","rgba(69,50,118,1)","rgba(55,103,140,1)","rgba(78,182,116,1)","rgba(66,177,122,1)","rgba(37,163,133,1)","rgba(51,110,141,1)","rgba(43,142,138,1)","rgba(42,170,130,1)","rgba(60,92,138,1)","rgba(120,208,83,1)","rgba(50,172,128,1)","rgba(76,181,117,1)","rgba(54,173,127,1)","rgba(44,134,139,1)","rgba(44,131,140,1)","rgba(55,103,140,1)","rgba(70,34,104,1)","rgba(43,141,138,1)","rgba(43,129,140,1)","rgba(43,146,137,1)","rgba(121,209,82,1)","rgba(44,135,139,1)","rgba(68,56,124,1)","rgba(39,158,134,1)","rgba(42,149,136,1)","rgba(59,93,139,1)","rgba(44,170,130,1)","rgba(44,134,139,1)","rgba(44,138,139,1)","rgba(41,153,136,1)","rgba(70,29,100,1)","rgba(41,153,135,1)","rgba(69,18,93,1)","rgba(51,172,127,1)","rgba(41,152,136,1)","rgba(43,140,138,1)","rgba(42,120,142,1)","rgba(43,144,137,1)","rgba(48,171,129,1)","rgba(49,113,141,1)","rgba(50,111,141,1)","rgba(38,169,131,1)","rgba(43,140,138,1)","rgba(43,141,138,1)","rgba(84,185,113,1)","rgba(37,163,133,1)","rgba(63,81,137,1)","rgba(43,145,137,1)","rgba(70,31,101,1)","rgba(63,80,137,1)","rgba(58,96,139,1)","rgba(54,105,140,1)","rgba(43,128,141,1)","rgba(37,164,133,1)","rgba(65,68,135,1)","rgba(94,191,106,1)","rgba(43,128,141,1)","rgba(43,127,141,1)","rgba(43,127,141,1)","rgba(93,190,106,1)","rgba(40,155,135,1)","rgba(82,184,114,1)","rgba(43,146,137,1)","rgba(43,119,142,1)","rgba(54,104,140,1)","rgba(44,135,139,1)","rgba(40,155,135,1)","rgba(37,163,133,1)","rgba(39,158,134,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(82,184,114,1)","rgba(43,140,138,1)","rgba(42,149,136,1)","rgba(43,123,142,1)","rgba(44,136,139,1)","rgba(69,46,114,1)","rgba(43,145,137,1)","rgba(44,131,140,1)","rgba(55,173,126,1)","rgba(44,138,139,1)","rgba(69,16,91,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(44,139,138,1)","rgba(43,119,142,1)","rgba(42,120,142,1)","rgba(44,133,140,1)","rgba(45,116,142,1)","rgba(44,133,140,1)","rgba(35,166,132,1)","rgba(43,143,138,1)","rgba(42,148,137,1)","rgba(67,178,121,1)","rgba(44,133,140,1)","rgba(47,171,129,1)","rgba(35,166,132,1)","rgba(47,171,129,1)","rgba(53,106,140,1)","rgba(108,200,95,1)","rgba(59,94,139,1)","rgba(39,158,134,1)","rgba(40,155,135,1)","rgba(37,164,133,1)","rgba(70,27,99,1)","rgba(43,119,142,1)","rgba(44,132,140,1)","rgba(54,104,140,1)","rgba(68,55,123,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(40,157,135,1)","rgba(54,105,140,1)","rgba(42,121,142,1)","rgba(59,94,139,1)","rgba(69,24,97,1)","rgba(44,134,139,1)","rgba(56,101,140,1)","rgba(42,120,142,1)","rgba(51,109,141,1)","rgba(72,180,119,1)","rgba(43,144,137,1)","rgba(43,125,141,1)","rgba(70,30,101,1)","rgba(38,161,134,1)","rgba(43,141,138,1)","rgba(41,153,135,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(43,128,140,1)","rgba(84,185,113,1)","rgba(64,176,123,1)","rgba(80,183,115,1)","rgba(49,112,141,1)","rgba(39,159,134,1)","rgba(57,99,139,1)","rgba(42,150,136,1)","rgba(43,144,137,1)","rgba(64,177,123,1)","rgba(41,153,135,1)","rgba(130,210,79,1)","rgba(50,111,141,1)","rgba(70,31,102,1)","rgba(44,135,139,1)","rgba(68,55,122,1)","rgba(59,94,139,1)","rgba(54,173,127,1)","rgba(43,143,138,1)","rgba(92,189,108,1)","rgba(51,109,141,1)","rgba(41,154,135,1)","rgba(69,178,120,1)","rgba(42,148,137,1)","rgba(56,101,140,1)","rgba(44,131,140,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(40,156,135,1)","rgba(44,138,139,1)","rgba(44,137,139,1)","rgba(40,156,135,1)","rgba(40,156,135,1)","rgba(66,177,122,1)","rgba(44,133,140,1)","rgba(43,140,138,1)","rgba(43,142,138,1)","rgba(43,146,137,1)","rgba(55,103,140,1)","rgba(67,178,121,1)","rgba(41,152,136,1)","rgba(44,131,140,1)","rgba(70,36,105,1)","rgba(44,131,140,1)","rgba(44,133,140,1)","rgba(39,159,134,1)","rgba(41,154,135,1)","rgba(44,132,140,1)","rgba(52,108,141,1)","rgba(40,155,135,1)","rgba(37,169,131,1)","rgba(43,146,137,1)","rgba(42,122,142,1)","rgba(43,123,142,1)","rgba(51,109,141,1)","rgba(43,144,137,1)","rgba(47,115,141,1)","rgba(41,170,130,1)","rgba(44,134,139,1)","rgba(149,213,75,1)","rgba(43,127,141,1)","rgba(42,120,142,1)","rgba(62,86,137,1)","rgba(65,177,122,1)","rgba(39,158,134,1)","rgba(38,161,134,1)","rgba(46,115,141,1)","rgba(57,98,139,1)","rgba(46,115,141,1)","rgba(42,120,142,1)","rgba(44,132,140,1)","rgba(42,121,142,1)","rgba(69,49,117,1)","rgba(36,165,133,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(76,181,117,1)","rgba(43,146,137,1)","rgba(43,144,137,1)","rgba(43,143,138,1)","rgba(43,123,141,1)","rgba(53,173,127,1)","rgba(44,133,140,1)","rgba(58,97,139,1)","rgba(88,187,110,1)","rgba(43,140,138,1)","rgba(43,129,140,1)","rgba(51,109,141,1)","rgba(43,140,138,1)","rgba(36,164,133,1)","rgba(64,77,136,1)","rgba(69,178,121,1)","rgba(63,79,137,1)","rgba(43,140,138,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(61,89,138,1)","rgba(70,38,107,1)","rgba(97,192,104,1)","rgba(43,143,138,1)","rgba(38,161,134,1)","rgba(37,164,133,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(43,140,138,1)","rgba(42,122,142,1)","rgba(56,100,139,1)","rgba(40,157,134,1)","rgba(69,46,115,1)","rgba(44,134,139,1)","rgba(62,85,137,1)","rgba(131,210,79,1)","rgba(44,136,139,1)","rgba(39,159,134,1)","rgba(49,113,141,1)","rgba(44,131,140,1)","rgba(35,167,132,1)","rgba(43,143,138,1)","rgba(42,151,136,1)","rgba(42,149,136,1)","rgba(57,99,139,1)","rgba(121,208,82,1)","rgba(121,209,82,1)","rgba(37,164,133,1)","rgba(43,146,137,1)","rgba(70,41,110,1)","rgba(39,159,134,1)","rgba(69,9,88,1)","rgba(51,109,141,1)","rgba(59,94,139,1)","rgba(43,147,137,1)","rgba(44,136,139,1)","rgba(44,131,140,1)","rgba(39,159,134,1)","rgba(38,161,134,1)","rgba(43,147,137,1)","rgba(48,113,141,1)","rgba(104,197,98,1)","rgba(40,155,135,1)","rgba(41,151,136,1)","rgba(38,161,134,1)","rgba(46,116,141,1)","rgba(42,150,136,1)","rgba(43,145,137,1)","rgba(41,154,135,1)","rgba(39,159,134,1)","rgba(43,142,138,1)","rgba(43,129,140,1)","rgba(60,90,138,1)","rgba(39,158,134,1)","rgba(51,109,141,1)","rgba(43,141,138,1)","rgba(88,187,110,1)","rgba(67,178,121,1)","rgba(105,197,97,1)","rgba(37,162,133,1)","rgba(41,154,135,1)","rgba(67,60,127,1)","rgba(43,141,138,1)","rgba(46,116,142,1)","rgba(78,182,116,1)","rgba(43,143,138,1)","rgba(44,135,139,1)","rgba(68,2,85,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(41,154,135,1)","rgba(97,193,104,1)","rgba(70,27,99,1)","rgba(55,103,140,1)","rgba(60,175,124,1)","rgba(43,128,140,1)","rgba(80,183,115,1)","rgba(69,11,89,1)","rgba(92,189,108,1)","rgba(41,152,136,1)","rgba(51,172,127,1)","rgba(58,97,139,1)","rgba(37,164,133,1)","rgba(37,164,133,1)","rgba(67,62,129,1)","rgba(66,63,130,1)","rgba(43,126,141,1)","rgba(58,97,139,1)","rgba(53,106,140,1)","rgba(35,167,132,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(53,106,140,1)","rgba(44,135,139,1)","rgba(47,115,141,1)","rgba(41,153,135,1)","rgba(92,189,108,1)","rgba(104,196,99,1)","rgba(90,188,109,1)","rgba(44,136,139,1)","rgba(68,178,121,1)","rgba(70,37,107,1)","rgba(46,116,142,1)","rgba(38,161,134,1)","rgba(71,179,119,1)","rgba(42,148,137,1)","rgba(41,154,135,1)","rgba(72,180,119,1)","rgba(68,3,85,1)","rgba(42,148,136,1)","rgba(43,146,137,1)","rgba(43,146,137,1)","rgba(132,211,79,1)","rgba(37,163,133,1)","rgba(41,154,135,1)","rgba(44,139,138,1)","rgba(70,35,105,1)","rgba(41,151,136,1)","rgba(40,156,135,1)","rgba(54,173,127,1)","rgba(67,177,122,1)","rgba(43,143,138,1)","rgba(39,159,134,1)","rgba(43,144,137,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(43,147,137,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(147,213,76,1)","rgba(44,131,140,1)","rgba(48,114,141,1)","rgba(36,165,133,1)","rgba(39,160,134,1)","rgba(44,131,140,1)","rgba(42,151,136,1)","rgba(69,25,97,1)","rgba(44,137,139,1)","rgba(69,14,91,1)","rgba(39,160,134,1)","rgba(38,162,133,1)","rgba(43,123,141,1)","rgba(44,133,140,1)","rgba(41,154,135,1)","rgba(41,153,135,1)","rgba(43,126,141,1)","rgba(44,131,140,1)","rgba(108,199,95,1)","rgba(40,155,135,1)","rgba(43,126,141,1)","rgba(47,114,141,1)","rgba(38,162,134,1)","rgba(39,160,134,1)","rgba(42,147,137,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(43,145,137,1)","rgba(79,183,115,1)","rgba(45,116,142,1)","rgba(43,143,138,1)","rgba(41,154,135,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(63,176,123,1)","rgba(43,142,138,1)","rgba(43,144,137,1)","rgba(36,165,133,1)","rgba(42,148,137,1)","rgba(35,166,132,1)","rgba(34,168,132,1)","rgba(60,92,138,1)","rgba(44,131,140,1)","rgba(64,76,136,1)","rgba(34,168,132,1)","rgba(44,132,140,1)","rgba(43,143,138,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(37,163,133,1)","rgba(69,47,115,1)","rgba(52,108,141,1)","rgba(62,84,137,1)","rgba(43,130,140,1)","rgba(43,125,141,1)","rgba(46,116,142,1)","rgba(40,155,135,1)","rgba(61,88,138,1)","rgba(61,87,138,1)","rgba(40,156,135,1)","rgba(50,111,141,1)","rgba(70,179,120,1)","rgba(38,161,134,1)","rgba(70,37,107,1)","rgba(43,123,142,1)","rgba(44,139,138,1)","rgba(43,130,140,1)","rgba(63,176,123,1)","rgba(65,71,135,1)","rgba(107,199,96,1)","rgba(44,132,140,1)","rgba(50,111,141,1)","rgba(42,151,136,1)","rgba(49,113,141,1)","rgba(52,172,127,1)","rgba(42,149,136,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(46,116,141,1)","rgba(44,136,139,1)","rgba(42,121,142,1)","rgba(45,116,142,1)","rgba(41,152,136,1)","rgba(43,145,137,1)","rgba(69,16,91,1)","rgba(70,26,98,1)","rgba(52,108,141,1)","rgba(62,83,137,1)","rgba(68,57,124,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(42,121,142,1)","rgba(90,189,109,1)","rgba(44,137,139,1)","rgba(44,138,139,1)","rgba(89,188,110,1)","rgba(42,121,142,1)","rgba(58,174,125,1)","rgba(41,154,135,1)","rgba(42,147,137,1)","rgba(41,154,135,1)","rgba(55,102,140,1)","rgba(42,121,142,1)","rgba(44,131,140,1)","rgba(42,148,137,1)","rgba(90,188,109,1)","rgba(43,129,140,1)","rgba(43,141,138,1)","rgba(43,119,142,1)","rgba(42,150,136,1)","rgba(42,120,142,1)","rgba(41,155,135,1)","rgba(43,124,141,1)","rgba(41,153,135,1)","rgba(68,58,126,1)","rgba(41,153,136,1)","rgba(76,182,117,1)","rgba(40,155,135,1)","rgba(34,168,132,1)","rgba(54,104,140,1)","rgba(43,139,138,1)","rgba(39,158,134,1)","rgba(43,146,137,1)","rgba(39,160,134,1)","rgba(41,153,135,1)","rgba(42,151,136,1)","rgba(44,135,139,1)","rgba(43,141,138,1)","rgba(43,130,140,1)","rgba(43,126,141,1)","rgba(44,133,140,1)","rgba(36,164,133,1)","rgba(43,129,140,1)","rgba(57,98,139,1)","rgba(58,95,139,1)","rgba(44,138,139,1)","rgba(69,23,96,1)","rgba(69,45,113,1)","rgba(63,78,136,1)","rgba(44,118,142,1)","rgba(43,127,141,1)","rgba(43,147,137,1)","rgba(75,181,118,1)","rgba(43,123,141,1)","rgba(44,134,139,1)","rgba(57,100,139,1)","rgba(63,82,137,1)","rgba(40,155,135,1)","rgba(56,100,139,1)","rgba(69,49,117,1)","rgba(44,136,139,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(39,160,134,1)","rgba(43,124,141,1)","rgba(47,171,129,1)","rgba(43,142,138,1)","rgba(43,127,141,1)","rgba(49,112,141,1)","rgba(43,128,141,1)","rgba(43,140,138,1)","rgba(44,132,140,1)","rgba(44,137,139,1)","rgba(51,110,141,1)","rgba(39,158,134,1)","rgba(53,173,127,1)","rgba(51,172,128,1)","rgba(51,109,141,1)","rgba(44,139,138,1)","rgba(42,120,142,1)","rgba(43,147,137,1)","rgba(41,152,136,1)","rgba(78,182,116,1)","rgba(39,159,134,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(122,209,81,1)","rgba(42,151,136,1)","rgba(62,84,137,1)","rgba(39,158,134,1)","rgba(43,139,138,1)","rgba(42,147,137,1)","rgba(35,167,132,1)","rgba(46,115,141,1)","rgba(42,148,137,1)","rgba(50,172,128,1)","rgba(53,106,140,1)","rgba(69,46,115,1)","rgba(43,129,140,1)","rgba(43,128,140,1)","rgba(43,143,138,1)","rgba(67,59,126,1)","rgba(44,133,140,1)","rgba(41,154,135,1)","rgba(43,127,141,1)","rgba(36,165,133,1)","rgba(53,107,140,1)","rgba(57,100,139,1)","rgba(35,166,133,1)","rgba(39,158,134,1)","rgba(44,138,139,1)","rgba(44,137,139,1)","rgba(41,155,135,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(108,199,95,1)","rgba(36,164,133,1)","rgba(44,132,140,1)","rgba(40,156,135,1)","rgba(42,150,136,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(42,149,136,1)","rgba(70,38,108,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(50,111,141,1)","rgba(44,132,140,1)","rgba(42,151,136,1)","rgba(60,90,138,1)","rgba(49,113,141,1)","rgba(61,89,138,1)","rgba(41,153,135,1)","rgba(39,158,134,1)","rgba(171,217,70,1)","rgba(69,43,111,1)","rgba(43,124,141,1)","rgba(42,170,130,1)","rgba(43,124,141,1)","rgba(43,147,137,1)","rgba(43,140,138,1)","rgba(44,137,139,1)","rgba(54,104,140,1)","rgba(68,54,121,1)","rgba(43,129,140,1)","rgba(54,105,140,1)","rgba(60,91,138,1)","rgba(44,135,139,1)","rgba(51,109,141,1)","rgba(43,146,137,1)","rgba(43,124,141,1)","rgba(40,156,135,1)","rgba(58,95,139,1)","rgba(43,130,140,1)","rgba(44,132,140,1)","rgba(42,120,142,1)","rgba(49,111,141,1)","rgba(44,118,142,1)","rgba(38,160,134,1)","rgba(44,139,138,1)","rgba(49,112,141,1)","rgba(42,122,142,1)","rgba(117,205,87,1)","rgba(43,142,138,1)","rgba(43,128,141,1)","rgba(38,162,133,1)","rgba(49,112,141,1)","rgba(79,183,115,1)","rgba(44,133,140,1)","rgba(80,183,115,1)","rgba(55,103,140,1)","rgba(63,82,137,1)","rgba(70,27,99,1)","rgba(42,122,142,1)","rgba(57,98,139,1)","rgba(41,155,135,1)","rgba(35,166,132,1)","rgba(67,60,127,1)","rgba(43,147,137,1)","rgba(42,147,137,1)","rgba(38,162,133,1)","rgba(50,110,141,1)","rgba(43,128,141,1)","rgba(64,73,136,1)","rgba(42,120,142,1)","rgba(52,107,140,1)","rgba(40,156,135,1)","rgba(42,121,142,1)","rgba(62,176,123,1)","rgba(44,133,140,1)","rgba(70,25,97,1)","rgba(41,152,136,1)","rgba(42,147,137,1)","rgba(70,37,106,1)","rgba(43,127,141,1)","rgba(43,139,138,1)","rgba(45,116,142,1)","rgba(41,154,135,1)","rgba(69,45,114,1)","rgba(45,171,129,1)","rgba(44,132,140,1)","rgba(45,117,142,1)","rgba(44,138,139,1)","rgba(62,85,137,1)","rgba(44,138,139,1)","rgba(54,105,140,1)","rgba(60,92,138,1)","rgba(43,146,137,1)","rgba(43,129,140,1)","rgba(43,143,138,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(63,83,137,1)","rgba(70,36,105,1)","rgba(39,159,134,1)","rgba(44,118,142,1)","rgba(95,191,105,1)","rgba(44,170,130,1)","rgba(90,188,109,1)","rgba(59,175,125,1)","rgba(44,137,139,1)","rgba(65,177,122,1)","rgba(64,75,136,1)","rgba(44,138,139,1)","rgba(63,78,136,1)","rgba(56,174,126,1)","rgba(43,144,137,1)","rgba(52,173,127,1)","rgba(61,88,138,1)","rgba(70,38,108,1)","rgba(43,139,138,1)","rgba(44,133,140,1)","rgba(56,174,126,1)","rgba(74,181,118,1)","rgba(44,170,130,1)","rgba(43,126,141,1)","rgba(44,138,139,1)","rgba(36,164,133,1)","rgba(43,145,137,1)","rgba(43,140,138,1)","rgba(44,133,140,1)","rgba(45,117,142,1)","rgba(43,140,138,1)","rgba(41,154,135,1)","rgba(42,150,136,1)","rgba(35,166,132,1)","rgba(43,123,141,1)","rgba(60,92,138,1)","rgba(58,96,139,1)","rgba(56,102,140,1)","rgba(43,142,138,1)","rgba(39,159,134,1)","rgba(55,103,140,1)","rgba(44,135,139,1)","rgba(43,123,141,1)","rgba(43,123,141,1)","rgba(62,85,137,1)","rgba(69,13,90,1)","rgba(43,123,142,1)","rgba(59,175,125,1)","rgba(66,177,122,1)","rgba(43,145,137,1)","rgba(49,113,141,1)","rgba(68,7,87,1)","rgba(114,203,90,1)","rgba(43,143,138,1)","rgba(42,151,136,1)","rgba(46,115,141,1)","rgba(43,144,137,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(43,125,141,1)","rgba(59,175,125,1)","rgba(44,138,139,1)","rgba(57,100,139,1)","rgba(44,135,139,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(44,134,139,1)","rgba(47,115,141,1)","rgba(70,29,100,1)","rgba(44,134,139,1)","rgba(65,71,135,1)","rgba(50,111,141,1)","rgba(43,141,138,1)","rgba(41,154,135,1)","rgba(57,174,125,1)","rgba(69,46,115,1)","rgba(51,109,141,1)","rgba(70,31,101,1)","rgba(43,124,141,1)","rgba(49,172,128,1)","rgba(99,194,102,1)","rgba(42,150,136,1)","rgba(40,157,135,1)","rgba(64,73,136,1)","rgba(42,148,137,1)","rgba(39,159,134,1)","rgba(43,128,141,1)","rgba(41,154,135,1)","rgba(44,132,140,1)","rgba(64,78,136,1)","rgba(49,172,128,1)","rgba(88,187,110,1)","rgba(70,30,101,1)","rgba(60,91,138,1)","rgba(79,183,115,1)","rgba(45,117,142,1)","rgba(43,143,138,1)","rgba(56,101,140,1)","rgba(42,121,142,1)","rgba(38,160,134,1)","rgba(43,145,137,1)","rgba(43,127,141,1)","rgba(44,137,139,1)","rgba(56,101,140,1)","rgba(43,128,140,1)","rgba(42,151,136,1)","rgba(52,107,140,1)","rgba(43,127,141,1)","rgba(49,113,141,1)","rgba(42,150,136,1)","rgba(36,164,133,1)","rgba(45,117,142,1)","rgba(186,220,66,1)","rgba(43,124,141,1)","rgba(49,112,141,1)","rgba(38,169,131,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(48,114,141,1)","rgba(43,145,137,1)","rgba(59,95,139,1)","rgba(44,132,140,1)","rgba(36,164,133,1)","rgba(50,111,141,1)","rgba(43,127,141,1)","rgba(90,188,109,1)","rgba(65,67,134,1)","rgba(53,106,140,1)","rgba(90,188,109,1)","rgba(37,162,133,1)","rgba(44,131,140,1)","rgba(43,142,138,1)","rgba(51,109,141,1)","rgba(43,123,141,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(43,142,138,1)","rgba(40,157,135,1)","rgba(61,175,124,1)","rgba(163,216,72,1)","rgba(44,135,139,1)","rgba(42,147,137,1)","rgba(43,129,140,1)","rgba(43,142,138,1)","rgba(78,182,116,1)","rgba(44,136,139,1)","rgba(57,174,125,1)","rgba(46,115,141,1)","rgba(39,158,134,1)","rgba(70,41,109,1)","rgba(61,86,138,1)","rgba(41,153,136,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(64,176,123,1)","rgba(46,116,141,1)","rgba(48,113,141,1)","rgba(34,167,132,1)","rgba(43,143,138,1)","rgba(46,116,142,1)","rgba(55,102,140,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(39,159,134,1)","rgba(64,77,136,1)","rgba(51,172,127,1)","rgba(43,129,140,1)","rgba(52,107,140,1)","rgba(49,112,141,1)","rgba(53,105,140,1)","rgba(46,116,141,1)","rgba(44,136,139,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(69,44,113,1)","rgba(43,146,137,1)","rgba(44,136,139,1)","rgba(51,110,141,1)","rgba(37,164,133,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(67,62,129,1)","rgba(51,110,141,1)","rgba(69,11,89,1)","rgba(48,114,141,1)","rgba(51,110,141,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(44,137,139,1)","rgba(43,128,141,1)","rgba(54,173,127,1)","rgba(68,56,123,1)","rgba(37,169,131,1)","rgba(42,151,136,1)","rgba(66,177,122,1)","rgba(54,105,140,1)","rgba(53,106,140,1)","rgba(40,157,134,1)","rgba(42,122,142,1)","rgba(34,167,132,1)","rgba(36,164,133,1)","rgba(36,164,133,1)","rgba(43,147,137,1)","rgba(43,146,137,1)","rgba(43,144,137,1)","rgba(76,182,117,1)","rgba(40,156,135,1)","rgba(43,128,141,1)","rgba(49,112,141,1)","rgba(43,124,141,1)","rgba(69,46,114,1)","rgba(46,116,142,1)","rgba(35,167,132,1)","rgba(71,179,119,1)","rgba(42,149,136,1)","rgba(39,160,134,1)","rgba(53,106,140,1)","rgba(77,182,116,1)","rgba(44,135,139,1)","rgba(43,125,141,1)","rgba(96,192,105,1)","rgba(43,170,130,1)","rgba(42,149,136,1)","rgba(46,116,141,1)","rgba(56,100,139,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(40,156,135,1)","rgba(43,141,138,1)","rgba(42,122,142,1)","rgba(44,138,139,1)","rgba(43,126,141,1)","rgba(69,43,112,1)","rgba(69,178,120,1)","rgba(73,180,119,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(41,152,136,1)","rgba(53,106,140,1)","rgba(43,139,138,1)","rgba(44,134,139,1)","rgba(44,137,139,1)","rgba(41,152,136,1)","rgba(42,121,142,1)","rgba(43,126,141,1)","rgba(44,133,140,1)","rgba(44,135,139,1)","rgba(41,154,135,1)","rgba(44,134,139,1)","rgba(50,110,141,1)","rgba(49,112,141,1)","rgba(43,123,141,1)","rgba(44,139,138,1)","rgba(68,55,123,1)","rgba(51,110,141,1)","rgba(40,169,131,1)","rgba(47,114,141,1)","rgba(43,146,137,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(44,139,138,1)","rgba(50,172,128,1)","rgba(56,100,140,1)","rgba(43,125,141,1)","rgba(69,25,97,1)","rgba(43,141,138,1)","rgba(42,149,136,1)","rgba(43,145,137,1)","rgba(53,106,140,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(43,143,138,1)","rgba(42,121,142,1)","rgba(43,145,137,1)","rgba(44,118,142,1)","rgba(69,11,89,1)","rgba(39,159,134,1)","rgba(70,33,103,1)","rgba(43,119,142,1)","rgba(45,117,142,1)","rgba(35,166,133,1)","rgba(40,156,135,1)","rgba(40,157,135,1)","rgba(69,51,118,1)","rgba(192,221,64,1)","rgba(44,137,139,1)","rgba(42,122,142,1)","rgba(68,55,123,1)","rgba(42,149,136,1)","rgba(43,146,137,1)","rgba(42,148,137,1)","rgba(42,150,136,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(65,68,135,1)","rgba(43,143,138,1)","rgba(67,177,122,1)","rgba(43,124,141,1)","rgba(42,147,137,1)","rgba(81,184,114,1)","rgba(44,135,139,1)","rgba(42,122,142,1)","rgba(82,184,114,1)","rgba(42,120,142,1)","rgba(43,124,141,1)","rgba(43,129,140,1)","rgba(40,156,135,1)","rgba(49,112,141,1)","rgba(47,171,129,1)","rgba(39,159,134,1)","rgba(44,133,140,1)","rgba(41,153,135,1)","rgba(35,166,132,1)","rgba(51,110,141,1)","rgba(40,155,135,1)","rgba(53,106,140,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(67,59,126,1)","rgba(35,167,132,1)","rgba(42,120,142,1)","rgba(62,86,138,1)","rgba(39,159,134,1)","rgba(42,120,142,1)","rgba(41,152,136,1)","rgba(58,95,139,1)","rgba(43,144,137,1)","rgba(43,145,137,1)","rgba(44,136,139,1)","rgba(69,52,120,1)","rgba(44,136,139,1)","rgba(63,78,136,1)","rgba(92,190,107,1)","rgba(40,169,131,1)","rgba(52,107,140,1)","rgba(43,145,137,1)","rgba(44,135,139,1)","rgba(61,87,138,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(40,155,135,1)","rgba(58,96,139,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(105,198,97,1)","rgba(44,133,140,1)","rgba(54,104,140,1)","rgba(86,186,111,1)","rgba(42,150,136,1)","rgba(42,121,142,1)","rgba(38,160,134,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(36,165,133,1)","rgba(50,172,128,1)","rgba(60,175,124,1)","rgba(41,152,136,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(40,157,135,1)","rgba(44,132,140,1)","rgba(50,111,141,1)","rgba(66,64,131,1)","rgba(44,136,139,1)","rgba(69,19,93,1)","rgba(43,125,141,1)","rgba(51,110,141,1)","rgba(77,182,116,1)","rgba(42,120,142,1)","rgba(58,96,139,1)","rgba(44,118,142,1)","rgba(53,106,140,1)","rgba(34,167,132,1)","rgba(44,137,139,1)","rgba(46,116,142,1)","rgba(54,104,140,1)","rgba(67,62,129,1)","rgba(43,123,141,1)","rgba(69,17,92,1)","rgba(40,156,135,1)","rgba(42,150,136,1)","rgba(44,133,140,1)","rgba(43,144,137,1)","rgba(46,171,129,1)","rgba(43,130,140,1)","rgba(44,139,139,1)","rgba(62,85,137,1)","rgba(43,129,140,1)","rgba(41,154,135,1)","rgba(62,84,137,1)","rgba(63,82,137,1)","rgba(54,105,140,1)","rgba(43,144,137,1)","rgba(43,142,138,1)","rgba(42,149,136,1)","rgba(44,135,139,1)","rgba(119,207,84,1)","rgba(41,153,135,1)","rgba(42,151,136,1)","rgba(43,142,138,1)","rgba(35,167,132,1)","rgba(40,156,135,1)","rgba(63,79,137,1)","rgba(42,148,137,1)","rgba(43,141,138,1)","rgba(42,120,142,1)","rgba(87,187,110,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(42,151,136,1)","rgba(54,104,140,1)","rgba(42,148,137,1)","rgba(57,98,139,1)","rgba(43,124,141,1)","rgba(43,129,140,1)","rgba(43,144,137,1)","rgba(65,69,135,1)","rgba(43,130,140,1)","rgba(42,150,136,1)","rgba(70,28,99,1)","rgba(44,131,140,1)","rgba(51,109,141,1)","rgba(50,111,141,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(60,90,138,1)","rgba(41,154,135,1)","rgba(43,145,137,1)","rgba(44,133,140,1)","rgba(44,136,139,1)","rgba(57,98,139,1)","rgba(43,125,141,1)","rgba(42,148,137,1)","rgba(69,11,89,1)","rgba(43,143,138,1)","rgba(44,118,142,1)","rgba(62,86,138,1)","rgba(36,164,133,1)","rgba(43,130,140,1)","rgba(39,159,134,1)","rgba(69,11,89,1)","rgba(44,133,140,1)","rgba(53,106,140,1)","rgba(49,112,141,1)","rgba(68,58,125,1)","rgba(38,161,134,1)","rgba(44,139,138,1)","rgba(59,93,138,1)","rgba(38,162,134,1)","rgba(51,172,128,1)","rgba(36,164,133,1)","rgba(55,103,140,1)","rgba(56,100,139,1)","rgba(43,142,138,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(35,166,133,1)","rgba(42,150,136,1)","rgba(38,161,134,1)","rgba(41,155,135,1)","rgba(43,141,138,1)","rgba(51,172,127,1)","rgba(43,140,138,1)","rgba(41,153,136,1)","rgba(35,166,132,1)","rgba(108,199,95,1)","rgba(46,116,142,1)","rgba(43,128,141,1)","rgba(68,54,121,1)","rgba(65,69,135,1)","rgba(52,107,140,1)","rgba(42,150,136,1)","rgba(50,172,128,1)","rgba(89,188,109,1)","rgba(63,80,137,1)","rgba(63,80,137,1)","rgba(69,50,118,1)","rgba(43,145,137,1)","rgba(61,88,138,1)","rgba(43,124,141,1)","rgba(43,124,141,1)","rgba(70,34,104,1)","rgba(58,96,139,1)","rgba(60,91,138,1)","rgba(44,132,140,1)","rgba(64,77,136,1)","rgba(61,87,138,1)","rgba(70,40,109,1)","rgba(68,7,87,1)","rgba(69,12,90,1)","rgba(67,59,126,1)","rgba(44,138,139,1)","rgba(55,103,140,1)","rgba(58,96,139,1)","rgba(48,114,141,1)","rgba(61,89,138,1)","rgba(63,82,137,1)","rgba(43,142,138,1)","rgba(69,44,113,1)","rgba(43,123,141,1)","rgba(70,30,101,1)","rgba(63,80,137,1)","rgba(41,154,135,1)","rgba(41,154,135,1)","rgba(65,69,135,1)","rgba(42,121,142,1)","rgba(43,124,141,1)","rgba(43,130,140,1)","rgba(41,170,130,1)","rgba(61,176,124,1)","rgba(43,124,141,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(53,107,140,1)","rgba(49,172,128,1)","rgba(45,117,142,1)","rgba(76,181,117,1)","rgba(44,137,139,1)","rgba(70,41,110,1)","rgba(42,149,136,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(66,64,131,1)","rgba(53,105,140,1)","rgba(41,152,136,1)","rgba(39,158,134,1)","rgba(43,145,137,1)","rgba(43,124,141,1)","rgba(43,145,137,1)","rgba(66,65,132,1)","rgba(86,186,112,1)","rgba(103,196,100,1)","rgba(44,133,140,1)","rgba(56,174,126,1)","rgba(38,162,133,1)","rgba(42,149,136,1)","rgba(57,98,139,1)","rgba(43,128,141,1)","rgba(55,102,140,1)","rgba(68,5,86,1)","rgba(42,148,136,1)","rgba(36,164,133,1)","rgba(40,155,135,1)","rgba(68,54,121,1)","rgba(43,141,138,1)","rgba(43,130,140,1)","rgba(44,132,140,1)","rgba(52,172,127,1)","rgba(43,145,137,1)","rgba(54,105,140,1)","rgba(50,111,141,1)","rgba(39,159,134,1)","rgba(129,210,80,1)","rgba(42,122,142,1)","rgba(46,116,141,1)","rgba(43,143,138,1)","rgba(100,194,102,1)","rgba(41,152,136,1)","rgba(62,84,137,1)","rgba(43,142,138,1)","rgba(55,103,140,1)","rgba(62,86,138,1)","rgba(67,62,129,1)","rgba(70,42,110,1)","rgba(70,35,105,1)","rgba(45,117,142,1)","rgba(65,68,135,1)","rgba(62,85,137,1)","rgba(64,72,136,1)","rgba(70,30,101,1)","rgba(69,43,111,1)","rgba(70,42,111,1)","rgba(69,20,94,1)","rgba(69,53,120,1)","rgba(42,121,142,1)","rgba(65,70,135,1)","rgba(44,131,140,1)","rgba(75,181,117,1)","rgba(68,6,86,1)","rgba(66,63,130,1)","rgba(70,38,107,1)","rgba(65,72,136,1)","rgba(63,80,137,1)","rgba(64,76,136,1)","rgba(42,122,142,1)","rgba(64,73,136,1)","rgba(43,130,140,1)","rgba(59,93,139,1)","rgba(43,124,141,1)","rgba(69,48,116,1)","rgba(70,40,109,1)","rgba(47,115,141,1)","rgba(54,104,140,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(38,160,134,1)","rgba(46,171,129,1)","rgba(46,116,141,1)","rgba(44,131,140,1)","rgba(69,16,91,1)","rgba(43,129,140,1)","rgba(43,142,138,1)","rgba(41,154,135,1)","rgba(68,55,122,1)","rgba(56,100,140,1)","rgba(69,48,116,1)","rgba(43,127,141,1)","rgba(53,107,140,1)","rgba(44,134,139,1)","rgba(50,110,141,1)","rgba(69,50,118,1)","rgba(70,33,103,1)","rgba(43,143,138,1)","rgba(43,147,137,1)","rgba(35,166,132,1)","rgba(69,25,97,1)","rgba(70,29,100,1)","rgba(43,123,141,1)","rgba(58,97,139,1)","rgba(69,44,113,1)","rgba(69,48,116,1)","rgba(43,145,137,1)","rgba(54,173,126,1)","rgba(70,32,103,1)","rgba(115,204,88,1)","rgba(43,126,141,1)","rgba(43,128,141,1)","rgba(62,85,137,1)","rgba(121,208,82,1)","rgba(43,119,142,1)","rgba(58,96,139,1)","rgba(70,35,105,1)","rgba(43,130,140,1)","rgba(64,77,136,1)","rgba(62,85,137,1)","rgba(42,151,136,1)","rgba(63,80,137,1)","rgba(68,7,87,1)","rgba(68,2,85,1)","rgba(56,101,140,1)","rgba(64,74,136,1)","rgba(42,147,137,1)","rgba(44,132,140,1)","rgba(43,147,137,1)","rgba(62,83,137,1)","rgba(44,131,140,1)","rgba(38,161,134,1)","rgba(49,113,141,1)","rgba(62,83,137,1)","rgba(69,22,95,1)","rgba(69,13,90,1)","rgba(43,126,141,1)","rgba(69,43,112,1)","rgba(43,143,138,1)","rgba(68,2,84,1)","rgba(70,32,103,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(68,54,122,1)","rgba(44,136,139,1)","rgba(42,120,142,1)","rgba(41,153,136,1)","rgba(43,130,140,1)","rgba(55,102,140,1)","rgba(44,132,140,1)","rgba(69,43,111,1)","rgba(41,152,136,1)","rgba(43,144,137,1)","rgba(38,162,133,1)","rgba(43,140,138,1)","rgba(44,118,142,1)","rgba(70,38,108,1)","rgba(40,157,135,1)","rgba(69,48,116,1)","rgba(70,35,105,1)","rgba(57,174,125,1)","rgba(43,143,138,1)","rgba(43,119,142,1)","rgba(42,149,136,1)","rgba(43,130,140,1)","rgba(63,80,137,1)","rgba(43,146,137,1)","rgba(63,82,137,1)","rgba(39,160,134,1)","rgba(48,113,141,1)","rgba(60,91,138,1)","rgba(43,125,141,1)","rgba(147,213,76,1)","rgba(68,4,85,1)","rgba(40,169,131,1)","rgba(54,173,126,1)","rgba(44,133,140,1)","rgba(69,12,89,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(61,87,138,1)","rgba(45,117,142,1)","rgba(44,118,142,1)","rgba(59,93,139,1)","rgba(52,108,140,1)","rgba(68,56,123,1)","rgba(45,117,142,1)","rgba(56,101,140,1)","rgba(53,105,140,1)","rgba(44,136,139,1)","rgba(69,10,88,1)","rgba(36,165,133,1)","rgba(55,103,140,1)","rgba(47,115,141,1)","rgba(43,128,141,1)","rgba(70,41,110,1)","rgba(69,43,112,1)","rgba(70,42,110,1)","rgba(44,135,139,1)","rgba(45,116,142,1)","rgba(45,117,142,1)","rgba(62,84,137,1)","rgba(43,124,141,1)","rgba(59,94,139,1)","rgba(44,138,139,1)","rgba(68,58,125,1)","rgba(36,165,133,1)","rgba(38,161,134,1)","rgba(66,63,130,1)","rgba(37,162,133,1)","rgba(47,114,141,1)","rgba(69,44,113,1)","rgba(69,16,91,1)","rgba(70,42,111,1)","rgba(43,140,138,1)","rgba(70,37,107,1)","rgba(42,148,137,1)","rgba(54,105,140,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(66,65,132,1)","rgba(43,129,140,1)","rgba(63,81,137,1)","rgba(60,90,138,1)","rgba(44,136,139,1)","rgba(54,105,140,1)","rgba(63,78,136,1)","rgba(69,49,117,1)","rgba(52,107,140,1)","rgba(42,122,142,1)","rgba(61,87,138,1)","rgba(42,150,136,1)","rgba(49,112,141,1)","rgba(54,105,140,1)","rgba(66,63,130,1)","rgba(68,2,84,1)","rgba(58,96,139,1)","rgba(42,148,137,1)","rgba(42,150,136,1)","rgba(44,134,139,1)","rgba(40,157,135,1)","rgba(55,103,140,1)","rgba(39,159,134,1)","rgba(62,83,137,1)","rgba(44,132,140,1)","rgba(37,163,133,1)","rgba(68,58,125,1)","rgba(81,184,114,1)","rgba(43,126,141,1)","rgba(37,163,133,1)","rgba(46,116,142,1)","rgba(42,150,136,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(44,133,140,1)","rgba(52,108,140,1)","rgba(41,153,136,1)","rgba(53,107,140,1)","rgba(40,157,135,1)","rgba(66,66,133,1)","rgba(43,145,137,1)","rgba(42,120,142,1)","rgba(64,77,136,1)","rgba(37,163,133,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(80,183,115,1)","rgba(37,163,133,1)","rgba(57,98,139,1)","rgba(38,162,134,1)","rgba(57,98,139,1)","rgba(43,141,138,1)","rgba(57,99,139,1)","rgba(40,156,135,1)","rgba(43,126,141,1)","rgba(45,116,142,1)","rgba(65,70,135,1)","rgba(64,76,136,1)","rgba(37,162,133,1)","rgba(43,142,138,1)","rgba(69,43,112,1)","rgba(42,122,142,1)","rgba(42,121,142,1)","rgba(114,203,89,1)","rgba(63,80,137,1)","rgba(44,134,139,1)","rgba(43,127,141,1)","rgba(44,131,140,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(69,50,118,1)","rgba(52,108,140,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(62,85,137,1)","rgba(62,85,137,1)","rgba(44,118,142,1)","rgba(70,37,107,1)","rgba(51,109,141,1)","rgba(103,196,99,1)","rgba(44,118,142,1)","rgba(43,143,138,1)","rgba(61,89,138,1)","rgba(60,91,138,1)","rgba(42,120,142,1)","rgba(44,136,139,1)","rgba(57,99,139,1)","rgba(40,169,131,1)","rgba(39,158,134,1)","rgba(48,113,141,1)","rgba(44,135,139,1)","rgba(43,146,137,1)","rgba(56,101,140,1)","rgba(40,155,135,1)","rgba(68,6,86,1)","rgba(38,160,134,1)","rgba(42,150,136,1)","rgba(69,15,91,1)","rgba(69,49,117,1)","rgba(70,39,108,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(45,170,129,1)","rgba(43,128,141,1)","rgba(69,13,90,1)","rgba(68,56,124,1)","rgba(69,11,89,1)","rgba(43,127,141,1)","rgba(41,153,135,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(61,87,138,1)","rgba(53,105,140,1)","rgba(41,152,136,1)","rgba(50,110,141,1)","rgba(44,138,139,1)","rgba(61,89,138,1)","rgba(43,140,138,1)","rgba(43,127,141,1)","rgba(43,143,138,1)","rgba(83,185,113,1)","rgba(39,159,134,1)","rgba(44,134,139,1)","rgba(43,129,140,1)","rgba(43,140,138,1)","rgba(65,70,135,1)","rgba(38,161,134,1)","rgba(69,21,94,1)","rgba(44,135,139,1)","rgba(35,167,132,1)","rgba(42,122,142,1)","rgba(58,97,139,1)","rgba(59,95,139,1)","rgba(42,121,142,1)","rgba(39,158,134,1)","rgba(44,138,139,1)","rgba(44,118,142,1)","rgba(43,143,138,1)","rgba(43,127,141,1)","rgba(93,190,107,1)","rgba(42,149,136,1)","rgba(43,143,138,1)","rgba(43,145,137,1)","rgba(43,129,140,1)","rgba(41,152,136,1)","rgba(42,150,136,1)","rgba(51,109,141,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(69,21,94,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(59,94,139,1)","rgba(51,110,141,1)","rgba(40,157,135,1)","rgba(62,83,137,1)","rgba(53,106,140,1)","rgba(69,51,119,1)","rgba(59,93,139,1)","rgba(79,183,116,1)","rgba(43,128,141,1)","rgba(44,136,139,1)","rgba(54,173,126,1)","rgba(64,76,136,1)","rgba(64,72,136,1)","rgba(50,111,141,1)","rgba(60,92,138,1)","rgba(44,118,142,1)","rgba(69,10,88,1)","rgba(68,57,124,1)","rgba(69,15,91,1)","rgba(42,149,136,1)","rgba(43,143,138,1)","rgba(54,104,140,1)","rgba(65,68,135,1)","rgba(61,88,138,1)","rgba(61,88,138,1)","rgba(42,148,137,1)","rgba(61,88,138,1)","rgba(45,117,142,1)","rgba(43,143,138,1)","rgba(56,101,140,1)","rgba(68,2,84,1)","rgba(43,126,141,1)","rgba(68,58,125,1)","rgba(43,125,141,1)","rgba(44,137,139,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(43,127,141,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(44,133,140,1)","rgba(43,126,141,1)","rgba(69,14,90,1)","rgba(42,122,142,1)","rgba(44,131,140,1)","rgba(36,165,133,1)","rgba(44,137,139,1)","rgba(41,154,135,1)","rgba(39,160,134,1)","rgba(43,141,138,1)","rgba(42,150,136,1)","rgba(51,109,141,1)","rgba(54,105,140,1)","rgba(43,123,142,1)","rgba(57,174,125,1)","rgba(43,146,137,1)","rgba(47,115,141,1)","rgba(38,160,134,1)","rgba(44,136,139,1)","rgba(44,131,140,1)","rgba(43,147,137,1)","rgba(36,164,133,1)","rgba(57,98,139,1)","rgba(35,167,132,1)","rgba(41,154,135,1)","rgba(42,120,142,1)","rgba(44,118,142,1)","rgba(69,15,91,1)","rgba(43,123,141,1)","rgba(55,102,140,1)","rgba(42,148,137,1)","rgba(41,153,135,1)","rgba(49,111,141,1)","rgba(43,143,138,1)","rgba(66,177,122,1)","rgba(66,177,122,1)","rgba(50,110,141,1)","rgba(69,18,92,1)","rgba(59,94,139,1)","rgba(90,188,109,1)","rgba(41,154,135,1)","rgba(39,158,134,1)","rgba(47,115,141,1)","rgba(43,126,141,1)","rgba(49,112,141,1)","rgba(53,106,140,1)","rgba(42,150,136,1)","rgba(67,62,129,1)","rgba(52,108,141,1)","rgba(46,116,141,1)","rgba(67,60,127,1)","rgba(70,37,106,1)","rgba(65,71,135,1)","rgba(49,112,141,1)","rgba(43,126,141,1)","rgba(43,142,138,1)","rgba(66,64,131,1)","rgba(48,113,141,1)","rgba(69,11,89,1)","rgba(55,102,140,1)","rgba(68,58,125,1)","rgba(70,30,101,1)","rgba(70,30,101,1)","rgba(70,29,100,1)","rgba(69,46,114,1)","rgba(69,24,96,1)","rgba(67,61,129,1)","rgba(37,163,133,1)","rgba(44,132,140,1)","rgba(44,132,140,1)","rgba(43,127,141,1)","rgba(69,49,117,1)","rgba(50,110,141,1)","rgba(60,92,138,1)","rgba(42,120,142,1)","rgba(69,25,97,1)","rgba(63,80,137,1)","rgba(121,209,82,1)","rgba(50,111,141,1)","rgba(43,123,141,1)","rgba(43,140,138,1)","rgba(43,127,141,1)","rgba(59,94,139,1)","rgba(43,129,140,1)","rgba(42,147,137,1)","rgba(69,14,90,1)","rgba(70,40,109,1)","rgba(43,141,138,1)","rgba(69,47,115,1)","rgba(69,21,94,1)","rgba(62,84,137,1)","rgba(53,106,140,1)","rgba(70,29,100,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(61,89,138,1)","rgba(48,113,141,1)","rgba(63,82,137,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(57,99,139,1)","rgba(69,51,119,1)","rgba(44,134,139,1)","rgba(70,30,101,1)","rgba(68,3,85,1)","rgba(70,33,103,1)","rgba(60,92,138,1)","rgba(52,108,140,1)","rgba(53,106,140,1)","rgba(44,131,140,1)","rgba(48,113,141,1)","rgba(75,181,118,1)","rgba(62,84,137,1)","rgba(93,190,107,1)","rgba(63,78,136,1)","rgba(69,24,97,1)","rgba(53,106,140,1)","rgba(43,123,142,1)","rgba(43,129,140,1)","rgba(51,109,141,1)","rgba(39,159,134,1)","rgba(43,124,141,1)","rgba(44,136,139,1)","rgba(69,51,118,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(53,106,140,1)","rgba(54,104,140,1)","rgba(69,48,116,1)","rgba(44,134,139,1)","rgba(65,69,135,1)","rgba(42,148,137,1)","rgba(40,155,135,1)","rgba(56,101,140,1)","rgba(70,38,107,1)","rgba(54,105,140,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(46,115,141,1)","rgba(43,142,138,1)","rgba(36,164,133,1)","rgba(42,148,137,1)","rgba(63,81,137,1)","rgba(69,51,118,1)","rgba(67,59,126,1)","rgba(42,122,142,1)","rgba(70,28,99,1)","rgba(42,149,136,1)","rgba(40,169,131,1)","rgba(43,146,137,1)","rgba(60,90,138,1)","rgba(43,130,140,1)","rgba(45,117,142,1)","rgba(46,171,129,1)","rgba(69,42,111,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(44,135,139,1)","rgba(67,61,128,1)","rgba(42,149,136,1)","rgba(43,140,138,1)","rgba(43,142,138,1)","rgba(44,170,130,1)","rgba(40,156,135,1)","rgba(60,92,138,1)","rgba(56,101,140,1)","rgba(42,150,136,1)","rgba(43,124,141,1)","rgba(42,122,142,1)","rgba(70,38,107,1)","rgba(44,138,139,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(44,132,140,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(52,108,141,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(44,137,139,1)","rgba(43,126,141,1)","rgba(64,77,136,1)","rgba(59,94,139,1)","rgba(94,190,106,1)","rgba(42,122,142,1)","rgba(59,94,139,1)","rgba(43,119,142,1)","rgba(60,92,138,1)","rgba(52,108,141,1)","rgba(42,120,142,1)","rgba(45,117,142,1)","rgba(50,111,141,1)","rgba(70,25,98,1)","rgba(36,164,133,1)","rgba(43,170,130,1)","rgba(39,158,134,1)","rgba(64,75,136,1)","rgba(61,87,138,1)","rgba(69,12,89,1)","rgba(69,43,112,1)","rgba(57,98,139,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(57,98,139,1)","rgba(43,147,137,1)","rgba(69,49,117,1)","rgba(43,142,138,1)","rgba(58,95,139,1)","rgba(68,54,122,1)","rgba(43,140,138,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(43,145,137,1)","rgba(69,23,96,1)","rgba(41,153,136,1)","rgba(43,127,141,1)","rgba(40,157,135,1)","rgba(44,133,140,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(55,102,140,1)","rgba(61,87,138,1)","rgba(44,135,139,1)","rgba(43,142,138,1)","rgba(56,100,139,1)","rgba(63,81,137,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(43,119,142,1)","rgba(40,157,135,1)","rgba(43,142,138,1)","rgba(70,30,101,1)","rgba(53,106,140,1)","rgba(69,25,97,1)","rgba(68,58,125,1)","rgba(70,41,110,1)","rgba(68,55,122,1)","rgba(69,9,88,1)","rgba(40,155,135,1)","rgba(69,49,117,1)","rgba(69,23,96,1)","rgba(68,4,86,1)","rgba(69,50,118,1)","rgba(42,149,136,1)","rgba(44,133,140,1)","rgba(43,128,141,1)","rgba(45,117,142,1)","rgba(37,162,133,1)","rgba(70,39,108,1)","rgba(56,174,126,1)","rgba(59,94,139,1)","rgba(69,51,119,1)","rgba(51,110,141,1)","rgba(51,109,141,1)","rgba(70,41,110,1)","rgba(61,87,138,1)","rgba(51,110,141,1)","rgba(61,88,138,1)","rgba(43,128,140,1)","rgba(43,123,141,1)","rgba(44,137,139,1)","rgba(42,150,136,1)","rgba(68,55,122,1)","rgba(69,46,114,1)","rgba(38,161,134,1)","rgba(47,171,129,1)","rgba(57,99,139,1)","rgba(47,171,129,1)","rgba(64,77,136,1)","rgba(61,87,138,1)","rgba(43,142,138,1)","rgba(69,24,96,1)","rgba(43,127,141,1)","rgba(47,115,141,1)","rgba(41,154,135,1)","rgba(44,133,140,1)","rgba(68,6,86,1)","rgba(59,175,125,1)","rgba(43,140,138,1)","rgba(60,91,138,1)","rgba(66,63,131,1)","rgba(39,160,134,1)","rgba(42,120,142,1)","rgba(52,108,140,1)","rgba(39,159,134,1)","rgba(60,175,124,1)","rgba(68,58,126,1)","rgba(65,68,135,1)","rgba(68,56,123,1)","rgba(63,81,137,1)","rgba(132,211,79,1)","rgba(44,118,142,1)","rgba(43,124,141,1)","rgba(69,47,115,1)","rgba(59,93,138,1)","rgba(64,73,136,1)","rgba(43,126,141,1)","rgba(43,123,142,1)","rgba(64,75,136,1)","rgba(41,152,136,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(40,156,135,1)","rgba(40,155,135,1)","rgba(43,119,142,1)","rgba(43,127,141,1)","rgba(35,167,132,1)","rgba(50,172,128,1)","rgba(62,85,137,1)","rgba(59,94,139,1)","rgba(46,116,141,1)","rgba(43,128,140,1)","rgba(43,124,141,1)","rgba(36,165,133,1)","rgba(42,122,142,1)","rgba(36,165,133,1)","rgba(44,136,139,1)","rgba(70,30,101,1)","rgba(42,148,136,1)","rgba(70,179,120,1)","rgba(51,172,127,1)","rgba(43,141,138,1)","rgba(70,32,102,1)","rgba(68,1,84,1)","rgba(53,106,140,1)","rgba(40,157,135,1)","rgba(43,128,141,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(54,104,140,1)","rgba(62,84,137,1)","rgba(43,123,141,1)","rgba(65,69,135,1)","rgba(43,124,141,1)","rgba(35,166,132,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(69,46,115,1)","rgba(46,116,141,1)","rgba(37,163,133,1)","rgba(51,110,141,1)","rgba(44,118,142,1)","rgba(69,22,95,1)","rgba(69,17,92,1)","rgba(66,64,131,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(40,156,135,1)","rgba(62,84,137,1)","rgba(36,165,133,1)","rgba(64,177,123,1)","rgba(44,136,139,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(44,131,140,1)","rgba(70,32,102,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(40,157,135,1)","rgba(70,36,105,1)","rgba(42,122,142,1)","rgba(63,82,137,1)","rgba(42,151,136,1)","rgba(36,164,133,1)","rgba(42,120,142,1)","rgba(58,96,139,1)","rgba(42,151,136,1)","rgba(42,121,142,1)","rgba(44,136,139,1)","rgba(56,100,139,1)","rgba(69,48,116,1)","rgba(65,70,135,1)","rgba(44,135,139,1)","rgba(69,15,91,1)","rgba(44,133,140,1)","rgba(44,137,139,1)","rgba(68,57,124,1)","rgba(36,164,133,1)","rgba(46,116,142,1)","rgba(61,88,138,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(55,103,140,1)","rgba(49,112,141,1)","rgba(52,108,141,1)","rgba(70,27,99,1)","rgba(68,4,85,1)","rgba(55,102,140,1)","rgba(55,103,140,1)","rgba(59,95,139,1)","rgba(43,124,141,1)","rgba(42,121,142,1)","rgba(56,101,140,1)","rgba(63,80,137,1)","rgba(68,57,125,1)","rgba(42,150,136,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(38,160,134,1)","rgba(36,165,133,1)","rgba(42,151,136,1)","rgba(68,5,86,1)","rgba(43,124,141,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(60,92,138,1)","rgba(67,59,126,1)","rgba(55,102,140,1)","rgba(59,93,138,1)","rgba(69,15,91,1)","rgba(56,100,139,1)","rgba(39,169,131,1)","rgba(47,115,141,1)","rgba(52,107,140,1)","rgba(70,37,106,1)","rgba(69,42,111,1)","rgba(43,140,138,1)","rgba(63,81,137,1)","rgba(43,142,138,1)","rgba(64,76,136,1)","rgba(41,153,136,1)","rgba(40,155,135,1)","rgba(52,107,140,1)","rgba(42,122,142,1)","rgba(44,138,139,1)","rgba(42,149,136,1)","rgba(42,150,136,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(44,134,139,1)","rgba(42,150,136,1)","rgba(44,132,140,1)","rgba(55,103,140,1)","rgba(50,111,141,1)","rgba(42,120,142,1)","rgba(44,135,139,1)","rgba(55,103,140,1)","rgba(63,82,137,1)","rgba(42,147,137,1)","rgba(51,110,141,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(61,88,138,1)","rgba(50,111,141,1)","rgba(39,158,134,1)","rgba(43,146,137,1)","rgba(43,130,140,1)","rgba(61,89,138,1)","rgba(43,144,137,1)","rgba(50,172,128,1)","rgba(62,83,137,1)","rgba(39,158,134,1)","rgba(37,162,133,1)","rgba(52,107,140,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(69,47,115,1)","rgba(62,86,138,1)","rgba(44,135,139,1)","rgba(44,138,139,1)","rgba(60,92,138,1)","rgba(56,100,139,1)","rgba(49,172,128,1)","rgba(61,87,138,1)","rgba(67,59,127,1)","rgba(42,149,136,1)","rgba(68,56,124,1)","rgba(43,130,140,1)","rgba(49,112,141,1)","rgba(64,73,136,1)","rgba(63,81,137,1)","rgba(69,16,91,1)","rgba(67,61,128,1)","rgba(67,61,128,1)","rgba(44,132,140,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(70,37,106,1)","rgba(43,126,141,1)","rgba(40,155,135,1)","rgba(43,125,141,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(42,148,137,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(68,54,122,1)","rgba(57,100,139,1)","rgba(67,59,126,1)","rgba(34,168,132,1)","rgba(68,4,85,1)","rgba(43,125,141,1)","rgba(48,114,141,1)","rgba(44,132,140,1)","rgba(43,128,141,1)","rgba(52,107,140,1)","rgba(54,105,140,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(43,130,140,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(62,85,137,1)","rgba(43,143,138,1)","rgba(69,42,111,1)","rgba(43,170,130,1)","rgba(59,94,139,1)","rgba(61,87,138,1)","rgba(44,132,140,1)","rgba(69,23,96,1)","rgba(69,19,93,1)","rgba(44,138,139,1)","rgba(68,8,87,1)","rgba(70,39,108,1)","rgba(64,72,136,1)","rgba(43,123,142,1)","rgba(46,116,142,1)","rgba(43,145,137,1)","rgba(45,117,142,1)","rgba(43,128,141,1)","rgba(43,130,140,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(50,111,141,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(43,126,141,1)","rgba(67,61,128,1)","rgba(66,63,130,1)","rgba(60,90,138,1)","rgba(51,109,141,1)","rgba(45,117,142,1)","rgba(70,26,98,1)","rgba(68,2,85,1)","rgba(41,153,136,1)","rgba(38,162,134,1)","rgba(60,91,138,1)","rgba(43,123,141,1)","rgba(59,93,139,1)","rgba(43,123,142,1)","rgba(57,99,139,1)","rgba(57,100,139,1)","rgba(43,144,137,1)","rgba(44,138,139,1)","rgba(44,133,140,1)","rgba(57,99,139,1)","rgba(53,106,140,1)","rgba(63,80,137,1)","rgba(51,110,141,1)","rgba(69,23,96,1)","rgba(62,83,137,1)","rgba(69,23,96,1)","rgba(61,89,138,1)","rgba(43,140,138,1)","rgba(68,4,86,1)","rgba(44,137,139,1)","rgba(43,146,137,1)","rgba(44,118,142,1)","rgba(50,111,141,1)","rgba(42,121,142,1)","rgba(39,158,134,1)","rgba(55,103,140,1)","rgba(45,116,142,1)","rgba(68,3,85,1)","rgba(42,120,142,1)","rgba(63,83,137,1)","rgba(53,107,140,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(43,130,140,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(44,136,139,1)","rgba(43,130,140,1)","rgba(43,123,141,1)","rgba(42,149,136,1)","rgba(41,154,135,1)","rgba(65,67,134,1)","rgba(50,111,141,1)","rgba(66,64,131,1)","rgba(40,156,135,1)","rgba(38,161,134,1)","rgba(45,117,142,1)","rgba(52,108,140,1)","rgba(42,122,142,1)","rgba(68,56,123,1)","rgba(50,111,141,1)","rgba(69,20,94,1)","rgba(53,106,140,1)","rgba(70,35,104,1)","rgba(70,29,100,1)","rgba(43,142,138,1)","rgba(51,172,127,1)","rgba(62,84,137,1)","rgba(68,5,86,1)","rgba(52,108,141,1)","rgba(45,117,142,1)","rgba(44,137,139,1)","rgba(44,131,140,1)","rgba(64,73,136,1)","rgba(68,4,85,1)","rgba(44,136,139,1)","rgba(69,15,91,1)","rgba(43,142,138,1)","rgba(54,105,140,1)","rgba(44,133,140,1)","rgba(69,13,90,1)","rgba(43,145,137,1)","rgba(70,41,110,1)","rgba(68,7,87,1)","rgba(68,58,125,1)","rgba(59,175,125,1)","rgba(43,124,141,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(43,119,142,1)","rgba(42,147,137,1)","rgba(40,155,135,1)","rgba(55,103,140,1)","rgba(57,99,139,1)","rgba(55,103,140,1)","rgba(69,20,94,1)","rgba(44,137,139,1)","rgba(63,82,137,1)","rgba(44,133,140,1)","rgba(43,147,137,1)","rgba(44,131,140,1)","rgba(44,134,139,1)","rgba(34,168,132,1)","rgba(62,83,137,1)","rgba(46,116,141,1)","rgba(42,121,142,1)","rgba(44,135,139,1)","rgba(44,131,140,1)","rgba(58,96,139,1)","rgba(58,97,139,1)","rgba(43,139,138,1)","rgba(52,107,140,1)","rgba(52,108,141,1)","rgba(69,9,88,1)","rgba(42,148,137,1)","rgba(70,33,103,1)","rgba(51,109,141,1)","rgba(63,79,137,1)","rgba(43,143,138,1)","rgba(38,162,133,1)","rgba(65,70,135,1)","rgba(43,130,140,1)","rgba(58,97,139,1)","rgba(69,24,96,1)","rgba(43,129,140,1)","rgba(70,40,109,1)","rgba(70,37,107,1)","rgba(43,141,138,1)","rgba(68,5,86,1)","rgba(43,127,141,1)","rgba(43,126,141,1)","rgba(58,97,139,1)","rgba(60,92,138,1)","rgba(43,128,141,1)","rgba(55,103,140,1)","rgba(60,91,138,1)","rgba(40,157,135,1)","rgba(70,41,109,1)","rgba(53,107,140,1)","rgba(48,113,141,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(42,121,142,1)","rgba(70,34,104,1)","rgba(36,165,133,1)","rgba(43,123,141,1)","rgba(58,96,139,1)","rgba(63,80,137,1)","rgba(44,132,140,1)","rgba(43,143,138,1)","rgba(43,124,141,1)","rgba(69,13,90,1)","rgba(49,112,141,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(42,121,142,1)","rgba(70,37,106,1)","rgba(69,21,95,1)","rgba(43,123,141,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(44,132,140,1)","rgba(44,118,142,1)","rgba(42,121,142,1)","rgba(70,31,101,1)","rgba(68,57,124,1)","rgba(68,6,86,1)","rgba(43,130,140,1)","rgba(63,79,137,1)","rgba(59,175,125,1)","rgba(70,38,107,1)","rgba(50,110,141,1)","rgba(45,117,142,1)","rgba(43,147,137,1)","rgba(64,77,136,1)","rgba(69,50,118,1)","rgba(64,75,136,1)","rgba(41,151,136,1)","rgba(64,73,136,1)","rgba(111,201,92,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(49,112,141,1)","rgba(40,156,135,1)","rgba(56,174,126,1)","rgba(55,102,140,1)","rgba(103,196,100,1)","rgba(43,146,137,1)","rgba(43,127,141,1)","rgba(48,113,141,1)","rgba(44,132,140,1)","rgba(63,79,137,1)","rgba(42,122,142,1)","rgba(46,115,141,1)","rgba(65,70,135,1)","rgba(70,39,108,1)","rgba(43,141,138,1)","rgba(54,173,126,1)","rgba(102,195,100,1)","rgba(68,58,125,1)","rgba(62,86,138,1)","rgba(43,128,141,1)","rgba(39,160,134,1)","rgba(43,123,142,1)","rgba(62,86,137,1)","rgba(41,154,135,1)","rgba(62,86,138,1)","rgba(43,126,141,1)","rgba(43,143,138,1)","rgba(51,109,141,1)","rgba(37,164,133,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(51,110,141,1)","rgba(51,110,141,1)","rgba(61,87,138,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(84,185,112,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(39,160,134,1)","rgba(43,145,137,1)","rgba(43,142,138,1)","rgba(49,111,141,1)","rgba(44,131,140,1)","rgba(47,115,141,1)","rgba(68,3,85,1)","rgba(63,176,123,1)","rgba(43,127,141,1)","rgba(69,22,95,1)","rgba(43,119,142,1)","rgba(69,52,120,1)","rgba(68,56,123,1)","rgba(70,25,97,1)","rgba(44,131,140,1)","rgba(43,129,140,1)","rgba(60,92,138,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(69,12,89,1)","rgba(42,121,142,1)","rgba(57,99,139,1)","rgba(47,114,141,1)","rgba(62,176,123,1)","rgba(61,87,138,1)","rgba(39,159,134,1)","rgba(70,41,110,1)","rgba(47,115,141,1)","rgba(46,115,141,1)","rgba(65,68,135,1)","rgba(69,50,118,1)","rgba(53,106,140,1)","rgba(42,150,136,1)","rgba(43,141,138,1)","rgba(43,129,140,1)","rgba(66,66,133,1)","rgba(62,84,137,1)","rgba(70,33,103,1)","rgba(48,113,141,1)","rgba(44,134,139,1)","rgba(54,104,140,1)","rgba(54,104,140,1)","rgba(44,136,139,1)","rgba(59,93,139,1)","rgba(41,154,135,1)","rgba(65,67,134,1)","rgba(57,98,139,1)","rgba(43,146,137,1)","rgba(66,65,132,1)","rgba(62,85,137,1)","rgba(43,127,141,1)","rgba(57,99,139,1)","rgba(45,117,142,1)","rgba(42,122,142,1)","rgba(39,158,134,1)","rgba(42,122,142,1)","rgba(43,125,141,1)","rgba(54,104,140,1)","rgba(62,86,138,1)","rgba(85,186,112,1)","rgba(44,118,142,1)","rgba(60,175,124,1)","rgba(42,121,142,1)","rgba(51,110,141,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(69,24,96,1)","rgba(66,65,132,1)","rgba(43,128,141,1)","rgba(43,128,141,1)","rgba(48,113,141,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(70,27,98,1)","rgba(49,112,141,1)","rgba(69,43,111,1)","rgba(42,122,142,1)","rgba(69,50,118,1)","rgba(43,129,140,1)","rgba(36,165,133,1)","rgba(41,154,135,1)","rgba(43,119,142,1)","rgba(50,110,141,1)","rgba(36,164,133,1)","rgba(62,83,137,1)","rgba(49,113,141,1)","rgba(62,86,138,1)","rgba(54,104,140,1)","rgba(61,88,138,1)","rgba(61,89,138,1)","rgba(45,117,142,1)","rgba(54,104,140,1)","rgba(43,144,137,1)","rgba(60,91,138,1)","rgba(59,94,139,1)","rgba(70,41,110,1)","rgba(42,148,137,1)","rgba(50,111,141,1)","rgba(44,135,139,1)","rgba(53,107,140,1)","rgba(43,143,138,1)","rgba(43,128,140,1)","rgba(56,101,140,1)","rgba(44,137,139,1)","rgba(41,154,135,1)","rgba(69,25,97,1)","rgba(43,128,140,1)","rgba(52,108,141,1)","rgba(52,108,141,1)","rgba(45,117,142,1)","rgba(43,123,141,1)","rgba(35,167,132,1)","rgba(43,147,137,1)","rgba(64,73,136,1)","rgba(69,18,92,1)","rgba(56,101,140,1)","rgba(43,144,137,1)","rgba(70,34,104,1)","rgba(57,99,139,1)","rgba(62,85,137,1)","rgba(49,112,141,1)","rgba(34,167,132,1)","rgba(64,76,136,1)","rgba(67,60,127,1)","rgba(39,158,134,1)","rgba(54,104,140,1)","rgba(43,123,142,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(58,95,139,1)","rgba(44,139,138,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(44,136,139,1)","rgba(43,146,137,1)","rgba(63,81,137,1)","rgba(43,123,142,1)","rgba(64,73,136,1)","rgba(66,65,132,1)","rgba(63,81,137,1)","rgba(43,140,138,1)","rgba(43,126,141,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(56,101,140,1)","rgba(66,64,131,1)","rgba(69,42,111,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(70,41,110,1)","rgba(70,40,109,1)","rgba(72,180,119,1)","rgba(54,105,140,1)","rgba(65,70,135,1)","rgba(65,66,134,1)","rgba(65,68,135,1)","rgba(43,119,142,1)","rgba(43,129,140,1)","rgba(67,61,128,1)","rgba(42,147,137,1)","rgba(38,162,133,1)","rgba(44,137,139,1)","rgba(84,185,112,1)","rgba(43,141,138,1)","rgba(41,154,135,1)","rgba(35,166,132,1)","rgba(69,46,114,1)","rgba(69,22,95,1)","rgba(47,115,141,1)","rgba(41,152,136,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(65,70,135,1)","rgba(60,175,124,1)","rgba(42,148,137,1)","rgba(44,131,140,1)","rgba(47,171,129,1)","rgba(70,27,99,1)","rgba(44,135,139,1)","rgba(57,99,139,1)","rgba(59,93,138,1)","rgba(43,127,141,1)","rgba(54,105,140,1)","rgba(64,72,136,1)","rgba(43,127,141,1)","rgba(69,25,97,1)","rgba(44,135,139,1)","rgba(44,137,139,1)","rgba(43,139,138,1)","rgba(44,132,140,1)","rgba(37,163,133,1)","rgba(54,104,140,1)","rgba(48,114,141,1)","rgba(43,128,141,1)","rgba(118,206,86,1)","rgba(64,76,136,1)","rgba(43,141,138,1)","rgba(48,114,141,1)","rgba(69,17,92,1)","rgba(64,76,136,1)","rgba(69,17,92,1)","rgba(48,171,128,1)","rgba(37,162,133,1)","rgba(63,79,137,1)","rgba(58,97,139,1)","rgba(43,146,137,1)","rgba(44,134,139,1)","rgba(47,115,141,1)","rgba(43,146,137,1)","rgba(51,109,141,1)","rgba(70,26,98,1)","rgba(51,109,141,1)","rgba(43,128,140,1)","rgba(49,112,141,1)","rgba(42,148,136,1)","rgba(38,161,134,1)","rgba(37,163,133,1)","rgba(43,119,142,1)","rgba(43,130,140,1)","rgba(66,64,131,1)","rgba(59,93,139,1)","rgba(39,159,134,1)","rgba(49,113,141,1)","rgba(75,181,117,1)","rgba(53,106,140,1)","rgba(44,136,139,1)","rgba(69,53,120,1)","rgba(43,124,141,1)","rgba(69,25,97,1)","rgba(64,75,136,1)","rgba(54,104,140,1)","rgba(36,168,132,1)","rgba(64,76,136,1)","rgba(64,74,136,1)","rgba(43,142,138,1)","rgba(68,4,85,1)","rgba(70,34,104,1)","rgba(43,130,140,1)","rgba(42,147,137,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(64,76,136,1)","rgba(40,156,135,1)","rgba(42,121,142,1)","rgba(41,153,135,1)","rgba(46,116,141,1)","rgba(43,128,141,1)","rgba(41,153,135,1)","rgba(53,106,140,1)","rgba(38,161,134,1)","rgba(42,121,142,1)","rgba(69,11,89,1)","rgba(57,98,139,1)","rgba(40,156,135,1)","rgba(69,46,115,1)","rgba(60,90,138,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(49,112,141,1)","rgba(49,112,141,1)","rgba(67,59,126,1)","rgba(53,106,140,1)","rgba(54,105,140,1)","rgba(47,114,141,1)","rgba(64,75,136,1)","rgba(55,173,126,1)","rgba(58,97,139,1)","rgba(62,84,137,1)","rgba(47,114,141,1)","rgba(40,155,135,1)","rgba(60,92,138,1)","rgba(64,73,136,1)","rgba(62,84,137,1)","rgba(61,87,138,1)","rgba(61,88,138,1)","rgba(37,163,133,1)","rgba(48,113,141,1)","rgba(35,168,132,1)","rgba(38,161,134,1)","rgba(63,82,137,1)","rgba(42,149,136,1)","rgba(47,114,141,1)","rgba(62,83,137,1)","rgba(54,105,140,1)","rgba(54,105,140,1)","rgba(58,97,139,1)","rgba(64,75,136,1)","rgba(43,119,142,1)","rgba(44,132,140,1)","rgba(67,59,126,1)","rgba(63,81,137,1)","rgba(44,131,140,1)","rgba(51,110,141,1)","rgba(54,105,140,1)","rgba(55,103,140,1)","rgba(68,54,121,1)","rgba(68,7,87,1)","rgba(63,79,137,1)","rgba(52,107,140,1)","rgba(43,129,140,1)","rgba(68,56,124,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(51,109,141,1)","rgba(36,165,133,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(38,161,134,1)","rgba(63,78,136,1)","rgba(46,171,129,1)","rgba(68,57,125,1)","rgba(61,88,138,1)","rgba(54,105,140,1)","rgba(43,127,141,1)","rgba(43,140,138,1)","rgba(48,114,141,1)","rgba(50,110,141,1)","rgba(43,170,130,1)","rgba(42,122,142,1)","rgba(94,190,106,1)","rgba(64,74,136,1)","rgba(43,126,141,1)","rgba(57,98,139,1)","rgba(40,157,134,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(58,96,139,1)","rgba(69,48,116,1)","rgba(49,172,128,1)","rgba(53,106,140,1)","rgba(44,135,139,1)","rgba(44,134,139,1)","rgba(43,143,138,1)","rgba(40,155,135,1)","rgba(37,169,131,1)","rgba(65,70,135,1)","rgba(69,21,94,1)","rgba(64,73,136,1)","rgba(43,144,137,1)","rgba(68,55,122,1)","rgba(36,164,133,1)","rgba(43,130,140,1)","rgba(42,149,136,1)","rgba(53,107,140,1)","rgba(67,61,129,1)","rgba(69,23,96,1)","rgba(41,152,136,1)","rgba(46,116,142,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(60,90,138,1)","rgba(40,155,135,1)","rgba(43,126,141,1)","rgba(43,146,137,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(56,101,140,1)","rgba(43,129,140,1)","rgba(42,151,136,1)","rgba(34,168,132,1)","rgba(43,129,140,1)","rgba(62,86,138,1)","rgba(88,187,110,1)","rgba(43,130,140,1)","rgba(36,166,133,1)","rgba(43,145,137,1)","rgba(34,168,132,1)","rgba(68,4,85,1)","rgba(62,83,137,1)","rgba(64,76,136,1)","rgba(61,88,138,1)","rgba(70,42,111,1)","rgba(69,46,114,1)","rgba(38,161,134,1)","rgba(39,169,131,1)","rgba(69,51,119,1)","rgba(36,165,133,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(152,214,75,1)","rgba(42,149,136,1)","rgba(42,122,142,1)","rgba(56,101,140,1)","rgba(43,144,137,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(43,130,140,1)","rgba(60,91,138,1)","rgba(42,149,136,1)","rgba(69,46,114,1)","rgba(41,152,136,1)","rgba(36,164,133,1)","rgba(48,114,141,1)","rgba(40,155,135,1)","rgba(44,132,140,1)","rgba(49,172,128,1)","rgba(50,111,141,1)","rgba(42,148,137,1)","rgba(44,135,139,1)","rgba(50,172,128,1)","rgba(44,139,138,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(70,32,103,1)","rgba(39,160,134,1)","rgba(68,55,123,1)","rgba(69,24,96,1)","rgba(35,166,132,1)","rgba(70,26,98,1)","rgba(44,134,139,1)","rgba(43,119,142,1)","rgba(43,143,138,1)","rgba(44,139,138,1)","rgba(48,114,141,1)","rgba(46,116,141,1)","rgba(56,101,140,1)","rgba(69,18,93,1)","rgba(60,91,138,1)","rgba(67,61,128,1)","rgba(42,151,136,1)","rgba(57,99,139,1)","rgba(44,136,139,1)","rgba(57,98,139,1)","rgba(44,131,140,1)","rgba(70,30,101,1)","rgba(39,159,134,1)","rgba(69,51,119,1)","rgba(43,119,142,1)","rgba(57,99,139,1)","rgba(53,107,140,1)","rgba(42,122,142,1)","rgba(43,129,140,1)","rgba(69,14,90,1)","rgba(43,141,138,1)","rgba(43,123,141,1)","rgba(68,6,86,1)","rgba(66,65,132,1)","rgba(70,27,99,1)","rgba(59,94,139,1)","rgba(70,30,101,1)","rgba(70,31,101,1)","rgba(51,110,141,1)","rgba(42,150,136,1)","rgba(48,171,129,1)","rgba(41,153,136,1)","rgba(43,142,138,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(67,62,129,1)","rgba(42,122,142,1)","rgba(42,147,137,1)","rgba(43,144,137,1)","rgba(38,162,134,1)","rgba(49,112,141,1)","rgba(43,144,137,1)","rgba(45,117,142,1)","rgba(48,114,141,1)","rgba(62,86,138,1)","rgba(44,133,140,1)","rgba(42,149,136,1)","rgba(41,152,136,1)","rgba(65,67,134,1)","rgba(44,132,140,1)","rgba(42,121,142,1)","rgba(40,169,131,1)","rgba(63,81,137,1)","rgba(40,155,135,1)","rgba(53,106,140,1)","rgba(46,116,142,1)","rgba(41,152,136,1)","rgba(43,124,141,1)","rgba(52,107,140,1)","rgba(44,118,142,1)","rgba(43,123,141,1)","rgba(38,162,133,1)","rgba(69,19,93,1)","rgba(70,36,106,1)","rgba(42,151,136,1)","rgba(44,134,139,1)","rgba(43,147,137,1)","rgba(43,123,142,1)","rgba(57,98,139,1)","rgba(70,41,110,1)","rgba(42,150,136,1)","rgba(44,136,139,1)","rgba(60,91,138,1)","rgba(39,160,134,1)","rgba(70,28,99,1)","rgba(44,134,139,1)","rgba(83,185,113,1)","rgba(43,131,140,1)","rgba(69,12,89,1)","rgba(70,34,104,1)","rgba(64,76,136,1)","rgba(43,141,138,1)","rgba(42,147,137,1)","rgba(53,106,140,1)","rgba(43,139,138,1)","rgba(50,111,141,1)","rgba(43,130,140,1)","rgba(63,81,137,1)","rgba(39,158,134,1)","rgba(44,134,139,1)","rgba(61,87,138,1)","rgba(65,71,135,1)","rgba(70,36,105,1)","rgba(68,4,85,1)","rgba(69,13,90,1)","rgba(70,36,106,1)","rgba(63,81,137,1)","rgba(44,137,139,1)","rgba(43,129,140,1)","rgba(65,67,134,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(69,48,116,1)","rgba(37,169,131,1)","rgba(57,99,139,1)","rgba(43,123,141,1)","rgba(43,142,138,1)","rgba(69,25,97,1)","rgba(70,27,99,1)","rgba(57,98,139,1)","rgba(62,176,123,1)","rgba(43,127,141,1)","rgba(43,119,142,1)","rgba(77,182,117,1)","rgba(69,19,93,1)","rgba(69,21,94,1)","rgba(56,101,140,1)","rgba(43,147,137,1)","rgba(43,124,141,1)","rgba(43,128,141,1)","rgba(39,158,134,1)","rgba(67,59,126,1)","rgba(44,131,140,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(43,119,142,1)","rgba(59,93,138,1)","rgba(64,77,136,1)","rgba(62,83,137,1)","rgba(65,68,135,1)","rgba(108,199,95,1)","rgba(64,75,136,1)","rgba(68,55,123,1)","rgba(43,123,142,1)","rgba(58,97,139,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(44,139,138,1)","rgba(42,120,142,1)","rgba(54,104,140,1)","rgba(42,122,142,1)","rgba(61,89,138,1)","rgba(58,97,139,1)","rgba(70,39,108,1)","rgba(42,150,136,1)","rgba(58,97,139,1)","rgba(37,169,131,1)","rgba(65,72,136,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(43,144,137,1)","rgba(68,7,87,1)","rgba(70,27,99,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(68,57,125,1)","rgba(69,50,118,1)","rgba(44,118,142,1)","rgba(56,100,140,1)","rgba(44,139,138,1)","rgba(63,79,136,1)","rgba(44,118,142,1)","rgba(43,141,138,1)","rgba(65,68,135,1)","rgba(42,150,136,1)","rgba(63,81,137,1)","rgba(44,137,139,1)","rgba(62,85,137,1)","rgba(65,69,135,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(44,136,139,1)","rgba(44,138,139,1)","rgba(53,107,140,1)","rgba(66,63,131,1)","rgba(63,81,137,1)","rgba(37,163,133,1)","rgba(41,154,135,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(70,41,110,1)","rgba(43,128,141,1)","rgba(58,95,139,1)","rgba(42,122,142,1)","rgba(43,119,142,1)","rgba(43,141,138,1)","rgba(50,111,141,1)","rgba(60,175,124,1)","rgba(43,144,137,1)","rgba(65,71,135,1)","rgba(50,111,141,1)","rgba(54,105,140,1)","rgba(69,17,92,1)","rgba(36,165,133,1)","rgba(65,177,122,1)","rgba(44,132,140,1)","rgba(44,137,139,1)","rgba(52,108,140,1)","rgba(61,89,138,1)","rgba(64,76,136,1)","rgba(68,2,84,1)","rgba(63,79,137,1)","rgba(39,159,134,1)","rgba(43,119,142,1)","rgba(69,24,97,1)","rgba(69,17,92,1)","rgba(61,89,138,1)","rgba(68,55,123,1)","rgba(43,126,141,1)","rgba(62,84,137,1)","rgba(43,123,141,1)","rgba(42,148,137,1)","rgba(43,129,140,1)","rgba(44,135,139,1)","rgba(42,120,142,1)","rgba(69,48,116,1)","rgba(63,82,137,1)","rgba(43,128,140,1)","rgba(60,90,138,1)","rgba(59,95,139,1)","rgba(65,70,135,1)","rgba(58,96,139,1)","rgba(44,135,139,1)","rgba(68,57,124,1)","rgba(70,179,120,1)","rgba(54,104,140,1)","rgba(43,123,141,1)","rgba(43,146,137,1)","rgba(48,114,141,1)","rgba(54,105,140,1)","rgba(43,128,140,1)","rgba(44,132,140,1)","rgba(68,2,85,1)","rgba(69,15,91,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(63,79,137,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(70,31,102,1)","rgba(49,112,141,1)","rgba(55,104,140,1)","rgba(43,128,141,1)","rgba(63,82,137,1)","rgba(70,34,104,1)","rgba(37,163,133,1)","rgba(80,183,115,1)","rgba(43,140,138,1)","rgba(43,130,140,1)","rgba(69,51,119,1)","rgba(65,71,135,1)","rgba(44,133,140,1)","rgba(46,115,141,1)","rgba(69,9,88,1)","rgba(44,134,139,1)","rgba(37,163,133,1)","rgba(115,204,88,1)","rgba(53,105,140,1)","rgba(69,8,88,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(52,107,140,1)","rgba(57,99,139,1)","rgba(43,123,141,1)","rgba(69,51,118,1)","rgba(63,82,137,1)","rgba(74,180,118,1)","rgba(52,107,140,1)","rgba(43,127,141,1)","rgba(43,119,142,1)","rgba(69,45,114,1)","rgba(70,41,110,1)","rgba(43,145,137,1)","rgba(130,210,79,1)","rgba(69,11,89,1)","rgba(69,47,115,1)","rgba(62,85,137,1)","rgba(42,122,142,1)","rgba(39,159,134,1)","rgba(51,109,141,1)","rgba(52,108,141,1)","rgba(73,180,119,1)","rgba(44,135,139,1)","rgba(69,15,91,1)","rgba(58,97,139,1)","rgba(69,50,117,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(44,131,140,1)","rgba(42,148,136,1)","rgba(44,131,140,1)","rgba(43,123,141,1)","rgba(38,160,134,1)","rgba(65,71,135,1)","rgba(70,42,111,1)","rgba(59,94,139,1)","rgba(54,104,140,1)","rgba(43,145,137,1)","rgba(53,107,140,1)","rgba(67,62,129,1)","rgba(41,155,135,1)","rgba(44,136,139,1)","rgba(54,105,140,1)","rgba(43,124,141,1)","rgba(43,126,141,1)","rgba(63,80,137,1)","rgba(64,75,136,1)","rgba(42,150,136,1)","rgba(43,143,138,1)","rgba(70,36,106,1)","rgba(61,88,138,1)","rgba(65,67,134,1)","rgba(43,140,138,1)","rgba(64,74,136,1)","rgba(56,101,140,1)","rgba(57,98,139,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(59,95,139,1)","rgba(37,164,133,1)","rgba(65,67,134,1)","rgba(70,41,110,1)","rgba(57,98,139,1)","rgba(44,135,139,1)","rgba(47,115,141,1)","rgba(62,83,137,1)","rgba(47,115,141,1)","rgba(51,109,141,1)","rgba(43,123,142,1)","rgba(61,87,138,1)","rgba(44,134,139,1)","rgba(43,125,141,1)","rgba(69,15,91,1)","rgba(52,108,140,1)","rgba(44,117,142,1)","rgba(65,177,122,1)","rgba(42,148,137,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(70,40,109,1)","rgba(44,138,139,1)","rgba(70,37,107,1)","rgba(64,77,136,1)","rgba(69,20,94,1)","rgba(40,157,134,1)","rgba(63,81,137,1)","rgba(40,156,135,1)","rgba(66,65,132,1)","rgba(43,125,141,1)","rgba(50,111,141,1)","rgba(57,99,139,1)","rgba(42,151,136,1)","rgba(70,26,98,1)","rgba(69,22,95,1)","rgba(74,180,118,1)","rgba(43,145,137,1)","rgba(57,99,139,1)","rgba(70,30,101,1)","rgba(42,149,136,1)","rgba(43,126,141,1)","rgba(59,93,139,1)","rgba(43,141,138,1)","rgba(79,183,116,1)","rgba(54,105,140,1)","rgba(55,104,140,1)","rgba(43,143,138,1)","rgba(53,107,140,1)","rgba(139,212,78,1)","rgba(68,178,121,1)","rgba(43,145,137,1)","rgba(44,132,140,1)","rgba(68,55,122,1)","rgba(40,155,135,1)","rgba(44,118,142,1)","rgba(68,58,125,1)","rgba(42,120,142,1)","rgba(53,106,140,1)","rgba(77,182,117,1)","rgba(44,134,139,1)","rgba(37,162,133,1)","rgba(43,144,137,1)","rgba(47,114,141,1)","rgba(70,32,102,1)","rgba(68,2,85,1)","rgba(64,78,136,1)","rgba(56,100,139,1)","rgba(69,43,112,1)","rgba(46,116,141,1)","rgba(69,42,111,1)","rgba(50,111,141,1)","rgba(58,97,139,1)","rgba(68,1,84,1)","rgba(43,119,142,1)","rgba(43,124,141,1)","rgba(60,90,138,1)","rgba(54,105,140,1)","rgba(70,31,102,1)","rgba(43,142,138,1)","rgba(69,18,92,1)","rgba(44,132,140,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(42,122,142,1)","rgba(57,98,139,1)","rgba(64,74,136,1)","rgba(59,94,139,1)","rgba(53,106,140,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(60,92,138,1)","rgba(60,91,138,1)","rgba(51,109,141,1)","rgba(52,108,140,1)","rgba(60,91,138,1)","rgba(43,143,138,1)","rgba(43,128,141,1)","rgba(66,65,132,1)","rgba(69,42,111,1)","rgba(41,152,136,1)","rgba(70,28,100,1)","rgba(44,137,139,1)","rgba(61,88,138,1)","rgba(43,124,141,1)","rgba(70,30,101,1)","rgba(52,108,140,1)","rgba(60,90,138,1)","rgba(43,123,141,1)","rgba(65,69,135,1)","rgba(43,141,138,1)","rgba(40,169,131,1)","rgba(44,135,139,1)","rgba(37,163,133,1)","rgba(69,17,92,1)","rgba(60,90,138,1)","rgba(43,143,138,1)","rgba(50,111,141,1)","rgba(41,153,136,1)","rgba(53,107,140,1)","rgba(61,87,138,1)","rgba(47,114,141,1)","rgba(59,95,139,1)","rgba(41,154,135,1)","rgba(59,95,139,1)","rgba(42,148,136,1)","rgba(40,155,135,1)","rgba(66,177,122,1)","rgba(43,126,141,1)","rgba(60,92,138,1)","rgba(68,6,86,1)","rgba(70,35,105,1)","rgba(41,152,136,1)","rgba(51,109,141,1)","rgba(69,13,90,1)","rgba(53,106,140,1)","rgba(41,170,130,1)","rgba(50,111,141,1)","rgba(41,153,135,1)","rgba(44,138,139,1)","rgba(57,99,139,1)","rgba(54,104,140,1)","rgba(69,45,114,1)","rgba(44,133,140,1)","rgba(53,106,140,1)","rgba(42,149,136,1)","rgba(42,120,142,1)","rgba(43,119,142,1)","rgba(44,139,138,1)","rgba(44,133,140,1)","rgba(42,122,142,1)","rgba(70,42,111,1)","rgba(69,24,97,1)","rgba(69,44,113,1)","rgba(111,201,92,1)","rgba(68,54,122,1)","rgba(70,35,105,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(68,4,86,1)","rgba(44,139,138,1)","rgba(61,175,124,1)","rgba(70,35,105,1)","rgba(66,64,131,1)","rgba(70,32,102,1)","rgba(65,66,134,1)","rgba(51,109,141,1)","rgba(65,70,135,1)","rgba(41,153,135,1)","rgba(62,86,138,1)","rgba(44,138,139,1)","rgba(39,158,134,1)","rgba(42,148,137,1)","rgba(63,82,137,1)","rgba(43,141,138,1)","rgba(45,117,142,1)","rgba(64,73,136,1)","rgba(59,93,139,1)","rgba(43,123,142,1)","rgba(40,156,135,1)","rgba(60,91,138,1)","rgba(61,86,138,1)","rgba(51,109,141,1)","rgba(57,99,139,1)","rgba(42,121,142,1)","rgba(62,83,137,1)","rgba(44,136,139,1)","rgba(59,95,139,1)","rgba(44,136,139,1)","rgba(65,68,135,1)","rgba(42,148,137,1)","rgba(63,81,137,1)","rgba(57,98,139,1)","rgba(89,188,109,1)","rgba(64,72,136,1)","rgba(64,73,136,1)","rgba(39,159,134,1)","rgba(57,99,139,1)","rgba(43,144,137,1)","rgba(42,121,142,1)","rgba(35,166,132,1)","rgba(47,115,141,1)","rgba(43,128,141,1)","rgba(45,117,142,1)","rgba(43,126,141,1)","rgba(98,193,104,1)","rgba(43,140,138,1)","rgba(48,113,141,1)","rgba(70,30,101,1)","rgba(50,111,141,1)","rgba(70,31,101,1)","rgba(44,138,139,1)","rgba(58,97,139,1)","rgba(43,125,141,1)","rgba(71,179,120,1)","rgba(70,35,105,1)","rgba(70,27,98,1)","rgba(38,162,133,1)","rgba(70,33,103,1)","rgba(53,107,140,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(53,106,140,1)","rgba(44,136,139,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(43,140,138,1)","rgba(62,84,137,1)","rgba(43,145,137,1)","rgba(69,51,119,1)","rgba(69,43,112,1)","rgba(42,151,136,1)","rgba(43,145,137,1)","rgba(52,108,141,1)","rgba(43,119,142,1)","rgba(69,25,97,1)","rgba(63,82,137,1)","rgba(53,106,140,1)","rgba(43,125,141,1)","rgba(43,119,142,1)","rgba(52,108,140,1)","rgba(44,134,139,1)","rgba(42,148,137,1)","rgba(44,135,139,1)","rgba(44,133,140,1)","rgba(44,135,139,1)","rgba(58,96,139,1)","rgba(53,106,140,1)","rgba(61,89,138,1)","rgba(44,136,139,1)","rgba(64,75,136,1)","rgba(43,145,137,1)","rgba(48,113,141,1)","rgba(64,75,136,1)","rgba(64,177,123,1)","rgba(44,134,139,1)","rgba(74,181,118,1)","rgba(37,164,133,1)","rgba(61,86,138,1)","rgba(64,77,136,1)","rgba(38,161,134,1)","rgba(69,51,119,1)","rgba(55,102,140,1)","rgba(43,128,141,1)","rgba(56,101,140,1)","rgba(55,102,140,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(52,172,127,1)","rgba(39,160,134,1)","rgba(55,102,140,1)","rgba(44,118,142,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(41,154,135,1)","rgba(63,79,137,1)","rgba(64,72,136,1)","rgba(52,107,140,1)","rgba(43,143,138,1)","rgba(44,132,140,1)","rgba(65,67,134,1)","rgba(43,125,141,1)","rgba(57,98,139,1)","rgba(70,26,98,1)","rgba(43,145,137,1)","rgba(70,31,102,1)","rgba(43,146,137,1)","rgba(45,117,142,1)","rgba(34,168,132,1)","rgba(54,104,140,1)","rgba(43,126,141,1)","rgba(69,50,118,1)","rgba(43,144,137,1)","rgba(54,105,140,1)","rgba(36,165,133,1)","rgba(48,113,141,1)","rgba(42,150,136,1)","rgba(43,128,141,1)","rgba(49,112,141,1)","rgba(43,123,142,1)","rgba(43,124,141,1)","rgba(46,116,141,1)","rgba(62,85,137,1)","rgba(69,52,120,1)","rgba(44,131,140,1)","rgba(59,93,138,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(43,126,141,1)","rgba(46,116,141,1)","rgba(70,27,99,1)","rgba(42,121,142,1)","rgba(37,169,131,1)","rgba(43,129,140,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(38,161,134,1)","rgba(66,66,133,1)","rgba(60,92,138,1)","rgba(41,154,135,1)","rgba(44,135,139,1)","rgba(41,153,135,1)","rgba(69,24,96,1)","rgba(38,161,134,1)","rgba(51,109,141,1)","rgba(69,24,96,1)","rgba(43,129,140,1)","rgba(69,20,94,1)","rgba(43,146,137,1)","rgba(70,33,103,1)","rgba(46,116,141,1)","rgba(53,106,140,1)","rgba(39,159,134,1)","rgba(42,121,142,1)","rgba(62,86,137,1)","rgba(43,130,140,1)","rgba(64,75,136,1)","rgba(47,115,141,1)","rgba(63,82,137,1)","rgba(70,36,106,1)","rgba(41,153,135,1)","rgba(70,41,110,1)","rgba(40,157,135,1)","rgba(42,148,137,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(44,136,139,1)","rgba(70,32,103,1)","rgba(51,109,141,1)","rgba(35,167,132,1)","rgba(68,55,123,1)","rgba(67,60,127,1)","rgba(43,140,138,1)","rgba(69,15,91,1)","rgba(43,141,138,1)","rgba(66,66,133,1)","rgba(69,25,97,1)","rgba(48,114,141,1)","rgba(45,117,142,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(43,144,137,1)","rgba(56,102,140,1)","rgba(47,114,141,1)","rgba(57,99,139,1)","rgba(69,49,117,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(43,125,141,1)","rgba(43,139,138,1)","rgba(70,35,105,1)","rgba(56,102,140,1)","rgba(43,125,141,1)","rgba(40,156,135,1)","rgba(64,75,136,1)","rgba(66,63,130,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(53,107,140,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(112,202,92,1)","rgba(43,140,138,1)","rgba(65,67,134,1)","rgba(45,117,142,1)","rgba(44,133,140,1)","rgba(111,201,93,1)","rgba(85,186,112,1)","rgba(65,69,135,1)","rgba(66,65,132,1)","rgba(69,14,90,1)","rgba(43,147,137,1)","rgba(43,123,141,1)","rgba(43,130,140,1)","rgba(69,21,94,1)","rgba(43,123,141,1)","rgba(40,157,135,1)","rgba(43,127,141,1)","rgba(59,94,139,1)","rgba(62,86,138,1)","rgba(64,76,136,1)","rgba(81,184,114,1)","rgba(51,110,141,1)","rgba(67,62,129,1)","rgba(50,111,141,1)","rgba(65,67,134,1)","rgba(43,127,141,1)","rgba(45,117,142,1)","rgba(68,3,85,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(65,72,136,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(44,132,140,1)","rgba(42,150,136,1)","rgba(42,149,136,1)","rgba(43,127,141,1)","rgba(69,47,115,1)","rgba(57,98,139,1)","rgba(75,181,118,1)","rgba(66,63,130,1)","rgba(55,103,140,1)","rgba(43,128,141,1)","rgba(44,139,138,1)","rgba(68,5,86,1)","rgba(44,118,142,1)","rgba(70,32,103,1)","rgba(43,124,141,1)","rgba(56,101,140,1)","rgba(68,7,87,1)","rgba(43,130,140,1)","rgba(34,168,132,1)","rgba(43,143,138,1)","rgba(54,105,140,1)","rgba(57,98,139,1)","rgba(42,120,142,1)","rgba(44,134,139,1)","rgba(42,121,142,1)","rgba(70,33,103,1)","rgba(46,116,142,1)","rgba(44,133,140,1)","rgba(70,27,99,1)","rgba(69,22,95,1)","rgba(50,111,141,1)","rgba(44,133,140,1)","rgba(69,51,118,1)","rgba(53,107,140,1)","rgba(43,127,141,1)","rgba(51,109,141,1)","rgba(58,96,139,1)","rgba(44,133,140,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(48,114,141,1)","rgba(66,63,130,1)","rgba(44,139,138,1)","rgba(45,117,142,1)","rgba(53,106,140,1)","rgba(44,133,140,1)","rgba(58,97,139,1)","rgba(39,159,134,1)","rgba(63,78,136,1)","rgba(64,74,136,1)","rgba(96,192,105,1)","rgba(121,208,83,1)","rgba(43,147,137,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(43,125,141,1)","rgba(44,134,139,1)","rgba(46,115,141,1)","rgba(47,115,141,1)","rgba(42,148,136,1)","rgba(52,107,140,1)","rgba(42,151,136,1)","rgba(67,178,121,1)","rgba(69,19,93,1)","rgba(87,187,110,1)","rgba(43,119,142,1)","rgba(42,148,137,1)","rgba(49,112,141,1)","rgba(48,113,141,1)","rgba(43,145,137,1)","rgba(43,144,137,1)","rgba(42,122,142,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(43,123,141,1)","rgba(42,151,136,1)","rgba(43,127,141,1)","rgba(42,122,142,1)","rgba(49,112,141,1)","rgba(45,116,142,1)","rgba(59,95,139,1)","rgba(43,125,141,1)","rgba(52,108,140,1)","rgba(38,161,134,1)","rgba(48,113,141,1)","rgba(37,163,133,1)","rgba(52,107,140,1)","rgba(49,113,141,1)","rgba(48,113,141,1)","rgba(43,125,141,1)","rgba(68,56,123,1)","rgba(70,31,102,1)","rgba(69,13,90,1)","rgba(51,109,141,1)","rgba(38,161,134,1)","rgba(60,91,138,1)","rgba(44,131,140,1)","rgba(67,61,128,1)","rgba(49,171,128,1)","rgba(43,129,140,1)","rgba(69,12,89,1)","rgba(42,122,142,1)","rgba(70,26,98,1)","rgba(44,137,139,1)","rgba(70,30,101,1)","rgba(42,149,136,1)","rgba(43,128,141,1)","rgba(35,166,133,1)","rgba(44,139,138,1)","rgba(58,96,139,1)","rgba(46,116,142,1)","rgba(43,140,138,1)","rgba(61,87,138,1)","rgba(43,119,142,1)","rgba(80,183,115,1)","rgba(42,120,142,1)","rgba(68,56,123,1)","rgba(43,142,138,1)","rgba(39,158,134,1)","rgba(69,9,88,1)","rgba(43,145,137,1)","rgba(43,143,138,1)","rgba(57,100,139,1)","rgba(43,127,141,1)","rgba(114,204,89,1)","rgba(69,21,95,1)","rgba(49,112,141,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(115,204,88,1)","rgba(43,141,138,1)","rgba(39,158,134,1)","rgba(37,164,133,1)","rgba(63,83,137,1)","rgba(53,106,140,1)","rgba(41,153,136,1)","rgba(61,86,138,1)","rgba(59,93,138,1)","rgba(69,44,112,1)","rgba(43,140,138,1)","rgba(53,106,140,1)","rgba(62,86,137,1)","rgba(64,78,136,1)","rgba(107,199,96,1)","rgba(42,120,142,1)","rgba(52,172,127,1)","rgba(44,118,142,1)","rgba(62,84,137,1)","rgba(69,50,118,1)","rgba(42,122,142,1)","rgba(70,39,108,1)","rgba(69,19,93,1)","rgba(43,126,141,1)","rgba(42,148,137,1)","rgba(53,105,140,1)","rgba(49,112,141,1)","rgba(40,156,135,1)","rgba(43,139,138,1)","rgba(44,137,139,1)","rgba(43,128,140,1)","rgba(36,165,133,1)","rgba(44,135,139,1)","rgba(35,167,132,1)","rgba(68,58,125,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(61,89,138,1)","rgba(43,143,138,1)","rgba(69,51,119,1)","rgba(41,152,136,1)","rgba(40,156,135,1)","rgba(42,149,136,1)","rgba(65,70,135,1)","rgba(41,154,135,1)","rgba(55,102,140,1)","rgba(69,49,117,1)","rgba(44,132,140,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(44,135,139,1)","rgba(65,67,134,1)","rgba(45,117,142,1)","rgba(41,152,136,1)","rgba(65,72,136,1)","rgba(43,130,140,1)","rgba(64,73,136,1)","rgba(50,110,141,1)","rgba(42,147,137,1)","rgba(43,130,140,1)","rgba(40,169,131,1)","rgba(56,100,139,1)","rgba(69,13,90,1)","rgba(45,117,142,1)","rgba(68,57,125,1)","rgba(66,63,130,1)","rgba(42,120,142,1)","rgba(67,59,127,1)","rgba(44,137,139,1)","rgba(43,129,140,1)","rgba(61,89,138,1)","rgba(53,107,140,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(66,63,130,1)","rgba(65,71,135,1)","rgba(43,145,137,1)","rgba(43,127,141,1)","rgba(43,143,138,1)","rgba(47,115,141,1)","rgba(44,133,140,1)","rgba(69,48,116,1)","rgba(56,101,140,1)","rgba(68,55,123,1)","rgba(53,106,140,1)","rgba(70,36,105,1)","rgba(43,128,141,1)","rgba(42,147,137,1)","rgba(50,111,141,1)","rgba(39,159,134,1)","rgba(43,130,140,1)","rgba(51,109,141,1)","rgba(42,120,142,1)","rgba(51,109,141,1)","rgba(44,138,139,1)","rgba(63,80,137,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(44,131,140,1)","rgba(61,88,138,1)","rgba(62,85,137,1)","rgba(62,83,137,1)","rgba(67,61,129,1)","rgba(65,72,136,1)","rgba(69,44,113,1)","rgba(69,13,90,1)","rgba(36,164,133,1)","rgba(40,156,135,1)","rgba(43,140,138,1)","rgba(44,132,140,1)","rgba(43,142,138,1)","rgba(63,176,123,1)","rgba(69,11,89,1)","rgba(67,62,129,1)","rgba(44,133,140,1)","rgba(42,150,136,1)","rgba(43,144,137,1)","rgba(64,76,136,1)","rgba(47,114,141,1)","rgba(35,166,132,1)","rgba(68,4,86,1)","rgba(41,152,136,1)","rgba(43,141,138,1)","rgba(44,136,139,1)","rgba(43,125,141,1)","rgba(43,142,138,1)","rgba(43,128,141,1)","rgba(60,92,138,1)","rgba(70,39,108,1)","rgba(46,116,141,1)","rgba(42,148,137,1)","rgba(56,101,140,1)","rgba(53,106,140,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(47,115,141,1)","rgba(122,209,81,1)","rgba(39,159,134,1)","rgba(58,174,125,1)","rgba(43,146,137,1)","rgba(51,172,128,1)","rgba(62,86,138,1)","rgba(36,165,133,1)","rgba(50,172,128,1)","rgba(43,125,141,1)","rgba(43,145,137,1)","rgba(58,97,139,1)","rgba(43,127,141,1)","rgba(63,81,137,1)","rgba(44,135,139,1)","rgba(43,124,141,1)","rgba(61,89,138,1)","rgba(55,103,140,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(59,93,138,1)","rgba(43,146,137,1)","rgba(43,139,138,1)","rgba(62,85,137,1)","rgba(70,38,107,1)","rgba(59,95,139,1)","rgba(56,101,140,1)","rgba(54,104,140,1)","rgba(57,99,139,1)","rgba(69,51,118,1)","rgba(43,127,141,1)","rgba(43,144,137,1)","rgba(52,108,141,1)","rgba(43,124,141,1)","rgba(43,142,138,1)","rgba(83,185,113,1)","rgba(59,175,125,1)","rgba(70,29,100,1)","rgba(44,136,139,1)","rgba(64,73,136,1)","rgba(70,36,106,1)","rgba(70,34,104,1)","rgba(44,133,140,1)","rgba(52,108,140,1)","rgba(48,114,141,1)","rgba(43,119,142,1)","rgba(43,130,140,1)","rgba(54,105,140,1)","rgba(65,67,134,1)","rgba(43,128,141,1)","rgba(42,149,136,1)","rgba(43,146,137,1)","rgba(55,103,140,1)","rgba(54,105,140,1)","rgba(40,169,131,1)","rgba(61,87,138,1)","rgba(69,50,118,1)","rgba(69,45,113,1)","rgba(53,106,140,1)","rgba(43,126,141,1)","rgba(43,127,141,1)","rgba(70,179,120,1)","rgba(69,23,96,1)","rgba(70,30,101,1)","rgba(44,133,140,1)","rgba(69,23,96,1)","rgba(52,109,141,1)","rgba(44,118,142,1)","rgba(54,104,140,1)","rgba(50,110,141,1)","rgba(49,112,141,1)","rgba(43,123,141,1)","rgba(34,168,132,1)","rgba(36,164,133,1)","rgba(43,128,141,1)","rgba(45,170,129,1)","rgba(70,32,102,1)","rgba(43,146,137,1)","rgba(40,155,135,1)","rgba(42,120,142,1)","rgba(61,89,138,1)","rgba(62,86,138,1)","rgba(57,99,139,1)","rgba(58,174,125,1)","rgba(44,138,139,1)","rgba(50,111,141,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(50,110,141,1)","rgba(55,103,140,1)","rgba(42,120,142,1)","rgba(64,73,136,1)","rgba(67,59,126,1)","rgba(47,115,141,1)","rgba(58,97,139,1)","rgba(69,43,111,1)","rgba(62,86,138,1)","rgba(43,127,141,1)","rgba(40,156,135,1)","rgba(47,115,141,1)","rgba(43,127,141,1)","rgba(66,66,133,1)","rgba(43,127,141,1)","rgba(68,54,121,1)","rgba(64,74,136,1)","rgba(58,97,139,1)","rgba(43,127,141,1)","rgba(43,139,138,1)","rgba(43,146,137,1)","rgba(70,40,109,1)","rgba(36,166,133,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(63,79,137,1)","rgba(71,179,119,1)","rgba(56,102,140,1)","rgba(59,93,139,1)","rgba(68,54,121,1)","rgba(51,109,141,1)","rgba(65,177,122,1)","rgba(64,75,136,1)","rgba(66,66,133,1)","rgba(112,202,92,1)","rgba(55,102,140,1)","rgba(65,71,135,1)","rgba(43,129,140,1)","rgba(42,122,142,1)","rgba(42,150,136,1)","rgba(43,128,141,1)","rgba(41,153,135,1)","rgba(44,134,139,1)","rgba(70,28,99,1)","rgba(44,137,139,1)","rgba(46,116,141,1)","rgba(48,171,128,1)","rgba(62,176,123,1)","rgba(68,6,86,1)","rgba(70,33,103,1)","rgba(34,167,132,1)","rgba(63,80,137,1)","rgba(52,108,140,1)","rgba(69,21,94,1)","rgba(44,134,139,1)","rgba(43,142,138,1)","rgba(44,136,139,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(49,112,141,1)","rgba(43,128,141,1)","rgba(43,146,137,1)","rgba(54,104,140,1)","rgba(62,86,138,1)","rgba(43,130,140,1)","rgba(59,95,139,1)","rgba(65,71,135,1)","rgba(42,122,142,1)","rgba(67,62,129,1)","rgba(46,116,142,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(44,133,140,1)","rgba(69,24,97,1)","rgba(44,134,139,1)","rgba(55,102,140,1)","rgba(70,36,106,1)","rgba(68,2,84,1)","rgba(51,109,141,1)","rgba(43,139,138,1)","rgba(62,84,137,1)","rgba(41,153,135,1)","rgba(38,169,131,1)","rgba(59,94,139,1)","rgba(48,114,141,1)","rgba(41,152,136,1)","rgba(42,149,136,1)","rgba(60,92,138,1)","rgba(57,99,139,1)","rgba(53,106,140,1)","rgba(59,93,139,1)","rgba(43,126,141,1)","rgba(56,174,126,1)","rgba(45,117,142,1)","rgba(69,51,119,1)","rgba(54,105,140,1)","rgba(51,109,141,1)","rgba(65,72,136,1)","rgba(61,87,138,1)","rgba(40,156,135,1)","rgba(44,138,139,1)","rgba(62,83,137,1)","rgba(51,172,128,1)","rgba(43,119,142,1)","rgba(49,112,141,1)","rgba(42,120,142,1)","rgba(43,129,140,1)","rgba(68,53,120,1)","rgba(69,25,97,1)","rgba(68,53,121,1)","rgba(43,139,138,1)","rgba(43,127,141,1)","rgba(70,30,101,1)","rgba(58,97,139,1)","rgba(44,137,139,1)","rgba(44,134,139,1)","rgba(43,144,137,1)","rgba(69,22,95,1)","rgba(68,56,123,1)","rgba(44,131,140,1)","rgba(43,130,140,1)","rgba(44,133,140,1)","rgba(70,34,104,1)","rgba(43,127,141,1)","rgba(66,65,132,1)","rgba(47,115,141,1)","rgba(63,78,136,1)","rgba(48,113,141,1)","rgba(70,31,102,1)","rgba(59,93,138,1)","rgba(47,115,141,1)","rgba(58,96,139,1)","rgba(44,136,139,1)","rgba(65,68,135,1)","rgba(43,124,141,1)","rgba(52,108,141,1)","rgba(70,37,107,1)","rgba(44,132,140,1)","rgba(44,138,139,1)","rgba(70,36,105,1)","rgba(46,116,142,1)","rgba(42,122,142,1)","rgba(43,128,141,1)","rgba(62,83,137,1)","rgba(44,136,139,1)","rgba(64,74,136,1)","rgba(61,87,138,1)","rgba(50,111,141,1)","rgba(40,157,134,1)","rgba(56,101,140,1)","rgba(43,147,137,1)","rgba(59,93,139,1)","rgba(70,37,106,1)","rgba(42,120,142,1)","rgba(69,43,112,1)","rgba(49,112,141,1)","rgba(59,94,139,1)","rgba(43,124,141,1)","rgba(44,136,139,1)","rgba(42,120,142,1)","rgba(63,79,137,1)","rgba(43,126,141,1)","rgba(48,113,141,1)","rgba(38,161,134,1)","rgba(49,112,141,1)","rgba(37,163,133,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(68,57,125,1)","rgba(62,83,137,1)","rgba(69,43,111,1)","rgba(70,33,103,1)","rgba(69,46,115,1)","rgba(55,103,140,1)","rgba(68,2,84,1)","rgba(69,16,91,1)","rgba(43,141,138,1)","rgba(44,139,138,1)","rgba(54,105,140,1)","rgba(44,131,140,1)","rgba(65,70,135,1)","rgba(59,93,139,1)","rgba(65,72,136,1)","rgba(37,163,133,1)","rgba(68,53,121,1)","rgba(57,98,139,1)","rgba(63,79,137,1)","rgba(42,151,136,1)","rgba(49,112,141,1)","rgba(44,134,139,1)","rgba(59,93,138,1)","rgba(44,132,140,1)","rgba(43,145,137,1)","rgba(42,120,142,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(64,73,136,1)","rgba(39,158,134,1)","rgba(69,48,116,1)","rgba(68,58,125,1)","rgba(41,153,135,1)","rgba(70,32,102,1)","rgba(55,103,140,1)","rgba(68,54,122,1)","rgba(69,47,115,1)","rgba(63,79,137,1)","rgba(45,117,142,1)","rgba(68,1,84,1)","rgba(43,124,141,1)","rgba(43,123,141,1)","rgba(44,137,139,1)","rgba(62,83,137,1)","rgba(53,106,140,1)","rgba(60,90,138,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(44,131,140,1)","rgba(69,53,120,1)","rgba(68,58,125,1)","rgba(43,140,138,1)","rgba(57,99,139,1)","rgba(61,87,138,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(62,83,137,1)","rgba(44,134,139,1)","rgba(69,45,114,1)","rgba(115,204,89,1)","rgba(70,40,109,1)","rgba(43,146,137,1)","rgba(43,147,137,1)","rgba(50,111,141,1)","rgba(49,113,141,1)","rgba(58,96,139,1)","rgba(43,119,142,1)","rgba(66,66,133,1)","rgba(43,143,138,1)","rgba(55,173,126,1)","rgba(69,52,120,1)","rgba(69,21,95,1)","rgba(45,117,142,1)","rgba(64,78,136,1)","rgba(43,125,141,1)","rgba(44,138,139,1)","rgba(59,94,139,1)","rgba(47,115,141,1)","rgba(43,140,138,1)","rgba(70,36,106,1)","rgba(43,130,140,1)","rgba(70,34,104,1)","rgba(62,86,137,1)","rgba(64,74,136,1)","rgba(43,145,137,1)","rgba(62,84,137,1)","rgba(56,101,140,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(44,118,142,1)","rgba(48,114,141,1)","rgba(42,148,136,1)","rgba(43,125,141,1)","rgba(49,113,141,1)","rgba(43,146,137,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(56,174,126,1)","rgba(43,127,141,1)","rgba(41,154,135,1)","rgba(69,14,91,1)","rgba(70,29,100,1)","rgba(65,71,135,1)","rgba(43,127,141,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(59,93,139,1)","rgba(47,115,141,1)","rgba(44,133,140,1)","rgba(69,47,115,1)","rgba(40,155,135,1)","rgba(44,134,139,1)","rgba(57,99,139,1)","rgba(42,121,142,1)","rgba(43,144,137,1)","rgba(60,92,138,1)","rgba(43,142,138,1)","rgba(55,102,140,1)","rgba(64,74,136,1)","rgba(55,103,140,1)","rgba(42,148,137,1)","rgba(51,172,127,1)","rgba(43,147,137,1)","rgba(38,160,134,1)","rgba(68,7,87,1)","rgba(59,94,139,1)","rgba(68,57,125,1)","rgba(51,110,141,1)","rgba(38,161,134,1)","rgba(64,75,136,1)","rgba(62,85,137,1)","rgba(52,108,140,1)","rgba(64,176,123,1)","rgba(41,152,136,1)","rgba(48,114,141,1)","rgba(62,85,137,1)","rgba(70,40,109,1)","rgba(68,5,86,1)","rgba(63,82,137,1)","rgba(58,97,139,1)","rgba(67,61,128,1)","rgba(43,127,141,1)","rgba(39,169,131,1)","rgba(41,152,136,1)","rgba(43,142,138,1)","rgba(54,173,126,1)","rgba(44,134,139,1)","rgba(44,132,140,1)","rgba(69,22,95,1)","rgba(43,130,140,1)","rgba(44,131,140,1)","rgba(50,111,141,1)","rgba(43,145,137,1)","rgba(43,130,140,1)","rgba(70,32,103,1)","rgba(43,119,142,1)","rgba(44,138,139,1)","rgba(43,145,137,1)","rgba(44,137,139,1)","rgba(42,121,142,1)","rgba(42,120,142,1)","rgba(59,93,139,1)","rgba(42,121,142,1)","rgba(43,126,141,1)","rgba(59,93,138,1)","rgba(42,148,137,1)","rgba(43,147,137,1)","rgba(43,124,141,1)","rgba(39,159,134,1)","rgba(42,122,142,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(43,140,138,1)","rgba(38,169,131,1)","rgba(49,112,141,1)","rgba(70,27,98,1)","rgba(43,124,141,1)","rgba(44,137,139,1)","rgba(37,162,133,1)","rgba(58,96,139,1)","rgba(42,120,142,1)","rgba(47,114,141,1)","rgba(68,2,85,1)","rgba(46,116,142,1)","rgba(54,105,140,1)","rgba(69,51,119,1)","rgba(62,176,123,1)","rgba(54,104,140,1)","rgba(61,87,138,1)","rgba(69,15,91,1)","rgba(44,137,139,1)","rgba(43,123,141,1)","rgba(49,171,128,1)","rgba(69,42,111,1)","rgba(42,120,142,1)","rgba(43,142,138,1)","rgba(35,167,132,1)","rgba(44,136,139,1)","rgba(44,118,142,1)","rgba(55,103,140,1)","rgba(42,120,142,1)","rgba(47,114,141,1)","rgba(62,83,137,1)","rgba(44,135,139,1)","rgba(70,42,111,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(49,112,141,1)","rgba(46,116,142,1)","rgba(49,112,141,1)","rgba(50,111,141,1)","rgba(55,102,140,1)","rgba(60,90,138,1)","rgba(66,63,130,1)","rgba(41,154,135,1)","rgba(64,77,136,1)","rgba(37,162,133,1)","rgba(44,134,139,1)","rgba(63,80,137,1)","rgba(50,111,141,1)","rgba(53,106,140,1)","rgba(50,110,141,1)","rgba(58,97,139,1)","rgba(55,103,140,1)","rgba(69,10,88,1)","rgba(69,22,95,1)","rgba(69,43,112,1)","rgba(52,107,140,1)","rgba(41,153,135,1)","rgba(44,118,142,1)","rgba(85,186,112,1)","rgba(41,153,136,1)","rgba(55,103,140,1)","rgba(40,157,135,1)","rgba(43,124,141,1)","rgba(43,139,138,1)","rgba(41,152,136,1)","rgba(42,151,136,1)","rgba(43,119,142,1)","rgba(60,90,138,1)","rgba(50,111,141,1)","rgba(45,117,142,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(34,168,132,1)","rgba(44,135,139,1)","rgba(42,120,142,1)","rgba(67,62,129,1)","rgba(43,124,141,1)","rgba(42,151,136,1)","rgba(37,162,133,1)","rgba(43,125,141,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(43,141,138,1)","rgba(43,143,138,1)","rgba(69,52,120,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(66,64,131,1)","rgba(74,180,118,1)","rgba(54,105,140,1)","rgba(67,61,129,1)","rgba(53,107,140,1)","rgba(43,140,138,1)","rgba(35,167,132,1)","rgba(43,126,141,1)","rgba(48,171,129,1)","rgba(53,107,140,1)","rgba(68,1,84,1)","rgba(41,154,135,1)","rgba(56,102,140,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(68,58,126,1)","rgba(56,101,140,1)","rgba(44,135,139,1)","rgba(42,121,142,1)","rgba(68,55,123,1)","rgba(56,100,139,1)","rgba(63,81,137,1)","rgba(43,126,141,1)","rgba(41,154,135,1)","rgba(43,125,141,1)","rgba(49,172,128,1)","rgba(68,57,124,1)","rgba(65,69,135,1)","rgba(67,63,130,1)","rgba(64,75,136,1)","rgba(42,122,142,1)","rgba(35,167,132,1)","rgba(137,211,78,1)","rgba(58,97,139,1)","rgba(50,111,141,1)","rgba(43,127,141,1)","rgba(64,76,136,1)","rgba(59,95,139,1)","rgba(43,145,137,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(43,123,141,1)","rgba(44,139,138,1)","rgba(42,121,142,1)","rgba(66,65,132,1)","rgba(58,96,139,1)","rgba(58,96,139,1)","rgba(43,124,141,1)","rgba(50,110,141,1)","rgba(68,3,85,1)","rgba(68,55,123,1)","rgba(48,114,141,1)","rgba(56,101,140,1)","rgba(41,153,136,1)","rgba(60,91,138,1)","rgba(36,164,133,1)","rgba(43,141,138,1)","rgba(57,99,139,1)","rgba(44,131,140,1)","rgba(51,110,141,1)","rgba(43,125,141,1)","rgba(65,67,134,1)","rgba(42,120,142,1)","rgba(62,83,137,1)","rgba(56,100,139,1)","rgba(40,156,135,1)","rgba(44,132,140,1)","rgba(54,104,140,1)","rgba(43,129,140,1)","rgba(44,139,138,1)","rgba(52,108,140,1)","rgba(37,163,133,1)","rgba(65,72,135,1)","rgba(66,66,133,1)","rgba(63,80,137,1)","rgba(43,144,137,1)","rgba(43,126,141,1)","rgba(65,70,135,1)","rgba(52,108,140,1)","rgba(69,47,115,1)","rgba(64,78,136,1)","rgba(41,153,136,1)","rgba(68,55,122,1)","rgba(55,102,140,1)","rgba(43,125,141,1)","rgba(44,135,139,1)","rgba(54,105,140,1)","rgba(39,160,134,1)","rgba(57,99,139,1)","rgba(69,18,93,1)","rgba(44,136,139,1)","rgba(62,86,138,1)","rgba(40,157,135,1)","rgba(56,100,139,1)","rgba(56,100,139,1)","rgba(53,106,140,1)","rgba(60,92,138,1)","rgba(69,47,115,1)","rgba(53,106,140,1)","rgba(70,30,101,1)","rgba(64,73,136,1)","rgba(57,98,139,1)","rgba(53,106,140,1)","rgba(64,74,136,1)","rgba(40,156,135,1)","rgba(69,9,88,1)","rgba(69,9,88,1)","rgba(70,26,98,1)","rgba(44,137,139,1)","rgba(62,86,137,1)","rgba(42,121,142,1)","rgba(44,138,139,1)","rgba(49,112,141,1)","rgba(59,95,139,1)","rgba(69,24,96,1)","rgba(47,114,141,1)","rgba(69,50,118,1)","rgba(55,102,140,1)","rgba(57,99,139,1)","rgba(44,118,142,1)","rgba(43,130,140,1)","rgba(61,87,138,1)","rgba(49,111,141,1)","rgba(44,138,139,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(46,115,141,1)","rgba(63,80,137,1)","rgba(68,1,84,1)","rgba(43,119,142,1)","rgba(62,83,137,1)","rgba(69,50,118,1)","rgba(58,97,139,1)","rgba(36,165,133,1)","rgba(45,116,142,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(70,36,106,1)","rgba(69,8,87,1)","rgba(54,105,140,1)","rgba(54,105,140,1)","rgba(43,141,138,1)","rgba(35,166,133,1)","rgba(57,99,139,1)","rgba(69,51,119,1)","rgba(44,131,140,1)","rgba(49,112,141,1)","rgba(56,101,140,1)","rgba(55,103,140,1)","rgba(59,94,139,1)","rgba(42,149,136,1)","rgba(42,149,136,1)","rgba(44,134,139,1)","rgba(41,151,136,1)","rgba(47,114,141,1)","rgba(49,113,141,1)","rgba(60,90,138,1)","rgba(43,143,138,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(56,100,139,1)","rgba(43,129,140,1)","rgba(50,111,141,1)","rgba(67,60,128,1)","rgba(43,123,141,1)","rgba(69,49,117,1)","rgba(62,86,138,1)","rgba(63,176,123,1)","rgba(37,164,133,1)","rgba(43,141,138,1)","rgba(70,28,99,1)","rgba(44,118,142,1)","rgba(43,123,141,1)","rgba(65,71,135,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(59,94,139,1)","rgba(61,87,138,1)","rgba(56,102,140,1)","rgba(42,150,136,1)","rgba(64,77,136,1)","rgba(60,91,138,1)","rgba(57,99,139,1)","rgba(70,31,101,1)","rgba(46,116,142,1)","rgba(51,109,141,1)","rgba(53,107,140,1)","rgba(45,116,142,1)","rgba(67,61,129,1)","rgba(43,127,141,1)","rgba(43,127,141,1)","rgba(45,171,129,1)","rgba(67,59,127,1)","rgba(43,127,141,1)","rgba(63,176,123,1)","rgba(65,70,135,1)","rgba(39,159,134,1)","rgba(42,151,136,1)","rgba(48,114,141,1)","rgba(69,50,118,1)","rgba(64,77,136,1)","rgba(102,195,100,1)","rgba(66,63,130,1)","rgba(70,29,100,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(43,130,140,1)","rgba(43,129,140,1)","rgba(43,141,138,1)","rgba(69,45,113,1)","rgba(44,135,139,1)","rgba(41,154,135,1)","rgba(44,131,140,1)","rgba(43,129,140,1)","rgba(50,111,141,1)","rgba(98,193,103,1)","rgba(40,155,135,1)","rgba(93,190,107,1)","rgba(52,107,140,1)","rgba(53,106,140,1)","rgba(40,155,135,1)","rgba(43,146,137,1)","rgba(70,29,100,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(70,26,98,1)","rgba(58,97,139,1)","rgba(71,179,120,1)","rgba(69,8,87,1)","rgba(43,142,138,1)","rgba(44,138,139,1)","rgba(64,76,136,1)","rgba(44,134,139,1)","rgba(48,114,141,1)","rgba(42,148,137,1)","rgba(43,124,141,1)","rgba(43,139,138,1)","rgba(69,12,89,1)","rgba(43,128,141,1)","rgba(68,58,126,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(40,157,134,1)","rgba(57,98,139,1)","rgba(43,140,138,1)","rgba(68,2,84,1)","rgba(52,172,127,1)","rgba(113,202,91,1)","rgba(43,128,141,1)","rgba(42,120,142,1)","rgba(42,122,142,1)","rgba(42,149,136,1)","rgba(51,109,141,1)","rgba(43,144,137,1)","rgba(57,99,139,1)","rgba(51,109,141,1)","rgba(43,119,142,1)","rgba(51,172,127,1)","rgba(59,93,138,1)","rgba(50,110,141,1)","rgba(61,88,138,1)","rgba(150,214,75,1)","rgba(69,13,90,1)","rgba(43,123,141,1)","rgba(52,107,140,1)","rgba(50,172,128,1)","rgba(44,134,139,1)","rgba(57,99,139,1)","rgba(40,155,135,1)","rgba(41,154,135,1)","rgba(42,120,142,1)","rgba(64,76,136,1)","rgba(49,112,141,1)","rgba(43,127,141,1)","rgba(44,138,139,1)","rgba(53,107,140,1)","rgba(43,143,138,1)","rgba(67,62,129,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(42,120,142,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(56,102,140,1)","rgba(54,104,140,1)","rgba(69,45,113,1)","rgba(70,36,105,1)","rgba(43,147,137,1)","rgba(66,64,131,1)","rgba(60,92,138,1)","rgba(68,4,85,1)","rgba(69,44,113,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(43,143,138,1)","rgba(52,108,140,1)","rgba(37,162,133,1)","rgba(42,150,136,1)","rgba(43,147,137,1)","rgba(44,138,139,1)","rgba(68,3,85,1)","rgba(69,44,112,1)","rgba(43,123,141,1)","rgba(43,127,141,1)","rgba(44,135,139,1)","rgba(35,168,132,1)","rgba(56,101,140,1)","rgba(44,139,138,1)","rgba(52,108,140,1)","rgba(52,107,140,1)","rgba(50,110,141,1)","rgba(56,101,140,1)","rgba(44,137,139,1)","rgba(69,17,92,1)","rgba(44,133,140,1)","rgba(65,72,136,1)","rgba(58,95,139,1)","rgba(65,70,135,1)","rgba(50,111,141,1)","rgba(65,68,135,1)","rgba(44,133,140,1)","rgba(54,104,140,1)","rgba(70,35,105,1)","rgba(57,98,139,1)","rgba(43,130,140,1)","rgba(35,167,132,1)","rgba(50,110,141,1)","rgba(58,174,125,1)","rgba(60,90,138,1)","rgba(43,140,138,1)","rgba(44,135,139,1)","rgba(43,126,141,1)","rgba(43,128,140,1)","rgba(60,91,138,1)","rgba(39,159,134,1)","rgba(56,101,140,1)","rgba(64,78,136,1)","rgba(59,95,139,1)","rgba(39,160,134,1)","rgba(52,108,140,1)","rgba(44,138,139,1)","rgba(39,158,134,1)","rgba(50,110,141,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(47,115,141,1)","rgba(69,18,92,1)","rgba(43,124,141,1)","rgba(58,97,139,1)","rgba(48,113,141,1)","rgba(43,141,138,1)","rgba(60,90,138,1)","rgba(53,106,140,1)","rgba(38,160,134,1)","rgba(64,78,136,1)","rgba(42,122,142,1)","rgba(70,36,106,1)","rgba(42,120,142,1)","rgba(65,71,135,1)","rgba(55,103,140,1)","rgba(50,111,141,1)","rgba(70,179,120,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(53,106,140,1)","rgba(64,73,136,1)","rgba(43,144,137,1)","rgba(60,93,138,1)","rgba(46,116,142,1)","rgba(69,43,112,1)","rgba(53,106,140,1)","rgba(60,91,138,1)","rgba(69,51,119,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(70,39,108,1)","rgba(41,153,136,1)","rgba(68,53,120,1)","rgba(82,184,114,1)","rgba(43,145,137,1)","rgba(57,98,139,1)","rgba(57,99,139,1)","rgba(109,200,94,1)","rgba(44,135,139,1)","rgba(43,142,138,1)","rgba(68,5,86,1)","rgba(38,161,134,1)","rgba(44,138,139,1)","rgba(67,62,130,1)","rgba(42,120,142,1)","rgba(42,121,142,1)","rgba(43,123,142,1)","rgba(44,132,140,1)","rgba(70,29,100,1)","rgba(40,157,135,1)","rgba(58,96,139,1)","rgba(43,142,138,1)","rgba(39,159,134,1)","rgba(42,122,142,1)","rgba(57,99,139,1)","rgba(70,31,102,1)","rgba(60,91,138,1)","rgba(58,96,139,1)","rgba(61,88,138,1)","rgba(51,172,127,1)","rgba(62,84,137,1)","rgba(67,59,127,1)","rgba(57,99,139,1)","rgba(42,122,142,1)","rgba(42,150,136,1)","rgba(68,55,123,1)","rgba(109,200,94,1)","rgba(51,110,141,1)","rgba(53,106,140,1)","rgba(60,90,138,1)","rgba(69,15,91,1)","rgba(44,135,139,1)","rgba(56,174,126,1)","rgba(43,123,141,1)","rgba(44,118,142,1)","rgba(44,132,140,1)","rgba(62,83,137,1)","rgba(49,112,141,1)","rgba(43,125,141,1)","rgba(64,76,136,1)","rgba(64,73,136,1)","rgba(66,64,131,1)","rgba(53,107,140,1)","rgba(44,118,142,1)","rgba(43,126,141,1)","rgba(36,164,133,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(69,10,88,1)","rgba(49,112,141,1)","rgba(52,107,140,1)","rgba(69,23,96,1)","rgba(42,170,130,1)","rgba(70,27,99,1)","rgba(44,139,138,1)","rgba(51,109,141,1)","rgba(44,118,142,1)","rgba(46,116,142,1)","rgba(62,85,137,1)","rgba(43,126,141,1)","rgba(43,126,141,1)","rgba(43,129,140,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(47,115,141,1)","rgba(56,101,140,1)","rgba(118,206,85,1)","rgba(59,93,139,1)","rgba(50,111,141,1)","rgba(43,126,141,1)","rgba(35,167,132,1)","rgba(43,145,137,1)","rgba(42,149,136,1)","rgba(42,121,142,1)","rgba(40,155,135,1)","rgba(44,137,139,1)","rgba(62,83,137,1)","rgba(66,66,133,1)","rgba(59,95,139,1)","rgba(46,116,141,1)","rgba(43,119,142,1)","rgba(64,77,136,1)","rgba(60,92,138,1)","rgba(44,131,140,1)","rgba(52,107,140,1)","rgba(69,11,89,1)","rgba(63,80,137,1)","rgba(43,141,138,1)","rgba(70,34,104,1)","rgba(68,178,121,1)","rgba(88,187,110,1)","rgba(57,98,139,1)","rgba(54,173,126,1)","rgba(69,42,111,1)","rgba(70,34,104,1)","rgba(43,128,141,1)","rgba(57,99,139,1)","rgba(69,50,118,1)","rgba(68,55,122,1)","rgba(44,138,139,1)","rgba(69,22,95,1)","rgba(43,124,141,1)","rgba(41,152,136,1)","rgba(70,26,98,1)","rgba(62,86,137,1)","rgba(51,110,141,1)","rgba(42,149,136,1)","rgba(51,109,141,1)","rgba(63,80,137,1)","rgba(51,110,141,1)","rgba(44,132,140,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(42,149,136,1)","rgba(43,129,140,1)","rgba(43,124,141,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(110,201,93,1)","rgba(44,137,139,1)","rgba(60,90,138,1)","rgba(43,119,142,1)","rgba(43,143,138,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(53,106,140,1)","rgba(63,78,136,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(43,128,141,1)","rgba(41,152,136,1)","rgba(54,105,140,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(63,78,136,1)","rgba(43,143,138,1)","rgba(70,39,108,1)","rgba(36,165,133,1)","rgba(47,114,141,1)","rgba(43,128,141,1)","rgba(63,82,137,1)","rgba(66,65,132,1)","rgba(55,103,140,1)","rgba(70,37,106,1)","rgba(43,125,141,1)","rgba(63,80,137,1)","rgba(66,66,133,1)","rgba(44,132,140,1)","rgba(43,146,137,1)","rgba(61,89,138,1)","rgba(68,57,124,1)","rgba(68,57,125,1)","rgba(43,141,138,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(44,118,142,1)","rgba(54,104,140,1)","rgba(64,76,136,1)","rgba(69,48,116,1)","rgba(68,4,86,1)","rgba(70,28,100,1)","rgba(57,98,139,1)","rgba(43,128,141,1)","rgba(44,139,138,1)","rgba(46,116,141,1)","rgba(70,32,102,1)","rgba(44,133,140,1)","rgba(69,16,91,1)","rgba(43,123,141,1)","rgba(49,112,141,1)","rgba(43,143,138,1)","rgba(50,111,141,1)","rgba(53,106,140,1)","rgba(47,115,141,1)","rgba(45,117,142,1)","rgba(44,136,139,1)","rgba(56,100,140,1)","rgba(58,97,139,1)","rgba(94,190,106,1)","rgba(44,135,139,1)","rgba(43,140,138,1)","rgba(46,116,142,1)","rgba(69,51,119,1)","rgba(40,157,135,1)","rgba(43,141,138,1)","rgba(42,121,142,1)","rgba(43,124,141,1)","rgba(43,128,140,1)","rgba(44,136,139,1)","rgba(43,141,138,1)","rgba(70,40,109,1)","rgba(43,119,142,1)","rgba(43,126,141,1)","rgba(63,78,136,1)","rgba(44,134,139,1)","rgba(44,133,140,1)","rgba(49,112,141,1)","rgba(44,118,142,1)","rgba(59,94,139,1)","rgba(51,110,141,1)","rgba(38,161,134,1)","rgba(70,31,101,1)","rgba(70,39,108,1)","rgba(52,108,140,1)","rgba(68,2,84,1)","rgba(53,106,140,1)","rgba(70,37,106,1)","rgba(69,25,97,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(44,135,139,1)","rgba(66,63,130,1)","rgba(43,130,140,1)","rgba(43,130,140,1)","rgba(42,147,137,1)","rgba(73,180,119,1)","rgba(67,62,129,1)","rgba(43,125,141,1)","rgba(42,150,136,1)","rgba(43,126,141,1)","rgba(60,91,138,1)","rgba(48,114,141,1)","rgba(68,54,121,1)","rgba(68,8,87,1)","rgba(76,182,117,1)","rgba(43,141,138,1)","rgba(43,141,138,1)","rgba(63,79,137,1)","rgba(43,119,142,1)","rgba(44,135,139,1)","rgba(44,134,139,1)","rgba(69,8,87,1)","rgba(52,108,141,1)","rgba(64,76,136,1)","rgba(41,154,135,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(43,130,140,1)","rgba(43,129,140,1)","rgba(69,17,92,1)","rgba(43,125,141,1)","rgba(43,144,138,1)","rgba(59,93,139,1)","rgba(57,99,139,1)","rgba(63,78,136,1)","rgba(69,45,113,1)","rgba(44,135,139,1)","rgba(38,160,134,1)","rgba(70,29,100,1)","rgba(50,111,141,1)","rgba(48,114,141,1)","rgba(43,143,138,1)","rgba(69,25,97,1)","rgba(67,59,126,1)","rgba(64,73,136,1)","rgba(68,54,122,1)","rgba(55,103,140,1)","rgba(44,133,140,1)","rgba(67,177,121,1)","rgba(43,129,140,1)","rgba(44,135,139,1)","rgba(49,112,141,1)","rgba(70,30,101,1)","rgba(41,153,136,1)","rgba(46,116,142,1)","rgba(43,141,138,1)","rgba(50,111,141,1)","rgba(68,7,87,1)","rgba(40,169,131,1)","rgba(52,107,140,1)","rgba(70,32,103,1)","rgba(43,126,141,1)","rgba(51,109,141,1)","rgba(47,115,141,1)","rgba(44,132,140,1)","rgba(43,129,140,1)","rgba(43,126,141,1)","rgba(64,74,136,1)","rgba(65,70,135,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(46,116,142,1)","rgba(40,157,135,1)","rgba(48,113,141,1)","rgba(66,177,122,1)","rgba(57,98,139,1)","rgba(64,76,136,1)","rgba(53,106,140,1)","rgba(46,116,141,1)","rgba(43,129,140,1)","rgba(47,115,141,1)","rgba(55,102,140,1)","rgba(43,131,140,1)","rgba(46,116,141,1)","rgba(42,122,142,1)","rgba(39,160,134,1)","rgba(44,134,139,1)","rgba(34,168,132,1)","rgba(61,89,138,1)","rgba(38,161,134,1)","rgba(62,86,137,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(69,15,91,1)","rgba(61,90,138,1)","rgba(49,172,128,1)","rgba(65,68,135,1)","rgba(69,47,115,1)","rgba(44,139,138,1)","rgba(63,82,137,1)","rgba(67,59,127,1)","rgba(38,160,134,1)","rgba(57,98,139,1)","rgba(46,115,141,1)","rgba(43,124,141,1)","rgba(54,104,140,1)","rgba(56,101,140,1)","rgba(42,122,142,1)","rgba(68,4,86,1)","rgba(68,6,87,1)","rgba(63,82,137,1)","rgba(51,109,141,1)","rgba(66,65,132,1)","rgba(40,156,135,1)","rgba(47,114,141,1)","rgba(68,3,85,1)","rgba(68,4,85,1)","rgba(44,133,140,1)","rgba(65,67,134,1)","rgba(67,58,126,1)","rgba(66,64,131,1)","rgba(54,104,140,1)","rgba(69,11,89,1)","rgba(69,23,96,1)","rgba(66,64,132,1)","rgba(43,130,140,1)","rgba(59,93,138,1)","rgba(56,101,140,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(65,68,135,1)","rgba(50,111,141,1)","rgba(66,65,132,1)","rgba(44,131,140,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(61,89,138,1)","rgba(44,136,139,1)","rgba(52,107,140,1)","rgba(69,22,95,1)","rgba(50,111,141,1)","rgba(44,117,142,1)","rgba(43,122,142,1)","rgba(43,143,138,1)","rgba(75,181,117,1)","rgba(70,26,98,1)","rgba(43,124,141,1)","rgba(69,22,95,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(53,107,140,1)","rgba(43,142,138,1)","rgba(51,109,141,1)","rgba(42,121,142,1)","rgba(42,149,136,1)","rgba(42,122,142,1)","rgba(68,53,120,1)","rgba(43,126,141,1)","rgba(62,83,137,1)","rgba(42,122,142,1)","rgba(44,137,139,1)","rgba(44,134,139,1)","rgba(69,178,120,1)","rgba(43,126,141,1)","rgba(55,104,140,1)","rgba(44,131,140,1)","rgba(42,120,142,1)","rgba(69,48,116,1)","rgba(58,97,139,1)","rgba(68,56,123,1)","rgba(43,123,141,1)","rgba(43,145,137,1)","rgba(68,178,121,1)","rgba(44,131,140,1)","rgba(69,9,88,1)","rgba(63,81,137,1)","rgba(43,129,140,1)","rgba(68,58,125,1)","rgba(60,90,138,1)","rgba(60,92,138,1)","rgba(43,124,141,1)","rgba(39,159,134,1)","rgba(66,63,130,1)","rgba(63,81,137,1)","rgba(43,128,141,1)","rgba(69,22,95,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(43,127,141,1)","rgba(69,15,91,1)","rgba(63,82,137,1)","rgba(56,101,140,1)","rgba(42,120,142,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(38,161,134,1)","rgba(58,96,139,1)","rgba(70,32,102,1)","rgba(62,85,137,1)","rgba(43,128,141,1)","rgba(58,96,139,1)","rgba(44,136,139,1)","rgba(69,43,111,1)","rgba(45,117,142,1)","rgba(69,44,113,1)","rgba(42,120,142,1)","rgba(42,120,142,1)","rgba(61,88,138,1)","rgba(50,111,141,1)","rgba(58,96,139,1)","rgba(70,42,111,1)","rgba(69,9,88,1)","rgba(69,46,115,1)","rgba(44,137,139,1)","rgba(48,113,141,1)","rgba(54,105,140,1)","rgba(46,116,141,1)","rgba(37,163,133,1)","rgba(52,108,141,1)","rgba(68,4,86,1)","rgba(69,9,88,1)","rgba(44,138,139,1)","rgba(41,153,135,1)","rgba(48,114,141,1)","rgba(45,117,142,1)","rgba(44,118,142,1)","rgba(59,94,139,1)","rgba(59,94,139,1)","rgba(69,13,90,1)","rgba(51,109,141,1)","rgba(57,99,139,1)","rgba(61,87,138,1)","rgba(44,137,139,1)","rgba(53,106,140,1)","rgba(67,59,126,1)","rgba(44,131,140,1)","rgba(42,121,142,1)","rgba(69,50,118,1)","rgba(68,2,84,1)","rgba(54,105,140,1)","rgba(64,74,136,1)","rgba(43,127,141,1)","rgba(66,63,130,1)","rgba(70,39,108,1)","rgba(51,110,141,1)","rgba(44,137,139,1)","rgba(43,127,141,1)","rgba(44,132,140,1)","rgba(68,55,122,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(58,97,139,1)","rgba(36,164,133,1)","rgba(43,119,142,1)","rgba(57,98,139,1)","rgba(43,140,138,1)","rgba(66,63,130,1)","rgba(42,151,136,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(53,106,140,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(63,82,137,1)","rgba(64,77,136,1)","rgba(50,111,141,1)","rgba(44,138,139,1)","rgba(49,113,141,1)","rgba(41,154,135,1)","rgba(46,115,141,1)","rgba(40,157,135,1)","rgba(68,8,87,1)","rgba(64,176,123,1)","rgba(43,127,141,1)","rgba(43,124,141,1)","rgba(61,89,138,1)","rgba(44,135,139,1)","rgba(50,110,141,1)","rgba(51,109,141,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(46,116,142,1)","rgba(56,100,139,1)","rgba(42,121,142,1)","rgba(68,57,124,1)","rgba(42,148,137,1)","rgba(43,124,141,1)","rgba(69,14,90,1)","rgba(44,134,139,1)","rgba(61,86,138,1)","rgba(48,114,141,1)","rgba(113,203,90,1)","rgba(41,152,136,1)","rgba(70,31,102,1)","rgba(69,45,114,1)","rgba(69,21,94,1)","rgba(39,158,134,1)","rgba(43,142,138,1)","rgba(44,133,140,1)","rgba(43,124,141,1)","rgba(42,120,142,1)","rgba(59,94,139,1)","rgba(55,103,140,1)","rgba(60,90,138,1)","rgba(44,135,139,1)","rgba(49,112,141,1)","rgba(66,63,130,1)","rgba(43,125,141,1)","rgba(43,125,141,1)","rgba(44,137,139,1)","rgba(58,96,139,1)","rgba(41,154,135,1)","rgba(69,46,114,1)","rgba(61,87,138,1)","rgba(51,109,141,1)","rgba(43,141,138,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(39,158,134,1)","rgba(43,128,140,1)","rgba(69,14,90,1)","rgba(62,84,137,1)","rgba(57,99,139,1)","rgba(43,129,140,1)","rgba(68,6,87,1)","rgba(59,95,139,1)","rgba(44,137,139,1)","rgba(57,98,139,1)","rgba(48,113,141,1)","rgba(43,130,140,1)","rgba(69,44,113,1)","rgba(44,139,138,1)","rgba(55,103,140,1)","rgba(40,156,135,1)","rgba(44,135,139,1)","rgba(43,127,141,1)","rgba(60,91,138,1)","rgba(55,103,140,1)","rgba(44,137,139,1)","rgba(66,66,133,1)","rgba(48,113,141,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(45,117,142,1)","rgba(43,144,137,1)","rgba(42,120,142,1)","rgba(69,46,114,1)","rgba(57,99,139,1)","rgba(70,28,99,1)","rgba(43,127,141,1)","rgba(69,18,93,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(42,120,142,1)","rgba(69,18,93,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(44,134,139,1)","rgba(70,28,99,1)","rgba(44,131,140,1)","rgba(55,103,140,1)","rgba(43,145,137,1)","rgba(59,95,139,1)","rgba(46,116,141,1)","rgba(50,110,141,1)","rgba(63,82,137,1)","rgba(63,81,137,1)","rgba(61,89,138,1)","rgba(60,91,138,1)","rgba(55,103,140,1)","rgba(35,166,132,1)","rgba(68,4,86,1)","rgba(50,110,141,1)","rgba(43,142,138,1)","rgba(52,108,141,1)","rgba(68,56,123,1)","rgba(70,29,100,1)","rgba(69,19,93,1)","rgba(43,143,138,1)","rgba(41,154,135,1)","rgba(67,59,126,1)","rgba(68,55,122,1)","rgba(62,83,137,1)","rgba(43,143,138,1)","rgba(70,40,109,1)","rgba(54,104,140,1)","rgba(44,138,139,1)","rgba(47,114,141,1)","rgba(64,77,136,1)","rgba(68,58,126,1)","rgba(69,24,97,1)","rgba(56,100,139,1)","rgba(70,37,106,1)","rgba(47,114,141,1)","rgba(66,65,132,1)","rgba(48,113,141,1)","rgba(69,12,89,1)","rgba(57,99,139,1)","rgba(43,125,141,1)","rgba(43,140,138,1)","rgba(55,103,140,1)","rgba(46,116,142,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(58,97,139,1)","rgba(59,94,139,1)","rgba(42,122,142,1)","rgba(66,66,133,1)","rgba(52,172,127,1)","rgba(43,144,137,1)","rgba(45,117,142,1)","rgba(65,69,135,1)","rgba(61,89,138,1)","rgba(68,56,124,1)","rgba(41,153,135,1)","rgba(42,121,142,1)","rgba(44,134,139,1)","rgba(43,143,138,1)","rgba(48,113,141,1)","rgba(52,108,140,1)","rgba(44,137,139,1)","rgba(44,133,140,1)","rgba(65,69,135,1)","rgba(64,73,136,1)","rgba(43,124,141,1)","rgba(69,14,90,1)","rgba(58,97,139,1)","rgba(42,122,142,1)","rgba(43,123,141,1)","rgba(61,87,138,1)","rgba(65,70,135,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(59,94,139,1)","rgba(42,122,142,1)","rgba(69,44,112,1)","rgba(42,151,136,1)","rgba(44,135,139,1)","rgba(70,33,104,1)","rgba(43,119,142,1)","rgba(69,23,96,1)","rgba(43,143,138,1)","rgba(43,141,138,1)","rgba(41,152,136,1)","rgba(43,143,138,1)","rgba(46,116,142,1)","rgba(59,95,139,1)","rgba(69,48,116,1)","rgba(43,128,140,1)","rgba(64,73,136,1)","rgba(44,135,139,1)","rgba(39,158,134,1)","rgba(60,175,124,1)","rgba(41,153,136,1)","rgba(49,113,141,1)","rgba(43,123,142,1)","rgba(41,154,135,1)","rgba(37,169,131,1)","rgba(43,147,137,1)","rgba(56,102,140,1)","rgba(42,121,142,1)","rgba(44,133,140,1)","rgba(64,73,136,1)","rgba(42,148,136,1)","rgba(41,153,136,1)","rgba(52,107,140,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(43,145,137,1)","rgba(68,54,122,1)","rgba(44,133,140,1)","rgba(54,105,140,1)","rgba(67,60,127,1)","rgba(43,127,141,1)","rgba(51,109,141,1)","rgba(62,85,137,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(43,126,141,1)","rgba(44,138,139,1)","rgba(51,110,141,1)","rgba(61,89,138,1)","rgba(62,83,137,1)","rgba(52,107,140,1)","rgba(55,103,140,1)","rgba(48,113,141,1)","rgba(63,80,137,1)","rgba(43,130,140,1)","rgba(67,59,127,1)","rgba(43,130,140,1)","rgba(69,18,92,1)","rgba(97,192,104,1)","rgba(52,108,141,1)","rgba(78,182,116,1)","rgba(41,153,135,1)","rgba(43,124,141,1)","rgba(58,97,139,1)","rgba(42,122,142,1)","rgba(60,90,138,1)","rgba(64,72,136,1)","rgba(57,99,139,1)","rgba(63,82,137,1)","rgba(54,105,140,1)","rgba(61,88,138,1)","rgba(57,98,139,1)","rgba(36,166,133,1)","rgba(54,105,140,1)","rgba(43,147,137,1)","rgba(69,46,114,1)","rgba(44,137,139,1)","rgba(42,120,142,1)","rgba(55,103,140,1)","rgba(43,124,141,1)","rgba(66,65,132,1)","rgba(62,85,137,1)","rgba(69,42,111,1)","rgba(64,78,136,1)","rgba(43,141,138,1)","rgba(115,204,88,1)","rgba(45,117,142,1)","rgba(70,29,100,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(70,30,101,1)","rgba(68,3,85,1)","rgba(43,146,137,1)","rgba(54,104,140,1)","rgba(37,169,131,1)","rgba(58,97,139,1)","rgba(69,48,116,1)","rgba(54,105,140,1)","rgba(41,152,136,1)","rgba(68,57,124,1)","rgba(44,131,140,1)","rgba(69,18,93,1)","rgba(43,126,141,1)","rgba(37,164,133,1)","rgba(49,112,141,1)","rgba(42,149,136,1)","rgba(41,152,136,1)","rgba(69,25,97,1)","rgba(54,104,140,1)","rgba(54,105,140,1)","rgba(63,82,137,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(40,155,135,1)","rgba(70,179,120,1)","rgba(68,56,123,1)","rgba(56,100,139,1)","rgba(64,73,136,1)","rgba(43,143,138,1)","rgba(42,121,142,1)","rgba(57,98,139,1)","rgba(69,19,93,1)","rgba(42,150,136,1)","rgba(38,160,134,1)","rgba(69,20,94,1)","rgba(69,44,113,1)","rgba(57,100,139,1)","rgba(70,36,106,1)","rgba(70,35,105,1)","rgba(49,113,141,1)","rgba(42,148,137,1)","rgba(39,159,134,1)","rgba(43,126,141,1)","rgba(68,56,124,1)","rgba(45,117,142,1)","rgba(67,59,126,1)","rgba(43,170,130,1)","rgba(64,77,136,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(69,16,91,1)","rgba(44,138,139,1)","rgba(61,87,138,1)","rgba(61,88,138,1)","rgba(43,119,142,1)","rgba(43,145,137,1)","rgba(44,139,138,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(46,115,141,1)","rgba(39,159,134,1)","rgba(42,150,136,1)","rgba(43,147,137,1)","rgba(114,203,89,1)","rgba(44,133,140,1)","rgba(65,67,134,1)","rgba(56,101,140,1)","rgba(43,142,138,1)","rgba(37,162,133,1)","rgba(38,160,134,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(42,150,136,1)","rgba(66,64,131,1)","rgba(70,28,100,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(69,17,92,1)","rgba(73,180,119,1)","rgba(35,166,133,1)","rgba(68,55,122,1)","rgba(69,11,89,1)","rgba(61,87,138,1)","rgba(43,139,138,1)","rgba(43,127,141,1)","rgba(43,129,140,1)","rgba(56,100,140,1)","rgba(69,20,94,1)","rgba(58,96,139,1)","rgba(43,145,137,1)","rgba(34,168,132,1)","rgba(43,146,137,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(34,168,132,1)","rgba(44,133,140,1)","rgba(67,62,129,1)","rgba(64,78,136,1)","rgba(60,90,138,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(69,10,89,1)","rgba(65,69,135,1)","rgba(63,79,137,1)","rgba(44,136,139,1)","rgba(69,20,94,1)","rgba(54,105,140,1)","rgba(63,81,137,1)","rgba(39,158,134,1)","rgba(70,34,104,1)","rgba(38,161,134,1)","rgba(68,1,84,1)","rgba(44,131,140,1)","rgba(70,38,108,1)","rgba(42,150,136,1)","rgba(59,94,139,1)","rgba(43,130,140,1)","rgba(69,20,94,1)","rgba(44,131,140,1)","rgba(49,111,141,1)","rgba(36,164,133,1)","rgba(62,85,137,1)","rgba(70,37,106,1)","rgba(44,134,139,1)","rgba(38,161,134,1)","rgba(43,126,141,1)","rgba(42,120,142,1)","rgba(69,23,96,1)","rgba(43,145,137,1)","rgba(44,138,139,1)","rgba(43,125,141,1)","rgba(43,140,138,1)","rgba(201,222,61,1)","rgba(38,161,134,1)","rgba(46,116,142,1)","rgba(43,146,137,1)","rgba(42,122,142,1)","rgba(61,89,138,1)","rgba(59,93,139,1)","rgba(44,132,140,1)","rgba(42,148,137,1)","rgba(68,54,121,1)","rgba(54,173,127,1)","rgba(44,131,140,1)","rgba(42,121,142,1)","rgba(43,127,141,1)","rgba(70,35,105,1)","rgba(43,143,138,1)","rgba(69,10,88,1)","rgba(54,105,140,1)","rgba(69,25,97,1)","rgba(41,154,135,1)","rgba(157,215,73,1)","rgba(70,34,104,1)","rgba(62,86,138,1)","rgba(61,88,138,1)","rgba(64,75,136,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(43,126,141,1)","rgba(38,169,131,1)","rgba(70,40,109,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(52,109,141,1)","rgba(67,59,127,1)","rgba(64,76,136,1)","rgba(58,96,139,1)","rgba(43,141,138,1)","rgba(43,141,138,1)","rgba(67,62,129,1)","rgba(43,129,140,1)","rgba(50,110,141,1)","rgba(44,137,139,1)","rgba(58,96,139,1)","rgba(95,191,105,1)","rgba(40,155,135,1)","rgba(69,15,91,1)","rgba(55,103,140,1)","rgba(65,69,135,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(68,54,122,1)","rgba(43,140,138,1)","rgba(52,107,140,1)","rgba(69,25,97,1)","rgba(70,36,106,1)","rgba(160,215,73,1)","rgba(113,203,90,1)","rgba(63,80,137,1)","rgba(58,97,139,1)","rgba(48,114,141,1)","rgba(68,55,123,1)","rgba(38,161,134,1)","rgba(46,116,142,1)","rgba(50,111,141,1)","rgba(41,153,135,1)","rgba(52,107,140,1)","rgba(44,139,138,1)","rgba(41,152,136,1)","rgba(49,112,141,1)","rgba(43,128,140,1)","rgba(65,72,136,1)","rgba(52,108,140,1)","rgba(62,83,137,1)","rgba(44,139,138,1)","rgba(44,118,142,1)","rgba(43,146,137,1)","rgba(53,107,140,1)","rgba(44,135,139,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(66,66,133,1)","rgba(43,146,137,1)","rgba(57,98,139,1)","rgba(41,153,135,1)","rgba(65,70,135,1)","rgba(43,130,140,1)","rgba(70,30,101,1)","rgba(70,41,110,1)","rgba(44,131,140,1)","rgba(44,138,139,1)","rgba(44,118,142,1)","rgba(69,24,96,1)","rgba(49,112,141,1)","rgba(42,147,137,1)","rgba(43,125,141,1)","rgba(42,121,142,1)","rgba(55,102,140,1)","rgba(35,167,132,1)","rgba(48,114,141,1)","rgba(44,118,142,1)","rgba(62,84,137,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(72,180,119,1)","rgba(44,118,142,1)","rgba(69,52,120,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(43,124,141,1)","rgba(44,170,130,1)","rgba(43,124,141,1)","rgba(43,142,138,1)","rgba(36,164,133,1)","rgba(69,24,97,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(44,138,139,1)","rgba(43,146,137,1)","rgba(48,171,128,1)","rgba(43,147,137,1)","rgba(44,138,139,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(39,158,134,1)","rgba(42,148,137,1)","rgba(42,149,136,1)","rgba(43,144,137,1)","rgba(43,139,138,1)","rgba(68,5,86,1)","rgba(62,84,137,1)","rgba(44,138,139,1)","rgba(43,142,138,1)","rgba(43,141,138,1)","rgba(43,127,141,1)","rgba(52,172,127,1)","rgba(54,105,140,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(43,128,141,1)","rgba(42,120,142,1)","rgba(34,167,132,1)","rgba(39,158,134,1)","rgba(40,157,135,1)","rgba(42,121,142,1)","rgba(61,89,138,1)","rgba(43,119,142,1)","rgba(68,55,123,1)","rgba(62,85,137,1)","rgba(70,34,104,1)","rgba(40,157,134,1)","rgba(45,117,142,1)","rgba(64,74,136,1)","rgba(39,158,134,1)","rgba(43,142,138,1)","rgba(41,153,136,1)","rgba(70,37,107,1)","rgba(68,56,124,1)","rgba(41,151,136,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(42,151,136,1)","rgba(49,112,141,1)","rgba(44,118,142,1)","rgba(43,128,140,1)","rgba(44,134,139,1)","rgba(102,196,100,1)","rgba(41,154,135,1)","rgba(40,155,135,1)","rgba(44,132,140,1)","rgba(43,119,142,1)","rgba(44,139,138,1)","rgba(65,67,134,1)","rgba(44,136,139,1)","rgba(50,172,128,1)","rgba(43,119,142,1)","rgba(55,102,140,1)","rgba(61,89,138,1)","rgba(43,139,138,1)","rgba(69,19,93,1)","rgba(43,123,141,1)","rgba(49,112,141,1)","rgba(63,79,137,1)","rgba(65,70,135,1)","rgba(69,52,119,1)","rgba(63,82,137,1)","rgba(65,67,134,1)","rgba(69,19,93,1)","rgba(44,131,140,1)","rgba(63,82,137,1)","rgba(42,120,142,1)","rgba(69,25,97,1)","rgba(70,37,107,1)","rgba(43,119,142,1)","rgba(43,119,142,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(46,116,141,1)","rgba(69,46,115,1)","rgba(70,37,106,1)","rgba(43,146,137,1)","rgba(37,162,133,1)","rgba(57,98,139,1)","rgba(55,102,140,1)","rgba(57,174,126,1)","rgba(66,64,131,1)","rgba(58,97,139,1)","rgba(67,62,130,1)","rgba(39,158,134,1)","rgba(58,97,139,1)","rgba(69,17,92,1)","rgba(39,159,134,1)","rgba(68,4,86,1)","rgba(68,58,125,1)","rgba(43,143,138,1)","rgba(69,23,96,1)","rgba(42,149,136,1)","rgba(70,29,100,1)","rgba(58,96,139,1)","rgba(69,50,118,1)","rgba(69,45,113,1)","rgba(70,27,99,1)","rgba(69,47,115,1)","rgba(69,22,95,1)","rgba(43,126,141,1)","rgba(45,117,142,1)","rgba(52,108,140,1)","rgba(66,65,132,1)","rgba(68,57,124,1)","rgba(70,33,103,1)","rgba(43,146,137,1)","rgba(50,110,141,1)","rgba(66,65,132,1)","rgba(64,72,136,1)","rgba(65,66,133,1)","rgba(65,177,122,1)","rgba(59,93,139,1)","rgba(70,31,102,1)","rgba(42,149,136,1)","rgba(70,36,105,1)","rgba(44,139,138,1)","rgba(69,178,121,1)","rgba(69,50,118,1)","rgba(67,59,126,1)","rgba(69,46,114,1)","rgba(70,42,111,1)","rgba(43,146,137,1)","rgba(163,216,72,1)","rgba(53,105,140,1)","rgba(55,103,140,1)","rgba(69,52,120,1)","rgba(60,91,138,1)","rgba(56,101,140,1)","rgba(52,108,140,1)","rgba(70,26,98,1)","rgba(59,93,139,1)","rgba(55,173,126,1)","rgba(66,66,133,1)","rgba(62,84,137,1)","rgba(50,111,141,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(59,93,138,1)","rgba(42,150,136,1)","rgba(43,140,138,1)","rgba(67,61,128,1)","rgba(42,149,136,1)","rgba(74,181,118,1)","rgba(40,157,135,1)","rgba(65,68,135,1)","rgba(59,94,139,1)","rgba(38,162,133,1)","rgba(58,96,139,1)","rgba(70,30,101,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(37,162,133,1)","rgba(63,81,137,1)","rgba(90,188,109,1)","rgba(38,162,134,1)","rgba(57,98,139,1)","rgba(44,137,139,1)","rgba(44,132,140,1)","rgba(67,178,121,1)","rgba(44,135,139,1)","rgba(40,156,135,1)","rgba(41,153,136,1)","rgba(86,186,111,1)","rgba(67,60,127,1)","rgba(69,9,88,1)","rgba(70,29,100,1)","rgba(68,5,86,1)","rgba(70,29,100,1)","rgba(49,172,128,1)","rgba(65,67,134,1)","rgba(44,139,138,1)","rgba(43,146,137,1)","rgba(44,138,139,1)","rgba(46,115,141,1)","rgba(66,63,130,1)","rgba(44,135,139,1)","rgba(69,18,93,1)","rgba(43,126,141,1)","rgba(104,196,99,1)","rgba(43,140,138,1)","rgba(51,110,141,1)","rgba(59,175,125,1)","rgba(61,87,138,1)","rgba(69,49,117,1)","rgba(65,70,135,1)","rgba(54,173,126,1)","rgba(39,158,134,1)","rgba(42,170,130,1)","rgba(44,133,140,1)","rgba(36,164,133,1)","rgba(58,97,139,1)","rgba(61,89,138,1)","rgba(69,24,96,1)","rgba(64,73,136,1)","rgba(69,52,119,1)","rgba(44,139,138,1)","rgba(54,105,140,1)","rgba(66,66,133,1)","rgba(70,25,97,1)","rgba(70,42,111,1)","rgba(45,117,142,1)","rgba(64,75,136,1)","rgba(62,176,123,1)","rgba(42,151,136,1)","rgba(43,142,138,1)","rgba(41,169,130,1)","rgba(58,95,139,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(49,112,141,1)","rgba(48,113,141,1)","rgba(65,70,135,1)","rgba(67,61,128,1)","rgba(70,32,103,1)","rgba(43,119,142,1)","rgba(62,83,137,1)","rgba(69,10,88,1)","rgba(42,150,136,1)","rgba(69,48,116,1)","rgba(58,96,139,1)","rgba(43,142,138,1)","rgba(58,97,139,1)","rgba(43,119,142,1)","rgba(57,100,139,1)","rgba(66,65,132,1)","rgba(43,143,138,1)","rgba(42,148,137,1)","rgba(43,146,137,1)","rgba(53,106,140,1)","rgba(34,168,132,1)","rgba(57,100,139,1)","rgba(40,155,135,1)","rgba(69,47,115,1)","rgba(69,46,114,1)","rgba(70,30,101,1)","rgba(43,119,142,1)","rgba(64,75,136,1)","rgba(42,150,136,1)","rgba(107,199,96,1)","rgba(70,27,99,1)","rgba(57,98,139,1)","rgba(99,194,102,1)","rgba(43,144,137,1)","rgba(40,155,135,1)","rgba(68,56,123,1)","rgba(58,97,139,1)","rgba(43,139,138,1)","rgba(58,96,139,1)","rgba(50,111,141,1)","rgba(47,171,129,1)","rgba(41,151,136,1)","rgba(38,161,134,1)","rgba(54,105,140,1)","rgba(55,103,140,1)","rgba(57,100,139,1)","rgba(37,162,133,1)","rgba(46,116,142,1)","rgba(68,57,124,1)","rgba(39,159,134,1)","rgba(36,164,133,1)","rgba(44,135,139,1)","rgba(40,155,135,1)","rgba(68,58,125,1)","rgba(68,58,125,1)","rgba(68,53,121,1)","rgba(69,20,94,1)","rgba(69,50,118,1)","rgba(44,136,139,1)","rgba(70,34,104,1)","rgba(62,85,137,1)","rgba(99,194,102,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(69,21,95,1)","rgba(42,122,142,1)","rgba(39,169,131,1)","rgba(44,136,139,1)","rgba(55,104,140,1)","rgba(43,124,141,1)","rgba(69,22,95,1)","rgba(68,4,85,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(55,103,140,1)","rgba(43,147,137,1)","rgba(41,154,135,1)","rgba(54,104,140,1)","rgba(66,65,132,1)","rgba(39,160,134,1)","rgba(38,160,134,1)","rgba(43,126,141,1)","rgba(44,137,139,1)","rgba(68,5,86,1)","rgba(59,93,139,1)","rgba(41,154,135,1)","rgba(57,99,139,1)","rgba(44,131,140,1)","rgba(44,131,140,1)","rgba(62,83,137,1)","rgba(54,104,140,1)","rgba(44,131,140,1)","rgba(69,23,96,1)","rgba(69,51,119,1)","rgba(62,176,124,1)","rgba(42,170,130,1)","rgba(42,148,137,1)","rgba(43,122,142,1)","rgba(70,36,105,1)","rgba(42,148,137,1)","rgba(69,10,88,1)","rgba(66,63,130,1)","rgba(43,143,138,1)","rgba(70,36,106,1)","rgba(64,78,136,1)","rgba(70,41,110,1)","rgba(56,101,140,1)","rgba(69,51,119,1)","rgba(41,152,136,1)","rgba(59,95,139,1)","rgba(67,59,126,1)","rgba(37,163,133,1)","rgba(70,29,100,1)","rgba(70,35,104,1)","rgba(43,128,140,1)","rgba(42,122,142,1)","rgba(63,81,137,1)","rgba(69,15,91,1)","rgba(67,62,129,1)","rgba(70,35,105,1)","rgba(63,82,137,1)","rgba(57,99,139,1)","rgba(43,141,138,1)","rgba(44,138,139,1)","rgba(78,182,116,1)","rgba(43,141,138,1)","rgba(65,72,136,1)","rgba(57,98,139,1)","rgba(66,66,133,1)","rgba(43,129,140,1)","rgba(55,103,140,1)","rgba(68,4,86,1)","rgba(45,117,142,1)","rgba(65,67,134,1)","rgba(43,123,141,1)","rgba(43,144,137,1)","rgba(69,19,93,1)","rgba(66,63,130,1)","rgba(58,97,139,1)","rgba(59,93,138,1)","rgba(69,25,97,1)","rgba(70,37,106,1)","rgba(69,23,96,1)","rgba(43,126,141,1)","rgba(54,104,140,1)","rgba(57,99,139,1)","rgba(65,70,135,1)","rgba(43,123,141,1)","rgba(59,94,139,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(64,75,136,1)","rgba(42,148,137,1)","rgba(69,16,91,1)","rgba(66,64,131,1)","rgba(66,66,133,1)","rgba(41,152,136,1)","rgba(53,107,140,1)","rgba(70,35,105,1)","rgba(60,92,138,1)","rgba(64,76,136,1)","rgba(35,167,132,1)","rgba(56,100,139,1)","rgba(54,104,140,1)","rgba(44,133,140,1)","rgba(68,6,87,1)","rgba(67,60,128,1)","rgba(44,132,140,1)","rgba(43,141,138,1)","rgba(63,80,137,1)","rgba(53,106,140,1)","rgba(41,154,135,1)","rgba(43,142,138,1)","rgba(69,14,90,1)","rgba(60,91,138,1)","rgba(53,106,140,1)","rgba(55,102,140,1)","rgba(50,110,141,1)","rgba(69,17,92,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(66,64,131,1)","rgba(64,76,136,1)","rgba(54,105,140,1)","rgba(61,88,138,1)","rgba(66,65,132,1)","rgba(69,48,116,1)","rgba(70,35,105,1)","rgba(67,63,130,1)","rgba(69,12,89,1)","rgba(67,62,130,1)","rgba(42,151,136,1)","rgba(62,86,138,1)","rgba(49,112,141,1)","rgba(80,183,115,1)","rgba(44,134,139,1)","rgba(64,76,136,1)","rgba(69,14,90,1)","rgba(47,114,141,1)","rgba(69,46,114,1)","rgba(69,16,91,1)","rgba(66,64,131,1)","rgba(64,72,136,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(59,94,139,1)","rgba(41,154,135,1)","rgba(69,49,117,1)","rgba(69,8,87,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(70,27,99,1)","rgba(67,61,128,1)","rgba(44,131,140,1)","rgba(40,155,135,1)","rgba(65,68,135,1)","rgba(68,3,85,1)","rgba(58,97,139,1)","rgba(57,99,139,1)","rgba(47,114,141,1)","rgba(63,78,136,1)","rgba(65,67,134,1)","rgba(70,26,98,1)","rgba(57,99,139,1)","rgba(66,64,131,1)","rgba(68,54,122,1)","rgba(44,136,139,1)","rgba(41,152,136,1)","rgba(69,52,120,1)","rgba(64,74,136,1)","rgba(62,85,137,1)","rgba(43,122,142,1)","rgba(69,48,116,1)","rgba(90,188,109,1)","rgba(38,162,134,1)","rgba(49,112,141,1)","rgba(53,107,140,1)","rgba(37,162,133,1)","rgba(59,94,139,1)","rgba(69,43,112,1)","rgba(70,29,100,1)","rgba(51,110,141,1)","rgba(69,9,88,1)","rgba(48,114,141,1)","rgba(43,140,138,1)","rgba(42,148,136,1)","rgba(69,46,114,1)","rgba(42,149,136,1)","rgba(64,76,136,1)","rgba(40,157,135,1)","rgba(56,101,140,1)","rgba(38,162,133,1)","rgba(53,106,140,1)","rgba(69,20,94,1)","rgba(69,20,94,1)","rgba(42,122,142,1)","rgba(53,107,140,1)","rgba(61,87,138,1)","rgba(70,36,106,1)","rgba(64,77,136,1)","rgba(70,41,109,1)","rgba(43,129,140,1)","rgba(68,58,125,1)","rgba(44,137,139,1)","rgba(70,37,107,1)","rgba(68,178,121,1)","rgba(47,115,141,1)","rgba(44,138,139,1)","rgba(69,45,113,1)","rgba(44,139,138,1)","rgba(38,162,133,1)","rgba(44,118,142,1)","rgba(70,39,108,1)","rgba(43,123,141,1)","rgba(43,123,142,1)","rgba(63,79,137,1)","rgba(42,148,136,1)","rgba(82,184,114,1)","rgba(43,125,141,1)","rgba(44,132,140,1)","rgba(43,140,138,1)","rgba(51,109,141,1)","rgba(70,28,99,1)","rgba(60,90,138,1)","rgba(70,42,110,1)","rgba(53,106,140,1)","rgba(60,92,138,1)","rgba(70,38,107,1)","rgba(38,161,134,1)","rgba(60,91,138,1)","rgba(69,16,91,1)","rgba(60,92,138,1)","rgba(65,68,135,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(59,94,139,1)","rgba(52,108,140,1)","rgba(62,84,137,1)","rgba(68,54,121,1)","rgba(64,73,136,1)","rgba(66,65,132,1)","rgba(64,72,136,1)","rgba(53,106,140,1)","rgba(70,38,107,1)","rgba(44,135,139,1)","rgba(62,176,124,1)","rgba(67,60,127,1)","rgba(69,22,95,1)","rgba(43,145,137,1)","rgba(44,136,139,1)","rgba(37,169,131,1)","rgba(70,30,101,1)","rgba(66,66,133,1)","rgba(70,32,102,1)","rgba(43,123,142,1)","rgba(43,127,141,1)","rgba(43,146,137,1)","rgba(70,37,106,1)","rgba(69,19,93,1)","rgba(64,76,136,1)","rgba(43,145,137,1)","rgba(61,89,138,1)","rgba(52,108,140,1)","rgba(62,84,137,1)","rgba(54,173,127,1)","rgba(68,56,123,1)","rgba(69,23,96,1)","rgba(63,80,137,1)","rgba(61,89,138,1)","rgba(70,29,100,1)","rgba(44,133,140,1)","rgba(36,165,133,1)","rgba(44,135,139,1)","rgba(42,150,136,1)","rgba(61,89,138,1)","rgba(51,109,141,1)","rgba(55,103,140,1)","rgba(69,12,89,1)","rgba(68,53,121,1)","rgba(42,150,136,1)","rgba(60,91,138,1)","rgba(67,62,129,1)","rgba(41,170,130,1)","rgba(70,38,108,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(68,5,86,1)","rgba(61,87,138,1)","rgba(43,126,141,1)","rgba(64,78,136,1)","rgba(49,112,141,1)","rgba(37,163,133,1)","rgba(44,131,140,1)","rgba(70,28,100,1)","rgba(60,92,138,1)","rgba(35,167,132,1)","rgba(37,162,133,1)","rgba(44,118,142,1)","rgba(61,87,138,1)","rgba(68,3,85,1)","rgba(64,76,136,1)","rgba(69,22,95,1)","rgba(45,117,142,1)","rgba(70,27,99,1)","rgba(59,94,139,1)","rgba(70,31,102,1)","rgba(66,64,131,1)","rgba(42,121,142,1)","rgba(67,60,127,1)","rgba(59,95,139,1)","rgba(68,5,86,1)","rgba(68,53,120,1)","rgba(66,64,131,1)","rgba(70,40,109,1)","rgba(64,72,136,1)","rgba(43,125,141,1)","rgba(46,116,142,1)","rgba(63,82,137,1)","rgba(70,35,105,1)","rgba(65,67,134,1)","rgba(70,34,104,1)","rgba(69,50,118,1)","rgba(69,44,112,1)","rgba(70,28,99,1)","rgba(66,65,132,1)","rgba(70,37,106,1)","rgba(64,74,136,1)","rgba(70,40,109,1)","rgba(41,153,136,1)","rgba(43,143,138,1)","rgba(67,60,127,1)","rgba(43,119,142,1)","rgba(61,89,138,1)","rgba(42,121,142,1)","rgba(61,89,138,1)","rgba(44,118,142,1)","rgba(64,73,136,1)","rgba(65,67,134,1)","rgba(43,127,141,1)","rgba(67,178,121,1)","rgba(43,130,140,1)","rgba(43,145,137,1)","rgba(42,148,137,1)","rgba(38,162,134,1)","rgba(70,34,104,1)","rgba(43,147,137,1)","rgba(42,120,142,1)","rgba(43,126,141,1)","rgba(50,110,141,1)","rgba(70,28,100,1)","rgba(62,86,138,1)","rgba(59,93,139,1)","rgba(69,50,118,1)","rgba(59,93,139,1)","rgba(51,110,141,1)","rgba(61,89,138,1)","rgba(70,29,100,1)","rgba(58,97,139,1)","rgba(43,146,137,1)","rgba(70,32,103,1)","rgba(68,55,122,1)","rgba(42,121,142,1)","rgba(69,19,93,1)","rgba(37,163,133,1)","rgba(43,128,141,1)","rgba(62,83,137,1)","rgba(54,105,140,1)","rgba(42,121,142,1)","rgba(68,54,122,1)","rgba(63,80,137,1)","rgba(66,65,132,1)","rgba(53,106,140,1)","rgba(59,95,139,1)","rgba(59,94,139,1)","rgba(39,159,134,1)","rgba(55,103,140,1)","rgba(59,95,139,1)","rgba(60,92,138,1)","rgba(44,138,139,1)","rgba(43,119,142,1)","rgba(69,49,117,1)","rgba(40,157,135,1)","rgba(52,108,141,1)","rgba(100,194,101,1)","rgba(69,47,115,1)","rgba(63,81,137,1)","rgba(69,9,88,1)","rgba(60,91,138,1)","rgba(64,77,136,1)","rgba(48,113,141,1)","rgba(42,122,142,1)","rgba(61,88,138,1)","rgba(66,65,132,1)","rgba(67,61,128,1)","rgba(69,49,117,1)","rgba(63,176,123,1)","rgba(68,3,85,1)","rgba(64,73,136,1)","rgba(70,37,107,1)","rgba(63,81,137,1)","rgba(62,83,137,1)","rgba(64,77,136,1)","rgba(43,144,137,1)","rgba(57,98,139,1)","rgba(43,127,141,1)","rgba(70,29,100,1)","rgba(44,131,140,1)","rgba(43,131,140,1)","rgba(54,105,140,1)","rgba(43,144,137,1)","rgba(61,89,138,1)","rgba(43,141,138,1)","rgba(53,107,140,1)","rgba(70,32,102,1)","rgba(70,39,108,1)","rgba(50,111,141,1)","rgba(44,118,142,1)","rgba(57,99,139,1)","rgba(44,133,140,1)","rgba(69,17,92,1)","rgba(42,150,136,1)","rgba(70,30,101,1)","rgba(68,4,86,1)","rgba(43,123,141,1)","rgba(43,147,137,1)","rgba(53,106,140,1)","rgba(68,54,122,1)","rgba(70,32,102,1)","rgba(70,38,107,1)","rgba(69,13,90,1)","rgba(42,121,142,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(38,160,134,1)","rgba(70,30,101,1)","rgba(43,144,137,1)","rgba(43,125,141,1)","rgba(34,168,132,1)","rgba(68,54,122,1)","rgba(55,102,140,1)","rgba(44,133,140,1)","rgba(43,144,137,1)","rgba(51,109,141,1)","rgba(50,172,128,1)","rgba(43,140,138,1)","rgba(48,113,141,1)","rgba(42,121,142,1)","rgba(44,138,139,1)","rgba(44,136,139,1)","rgba(64,73,136,1)","rgba(69,12,89,1)","rgba(59,93,138,1)","rgba(64,73,136,1)","rgba(43,147,137,1)","rgba(52,107,140,1)","rgba(69,16,91,1)","rgba(60,91,138,1)","rgba(43,129,140,1)","rgba(70,29,100,1)","rgba(106,198,97,1)","rgba(79,183,115,1)","rgba(69,21,95,1)","rgba(65,67,134,1)","rgba(43,140,138,1)","rgba(54,104,140,1)","rgba(40,155,135,1)","rgba(66,64,131,1)","rgba(70,29,100,1)","rgba(43,139,138,1)","rgba(36,165,133,1)","rgba(41,154,135,1)","rgba(95,191,105,1)","rgba(67,62,129,1)","rgba(58,97,139,1)","rgba(67,60,127,1)","rgba(52,108,141,1)","rgba(37,164,133,1)","rgba(64,77,136,1)","rgba(67,61,128,1)","rgba(67,61,129,1)","rgba(66,65,132,1)","rgba(49,112,141,1)","rgba(50,111,141,1)","rgba(49,112,141,1)","rgba(68,57,125,1)","rgba(64,75,136,1)","rgba(62,86,138,1)","rgba(36,165,133,1)","rgba(56,100,140,1)","rgba(69,15,91,1)","rgba(65,70,135,1)","rgba(58,97,139,1)","rgba(38,160,134,1)","rgba(59,93,138,1)","rgba(64,78,136,1)","rgba(41,152,136,1)","rgba(70,39,108,1)","rgba(69,44,112,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(65,72,136,1)","rgba(37,163,133,1)","rgba(69,21,94,1)","rgba(42,150,136,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(70,42,111,1)","rgba(70,42,110,1)","rgba(38,162,133,1)","rgba(36,165,133,1)","rgba(40,156,135,1)","rgba(65,66,133,1)","rgba(57,98,139,1)","rgba(34,167,132,1)","rgba(69,52,120,1)","rgba(46,116,141,1)","rgba(42,150,136,1)","rgba(69,52,120,1)","rgba(44,133,140,1)","rgba(69,11,89,1)","rgba(50,111,141,1)","rgba(57,99,139,1)","rgba(63,81,137,1)","rgba(64,72,136,1)","rgba(69,16,91,1)","rgba(46,116,142,1)","rgba(40,169,131,1)","rgba(69,22,95,1)","rgba(64,74,136,1)","rgba(39,160,134,1)","rgba(39,158,134,1)","rgba(43,126,141,1)","rgba(68,1,84,1)","rgba(62,83,137,1)","rgba(67,61,128,1)","rgba(47,115,141,1)","rgba(68,54,122,1)","rgba(57,99,139,1)","rgba(69,11,89,1)","rgba(68,6,86,1)","rgba(65,72,136,1)","rgba(69,12,89,1)","rgba(68,4,86,1)","rgba(50,111,141,1)","rgba(63,79,137,1)","rgba(68,5,86,1)","rgba(70,38,107,1)","rgba(69,48,116,1)","rgba(42,149,136,1)","rgba(60,90,138,1)","rgba(63,78,136,1)","rgba(50,111,141,1)","rgba(61,90,138,1)","rgba(69,20,94,1)","rgba(60,90,138,1)","rgba(65,70,135,1)","rgba(45,117,142,1)","rgba(61,88,138,1)","rgba(65,72,136,1)","rgba(70,25,97,1)","rgba(68,178,121,1)","rgba(42,151,136,1)","rgba(43,126,141,1)","rgba(42,120,142,1)","rgba(43,142,138,1)","rgba(69,14,90,1)","rgba(43,124,141,1)","rgba(48,113,141,1)","rgba(56,100,139,1)","rgba(68,56,123,1)","rgba(55,103,140,1)","rgba(39,169,131,1)","rgba(44,138,139,1)","rgba(67,61,129,1)","rgba(37,163,133,1)","rgba(66,66,133,1)","rgba(69,20,94,1)","rgba(36,165,133,1)","rgba(69,16,91,1)","rgba(41,152,136,1)","rgba(43,146,137,1)","rgba(42,148,137,1)","rgba(42,121,142,1)","rgba(56,100,140,1)","rgba(43,130,140,1)","rgba(68,53,121,1)","rgba(66,64,131,1)","rgba(56,101,140,1)","rgba(60,92,138,1)","rgba(64,76,136,1)","rgba(69,12,89,1)","rgba(70,37,107,1)","rgba(44,138,139,1)","rgba(48,114,141,1)","rgba(43,130,140,1)","rgba(46,116,142,1)","rgba(65,70,135,1)","rgba(41,153,135,1)","rgba(58,97,139,1)","rgba(70,34,104,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(56,102,140,1)","rgba(35,166,132,1)","rgba(63,79,137,1)","rgba(65,70,135,1)","rgba(44,118,142,1)","rgba(42,120,142,1)","rgba(70,39,108,1)","rgba(70,37,106,1)","rgba(65,70,135,1)","rgba(69,12,89,1)","rgba(69,23,96,1)","rgba(64,73,136,1)","rgba(69,16,91,1)","rgba(68,178,121,1)","rgba(43,142,138,1)","rgba(70,33,104,1)","rgba(43,123,141,1)","rgba(37,163,133,1)","rgba(43,130,140,1)","rgba(57,99,139,1)","rgba(70,30,101,1)","rgba(40,156,135,1)","rgba(67,62,129,1)","rgba(43,144,137,1)","rgba(48,114,141,1)","rgba(69,25,97,1)","rgba(70,33,104,1)","rgba(69,14,90,1)","rgba(43,144,137,1)","rgba(68,3,85,1)","rgba(65,71,135,1)","rgba(47,115,141,1)","rgba(56,102,140,1)","rgba(64,72,136,1)","rgba(64,75,136,1)","rgba(69,44,112,1)","rgba(70,32,103,1)","rgba(64,75,136,1)","rgba(44,134,139,1)","rgba(53,107,140,1)","rgba(50,111,141,1)","rgba(43,143,138,1)","rgba(44,137,139,1)","rgba(39,159,134,1)","rgba(68,2,85,1)","rgba(65,66,133,1)","rgba(68,7,87,1)","rgba(35,167,132,1)","rgba(64,75,136,1)","rgba(53,106,140,1)","rgba(35,167,132,1)","rgba(63,81,137,1)","rgba(70,39,108,1)","rgba(69,46,115,1)","rgba(69,43,112,1)","rgba(70,34,104,1)","rgba(70,36,106,1)","rgba(70,27,99,1)","rgba(63,79,137,1)","rgba(68,3,85,1)","rgba(69,24,96,1)","rgba(63,83,137,1)","rgba(65,69,135,1)","rgba(70,34,104,1)","rgba(59,95,139,1)","rgba(68,4,86,1)","rgba(44,136,139,1)","rgba(80,183,115,1)","rgba(64,75,136,1)","rgba(67,59,126,1)","rgba(39,169,131,1)","rgba(45,117,142,1)","rgba(69,44,112,1)","rgba(57,99,139,1)","rgba(61,90,138,1)","rgba(53,105,140,1)","rgba(69,12,89,1)","rgba(62,85,137,1)","rgba(70,28,100,1)","rgba(67,61,129,1)","rgba(67,60,128,1)","rgba(54,104,140,1)","rgba(68,57,124,1)","rgba(64,78,136,1)","rgba(65,69,135,1)","rgba(69,16,91,1)","rgba(63,79,137,1)","rgba(68,56,123,1)","rgba(69,13,90,1)","rgba(62,83,137,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(70,29,100,1)","rgba(43,127,141,1)","rgba(67,60,127,1)","rgba(62,85,137,1)","rgba(64,76,136,1)","rgba(52,108,141,1)","rgba(54,173,127,1)","rgba(43,142,138,1)","rgba(43,124,141,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(44,137,139,1)","rgba(48,113,141,1)","rgba(43,141,138,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(65,69,135,1)","rgba(68,4,85,1)","rgba(64,73,136,1)","rgba(70,40,109,1)","rgba(61,87,138,1)","rgba(57,174,125,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(48,114,141,1)","rgba(41,153,135,1)","rgba(69,25,97,1)","rgba(47,115,141,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(70,31,102,1)","rgba(215,225,55,1)","rgba(42,149,136,1)","rgba(69,16,92,1)","rgba(42,120,142,1)","rgba(64,77,136,1)","rgba(69,11,89,1)","rgba(69,50,117,1)","rgba(65,67,134,1)","rgba(70,38,107,1)","rgba(68,53,120,1)","rgba(46,116,141,1)","rgba(107,198,96,1)","rgba(69,23,96,1)","rgba(43,129,140,1)","rgba(70,29,100,1)","rgba(44,136,139,1)","rgba(66,65,132,1)","rgba(61,88,138,1)","rgba(65,72,136,1)","rgba(43,129,140,1)","rgba(65,72,135,1)","rgba(67,59,126,1)","rgba(43,140,138,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(70,42,111,1)","rgba(70,29,100,1)","rgba(68,1,84,1)","rgba(58,97,139,1)","rgba(43,147,137,1)","rgba(55,103,140,1)","rgba(36,166,133,1)","rgba(39,158,134,1)","rgba(69,11,89,1)","rgba(69,12,89,1)","rgba(69,14,90,1)","rgba(70,31,101,1)","rgba(41,152,136,1)","rgba(69,43,112,1)","rgba(69,24,97,1)","rgba(62,84,137,1)","rgba(64,74,136,1)","rgba(43,131,140,1)","rgba(38,169,131,1)","rgba(69,43,111,1)","rgba(57,99,139,1)","rgba(42,150,136,1)","rgba(67,60,127,1)","rgba(48,114,141,1)","rgba(44,170,130,1)","rgba(59,93,139,1)","rgba(39,158,134,1)","rgba(62,85,137,1)","rgba(69,44,113,1)","rgba(70,38,108,1)","rgba(70,26,98,1)","rgba(52,107,140,1)","rgba(62,84,137,1)","rgba(67,60,128,1)","rgba(63,83,137,1)","rgba(64,74,136,1)","rgba(70,41,110,1)","rgba(65,67,134,1)","rgba(65,69,135,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(66,65,132,1)","rgba(64,74,136,1)","rgba(53,107,140,1)","rgba(69,46,114,1)","rgba(56,100,139,1)","rgba(65,67,134,1)","rgba(61,89,138,1)","rgba(43,123,141,1)","rgba(55,103,140,1)","rgba(67,63,130,1)","rgba(52,107,140,1)","rgba(64,72,136,1)","rgba(70,36,106,1)","rgba(58,96,139,1)","rgba(67,60,127,1)","rgba(65,71,135,1)","rgba(64,73,136,1)","rgba(69,20,94,1)","rgba(68,53,121,1)","rgba(68,53,120,1)","rgba(69,51,119,1)","rgba(68,57,125,1)","rgba(70,36,106,1)","rgba(68,58,125,1)","rgba(66,65,132,1)","rgba(58,96,139,1)","rgba(67,59,126,1)","rgba(61,88,138,1)","rgba(65,69,135,1)","rgba(69,52,119,1)","rgba(68,2,85,1)","rgba(69,52,120,1)","rgba(58,96,139,1)","rgba(69,16,91,1)","rgba(64,77,136,1)","rgba(44,133,140,1)","rgba(44,132,140,1)","rgba(34,168,132,1)","rgba(42,149,136,1)","rgba(48,113,141,1)","rgba(55,102,140,1)","rgba(43,129,140,1)","rgba(69,52,120,1)","rgba(67,62,129,1)","rgba(60,91,138,1)","rgba(40,157,135,1)","rgba(65,71,135,1)","rgba(62,84,137,1)","rgba(70,33,103,1)","rgba(69,178,120,1)","rgba(53,105,140,1)","rgba(47,114,141,1)","rgba(69,47,116,1)","rgba(68,56,123,1)","rgba(68,6,86,1)","rgba(64,74,136,1)","rgba(69,19,93,1)","rgba(70,40,109,1)","rgba(69,22,95,1)","rgba(66,63,130,1)","rgba(40,156,135,1)","rgba(63,81,137,1)","rgba(70,35,105,1)","rgba(69,52,120,1)","rgba(69,48,116,1)","rgba(39,159,134,1)","rgba(43,139,138,1)","rgba(46,116,142,1)","rgba(43,126,141,1)","rgba(65,67,134,1)","rgba(107,198,96,1)","rgba(43,145,137,1)","rgba(64,73,136,1)","rgba(70,29,100,1)","rgba(62,176,124,1)","rgba(35,166,133,1)","rgba(43,147,137,1)","rgba(70,37,106,1)","rgba(54,173,127,1)","rgba(42,149,136,1)","rgba(35,168,132,1)","rgba(65,71,135,1)","rgba(60,92,138,1)","rgba(55,103,140,1)","rgba(60,91,138,1)","rgba(70,38,107,1)","rgba(52,108,141,1)","rgba(60,91,138,1)","rgba(42,151,136,1)","rgba(65,70,135,1)","rgba(50,110,141,1)","rgba(68,6,87,1)","rgba(70,28,99,1)","rgba(51,109,141,1)","rgba(69,49,117,1)","rgba(70,37,107,1)","rgba(67,61,128,1)","rgba(70,35,105,1)","rgba(43,143,138,1)","rgba(49,112,141,1)","rgba(36,168,131,1)","rgba(41,152,136,1)","rgba(63,79,137,1)","rgba(43,143,138,1)","rgba(44,134,139,1)","rgba(42,149,136,1)","rgba(52,173,127,1)","rgba(73,180,119,1)","rgba(69,12,89,1)","rgba(55,103,140,1)","rgba(56,174,126,1)","rgba(39,158,134,1)","rgba(65,177,122,1)","rgba(42,150,136,1)","rgba(50,111,141,1)","rgba(35,168,132,1)","rgba(47,115,141,1)","rgba(43,144,137,1)","rgba(43,170,130,1)","rgba(69,25,97,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(43,139,138,1)","rgba(43,142,138,1)","rgba(41,153,135,1)","rgba(36,164,133,1)","rgba(60,175,124,1)","rgba(44,133,140,1)","rgba(49,113,141,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(64,77,136,1)","rgba(71,179,119,1)","rgba(41,153,136,1)","rgba(125,209,80,1)","rgba(44,136,139,1)","rgba(41,153,135,1)","rgba(36,165,133,1)","rgba(43,129,140,1)","rgba(69,43,112,1)","rgba(43,145,137,1)","rgba(43,123,141,1)","rgba(40,156,135,1)","rgba(43,126,141,1)","rgba(68,4,86,1)","rgba(69,14,90,1)","rgba(68,5,86,1)","rgba(41,153,135,1)","rgba(67,178,121,1)","rgba(44,135,139,1)","rgba(69,48,116,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(37,164,133,1)","rgba(89,188,110,1)","rgba(42,150,136,1)","rgba(47,171,129,1)","rgba(56,101,140,1)","rgba(67,59,126,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(42,121,142,1)","rgba(59,95,139,1)","rgba(69,13,90,1)","rgba(78,182,116,1)","rgba(38,160,134,1)","rgba(52,108,141,1)","rgba(91,189,108,1)","rgba(44,138,139,1)","rgba(43,139,138,1)","rgba(43,130,140,1)","rgba(71,179,120,1)","rgba(44,136,139,1)","rgba(69,25,97,1)","rgba(42,149,136,1)","rgba(55,103,140,1)","rgba(69,49,117,1)","rgba(43,140,138,1)","rgba(43,119,142,1)","rgba(43,126,141,1)","rgba(60,92,138,1)","rgba(44,134,139,1)","rgba(82,184,114,1)","rgba(69,43,112,1)","rgba(50,172,128,1)","rgba(59,175,125,1)","rgba(104,196,99,1)","rgba(90,188,109,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(54,105,140,1)","rgba(62,83,137,1)","rgba(48,113,141,1)","rgba(43,119,142,1)","rgba(43,123,142,1)","rgba(59,93,139,1)","rgba(59,175,125,1)","rgba(94,191,106,1)","rgba(49,112,141,1)","rgba(91,189,108,1)","rgba(48,114,141,1)","rgba(39,158,134,1)","rgba(42,149,136,1)","rgba(39,158,134,1)","rgba(68,57,125,1)","rgba(75,181,118,1)","rgba(42,150,136,1)","rgba(42,122,142,1)","rgba(43,145,137,1)","rgba(41,153,136,1)","rgba(43,144,137,1)","rgba(77,182,117,1)","rgba(69,178,121,1)","rgba(58,96,139,1)","rgba(63,81,137,1)","rgba(42,147,137,1)","rgba(34,167,132,1)","rgba(100,194,102,1)","rgba(119,207,84,1)","rgba(41,152,136,1)","rgba(34,168,132,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(67,59,126,1)","rgba(44,137,139,1)","rgba(65,71,135,1)","rgba(37,163,133,1)","rgba(69,44,113,1)","rgba(63,79,137,1)","rgba(58,97,139,1)","rgba(38,160,134,1)","rgba(42,151,136,1)","rgba(39,158,134,1)","rgba(97,192,104,1)","rgba(41,154,135,1)","rgba(35,166,133,1)","rgba(43,123,142,1)","rgba(65,68,135,1)","rgba(43,123,141,1)","rgba(63,80,137,1)","rgba(70,38,107,1)","rgba(38,162,134,1)","rgba(48,114,141,1)","rgba(68,55,123,1)","rgba(36,164,133,1)","rgba(42,120,142,1)","rgba(44,118,142,1)","rgba(69,8,87,1)","rgba(45,117,142,1)","rgba(60,92,138,1)","rgba(44,132,140,1)","rgba(38,161,134,1)","rgba(66,66,133,1)","rgba(41,153,136,1)","rgba(38,161,134,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(43,125,141,1)","rgba(62,83,137,1)","rgba(70,40,109,1)","rgba(69,9,88,1)","rgba(43,125,141,1)","rgba(62,86,138,1)","rgba(64,73,136,1)","rgba(44,135,139,1)","rgba(61,88,138,1)","rgba(36,164,133,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(53,106,140,1)","rgba(36,165,133,1)","rgba(62,86,138,1)","rgba(76,182,117,1)","rgba(56,101,140,1)","rgba(69,23,96,1)","rgba(70,40,109,1)","rgba(68,54,122,1)","rgba(67,62,129,1)","rgba(68,53,121,1)","rgba(41,170,130,1)","rgba(35,167,132,1)","rgba(70,26,98,1)","rgba(70,38,108,1)","rgba(83,185,113,1)","rgba(60,175,124,1)","rgba(45,117,142,1)","rgba(51,109,141,1)","rgba(56,101,140,1)","rgba(69,49,117,1)","rgba(44,170,130,1)","rgba(77,182,116,1)","rgba(46,116,142,1)","rgba(44,135,139,1)","rgba(68,2,85,1)","rgba(45,116,142,1)","rgba(36,166,133,1)","rgba(70,31,102,1)","rgba(44,134,139,1)","rgba(45,117,142,1)","rgba(44,138,139,1)","rgba(42,170,130,1)","rgba(57,98,139,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(61,87,138,1)","rgba(69,20,94,1)","rgba(70,34,104,1)","rgba(60,90,138,1)","rgba(43,119,142,1)","rgba(69,21,95,1)","rgba(50,111,141,1)","rgba(62,84,137,1)","rgba(70,29,100,1)","rgba(69,48,116,1)","rgba(61,89,138,1)","rgba(68,56,123,1)","rgba(60,90,138,1)","rgba(44,137,139,1)","rgba(68,2,84,1)","rgba(64,74,136,1)","rgba(50,111,141,1)","rgba(91,189,108,1)","rgba(53,107,140,1)","rgba(69,46,114,1)","rgba(65,67,134,1)","rgba(68,58,125,1)","rgba(56,101,140,1)","rgba(37,163,133,1)","rgba(69,11,89,1)","rgba(41,151,136,1)","rgba(42,149,136,1)","rgba(52,108,140,1)","rgba(37,162,133,1)","rgba(68,55,123,1)","rgba(50,110,141,1)","rgba(65,70,135,1)","rgba(69,50,118,1)","rgba(43,147,137,1)","rgba(66,63,131,1)","rgba(45,116,142,1)","rgba(41,153,135,1)","rgba(69,45,113,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(69,51,119,1)","rgba(64,76,136,1)","rgba(68,7,87,1)","rgba(66,66,133,1)","rgba(55,103,140,1)","rgba(47,171,129,1)","rgba(43,143,138,1)","rgba(66,65,132,1)","rgba(43,142,138,1)","rgba(43,145,137,1)","rgba(58,95,139,1)","rgba(81,184,114,1)","rgba(58,96,139,1)","rgba(68,55,123,1)","rgba(69,10,88,1)","rgba(63,81,137,1)","rgba(58,96,139,1)","rgba(44,170,129,1)","rgba(59,93,139,1)","rgba(66,66,133,1)","rgba(55,102,140,1)","rgba(45,170,129,1)","rgba(62,83,137,1)","rgba(40,156,135,1)","rgba(69,46,114,1)","rgba(70,30,101,1)","rgba(40,169,131,1)","rgba(43,123,142,1)","rgba(70,31,102,1)","rgba(43,126,141,1)","rgba(43,126,141,1)","rgba(43,125,141,1)","rgba(54,105,140,1)","rgba(56,101,140,1)","rgba(64,77,136,1)","rgba(68,54,122,1)","rgba(78,182,116,1)","rgba(68,55,123,1)","rgba(94,191,106,1)","rgba(54,173,127,1)","rgba(43,125,141,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(38,160,134,1)","rgba(43,119,142,1)","rgba(72,179,119,1)","rgba(63,81,137,1)","rgba(42,149,136,1)","rgba(52,108,140,1)","rgba(68,5,86,1)","rgba(64,73,136,1)","rgba(70,30,101,1)","rgba(57,99,139,1)","rgba(39,158,134,1)","rgba(39,158,134,1)","rgba(39,160,134,1)","rgba(90,189,109,1)","rgba(43,139,138,1)","rgba(68,56,124,1)","rgba(58,97,139,1)","rgba(69,21,94,1)","rgba(44,136,139,1)","rgba(69,51,119,1)","rgba(64,72,136,1)","rgba(43,125,141,1)","rgba(69,25,97,1)","rgba(69,17,92,1)","rgba(45,117,142,1)","rgba(66,63,130,1)","rgba(96,192,104,1)","rgba(56,100,139,1)","rgba(41,153,135,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(75,181,117,1)","rgba(43,145,137,1)","rgba(38,161,134,1)","rgba(61,175,124,1)","rgba(44,134,139,1)","rgba(56,101,140,1)","rgba(69,46,114,1)","rgba(42,120,142,1)","rgba(63,81,137,1)","rgba(43,145,137,1)","rgba(70,28,99,1)","rgba(69,24,96,1)","rgba(49,113,141,1)","rgba(60,92,138,1)","rgba(51,110,141,1)","rgba(46,116,141,1)","rgba(69,45,114,1)","rgba(43,144,137,1)","rgba(68,56,123,1)","rgba(78,182,116,1)","rgba(69,23,96,1)","rgba(48,113,141,1)","rgba(43,143,138,1)","rgba(49,112,141,1)","rgba(43,130,140,1)","rgba(49,113,141,1)","rgba(49,112,141,1)","rgba(44,134,139,1)","rgba(59,93,138,1)","rgba(53,106,140,1)","rgba(54,105,140,1)","rgba(61,87,138,1)","rgba(65,72,136,1)","rgba(57,99,139,1)","rgba(68,55,123,1)","rgba(44,135,139,1)","rgba(71,179,119,1)","rgba(43,120,142,1)","rgba(36,164,133,1)","rgba(42,151,136,1)","rgba(36,165,133,1)","rgba(43,130,140,1)","rgba(51,109,141,1)","rgba(43,145,137,1)","rgba(44,132,140,1)","rgba(42,149,136,1)","rgba(42,148,137,1)","rgba(118,206,86,1)","rgba(42,149,136,1)","rgba(34,167,132,1)","rgba(35,166,132,1)","rgba(69,21,94,1)","rgba(44,136,139,1)","rgba(44,135,139,1)","rgba(35,166,132,1)","rgba(44,136,139,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(52,173,127,1)","rgba(38,161,134,1)","rgba(43,140,138,1)","rgba(70,27,98,1)","rgba(42,150,136,1)","rgba(89,188,109,1)","rgba(43,122,142,1)","rgba(65,69,135,1)","rgba(42,147,137,1)","rgba(42,150,136,1)","rgba(51,110,141,1)","rgba(62,85,137,1)","rgba(44,134,139,1)","rgba(44,135,139,1)","rgba(55,103,140,1)","rgba(42,121,142,1)","rgba(69,23,96,1)","rgba(70,36,106,1)","rgba(69,44,113,1)","rgba(69,23,96,1)","rgba(44,131,140,1)","rgba(44,118,142,1)","rgba(53,106,140,1)","rgba(42,121,142,1)","rgba(105,197,98,1)","rgba(43,147,137,1)","rgba(43,139,138,1)","rgba(44,132,140,1)","rgba(42,150,136,1)","rgba(53,173,127,1)","rgba(42,151,136,1)","rgba(70,34,104,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(77,182,116,1)","rgba(43,145,137,1)","rgba(69,179,120,1)","rgba(61,89,138,1)","rgba(39,158,134,1)","rgba(53,105,140,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(36,166,133,1)","rgba(69,19,93,1)","rgba(43,142,138,1)","rgba(44,138,139,1)","rgba(111,201,93,1)","rgba(65,69,135,1)","rgba(42,122,142,1)","rgba(43,139,138,1)","rgba(69,21,94,1)","rgba(69,43,111,1)","rgba(70,33,104,1)","rgba(73,180,119,1)","rgba(43,131,140,1)","rgba(66,65,132,1)","rgba(45,116,142,1)","rgba(43,145,137,1)","rgba(60,175,124,1)","rgba(43,142,138,1)","rgba(70,30,101,1)","rgba(41,154,135,1)","rgba(39,160,134,1)","rgba(40,155,135,1)","rgba(40,155,135,1)","rgba(69,21,95,1)","rgba(70,26,98,1)","rgba(44,134,139,1)","rgba(55,103,140,1)","rgba(36,164,133,1)","rgba(44,134,139,1)","rgba(79,183,115,1)","rgba(65,72,136,1)","rgba(97,192,104,1)","rgba(44,137,139,1)","rgba(74,180,118,1)","rgba(38,160,134,1)","rgba(43,146,137,1)","rgba(43,123,141,1)","rgba(67,61,128,1)","rgba(41,154,135,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(41,153,135,1)","rgba(68,53,121,1)","rgba(44,132,140,1)","rgba(41,154,135,1)","rgba(43,128,140,1)","rgba(93,190,107,1)","rgba(68,178,121,1)","rgba(67,178,121,1)","rgba(42,148,136,1)","rgba(43,144,137,1)","rgba(64,76,136,1)","rgba(35,167,132,1)","rgba(41,155,135,1)","rgba(63,79,137,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(43,145,137,1)","rgba(43,131,140,1)","rgba(37,169,131,1)","rgba(68,1,84,1)","rgba(38,160,134,1)","rgba(65,177,122,1)","rgba(69,25,97,1)"],"opacity":1,"size":5.66929133858268,"symbol":"circle","line":{"width":1.88976377952756,"color":["rgba(62,85,137,1)","rgba(59,175,125,1)","rgba(42,122,142,1)","rgba(68,3,85,1)","rgba(43,141,138,1)","rgba(70,34,104,1)","rgba(202,223,60,1)","rgba(140,212,78,1)","rgba(69,52,120,1)","rgba(68,4,85,1)","rgba(69,49,117,1)","rgba(69,20,94,1)","rgba(38,161,134,1)","rgba(39,159,134,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(42,121,142,1)","rgba(86,186,111,1)","rgba(45,117,142,1)","rgba(53,106,140,1)","rgba(68,54,122,1)","rgba(57,98,139,1)","rgba(70,179,120,1)","rgba(41,154,135,1)","rgba(70,30,101,1)","rgba(69,45,114,1)","rgba(34,168,132,1)","rgba(124,209,81,1)","rgba(65,69,135,1)","rgba(59,93,139,1)","rgba(43,129,140,1)","rgba(44,132,140,1)","rgba(97,192,104,1)","rgba(44,131,140,1)","rgba(69,19,93,1)","rgba(44,132,140,1)","rgba(43,140,138,1)","rgba(40,157,135,1)","rgba(41,152,136,1)","rgba(42,121,142,1)","rgba(91,189,108,1)","rgba(43,126,141,1)","rgba(69,50,118,1)","rgba(44,139,138,1)","rgba(69,14,90,1)","rgba(80,183,115,1)","rgba(41,152,136,1)","rgba(68,53,120,1)","rgba(104,197,98,1)","rgba(36,164,133,1)","rgba(41,152,136,1)","rgba(39,169,131,1)","rgba(54,105,140,1)","rgba(44,136,139,1)","rgba(43,139,138,1)","rgba(37,163,133,1)","rgba(69,15,91,1)","rgba(57,98,139,1)","rgba(43,126,141,1)","rgba(56,100,140,1)","rgba(36,164,133,1)","rgba(38,160,134,1)","rgba(95,191,105,1)","rgba(39,160,134,1)","rgba(44,135,139,1)","rgba(43,143,138,1)","rgba(39,158,134,1)","rgba(40,157,135,1)","rgba(39,159,134,1)","rgba(64,77,136,1)","rgba(68,57,124,1)","rgba(66,66,133,1)","rgba(62,86,138,1)","rgba(45,170,129,1)","rgba(43,125,141,1)","rgba(43,170,130,1)","rgba(173,217,69,1)","rgba(41,153,135,1)","rgba(111,201,92,1)","rgba(64,73,136,1)","rgba(42,150,136,1)","rgba(43,139,138,1)","rgba(106,198,97,1)","rgba(50,111,141,1)","rgba(53,107,140,1)","rgba(69,51,118,1)","rgba(40,156,135,1)","rgba(43,128,140,1)","rgba(62,85,137,1)","rgba(67,59,126,1)","rgba(44,137,139,1)","rgba(98,193,103,1)","rgba(43,147,137,1)","rgba(44,139,138,1)","rgba(44,134,139,1)","rgba(43,130,140,1)","rgba(43,143,138,1)","rgba(44,131,140,1)","rgba(43,126,141,1)","rgba(37,162,133,1)","rgba(38,161,134,1)","rgba(50,111,141,1)","rgba(57,174,126,1)","rgba(36,166,133,1)","rgba(36,165,133,1)","rgba(38,161,134,1)","rgba(154,214,74,1)","rgba(68,5,86,1)","rgba(44,133,140,1)","rgba(43,131,140,1)","rgba(69,12,89,1)","rgba(36,165,133,1)","rgba(43,127,141,1)","rgba(41,152,136,1)","rgba(36,164,133,1)","rgba(70,38,107,1)","rgba(44,139,138,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(54,104,140,1)","rgba(43,141,138,1)","rgba(68,8,87,1)","rgba(43,130,140,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(68,5,86,1)","rgba(56,101,140,1)","rgba(44,138,139,1)","rgba(65,177,122,1)","rgba(57,98,139,1)","rgba(44,134,139,1)","rgba(43,140,138,1)","rgba(45,117,142,1)","rgba(253,231,37,1)","rgba(66,65,132,1)","rgba(218,225,54,1)","rgba(42,150,136,1)","rgba(43,130,140,1)","rgba(44,136,139,1)","rgba(50,110,141,1)","rgba(47,171,129,1)","rgba(42,150,136,1)","rgba(70,179,120,1)","rgba(90,188,109,1)","rgba(43,170,130,1)","rgba(45,117,142,1)","rgba(69,46,114,1)","rgba(42,149,136,1)","rgba(42,120,142,1)","rgba(44,137,139,1)","rgba(58,97,139,1)","rgba(43,144,137,1)","rgba(69,45,113,1)","rgba(69,50,118,1)","rgba(44,118,142,1)","rgba(58,97,139,1)","rgba(65,69,135,1)","rgba(43,145,137,1)","rgba(42,151,136,1)","rgba(38,161,134,1)","rgba(43,143,138,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(39,158,134,1)","rgba(59,93,139,1)","rgba(48,113,141,1)","rgba(60,92,138,1)","rgba(67,58,126,1)","rgba(44,133,140,1)","rgba(38,162,133,1)","rgba(43,146,137,1)","rgba(66,65,133,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(55,102,140,1)","rgba(64,77,136,1)","rgba(69,22,95,1)","rgba(43,145,137,1)","rgba(41,154,135,1)","rgba(65,70,135,1)","rgba(66,64,131,1)","rgba(58,174,125,1)","rgba(91,189,108,1)","rgba(43,125,141,1)","rgba(80,183,115,1)","rgba(56,174,126,1)","rgba(38,161,134,1)","rgba(155,214,74,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(43,147,137,1)","rgba(37,162,133,1)","rgba(50,111,141,1)","rgba(43,125,141,1)","rgba(69,16,91,1)","rgba(43,125,141,1)","rgba(54,173,127,1)","rgba(43,119,142,1)","rgba(42,122,142,1)","rgba(39,158,134,1)","rgba(66,66,133,1)","rgba(35,166,133,1)","rgba(42,150,136,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(37,163,133,1)","rgba(69,20,94,1)","rgba(42,148,136,1)","rgba(39,169,131,1)","rgba(44,134,139,1)","rgba(42,120,142,1)","rgba(42,122,142,1)","rgba(43,130,140,1)","rgba(76,181,117,1)","rgba(138,212,78,1)","rgba(205,223,60,1)","rgba(75,181,117,1)","rgba(43,143,138,1)","rgba(43,130,140,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(68,56,124,1)","rgba(55,102,140,1)","rgba(165,216,72,1)","rgba(43,142,138,1)","rgba(43,147,137,1)","rgba(35,167,132,1)","rgba(66,63,130,1)","rgba(69,43,111,1)","rgba(87,187,111,1)","rgba(42,151,136,1)","rgba(70,39,108,1)","rgba(66,63,131,1)","rgba(57,98,139,1)","rgba(43,127,141,1)","rgba(43,146,137,1)","rgba(64,77,136,1)","rgba(98,193,103,1)","rgba(43,144,137,1)","rgba(54,173,126,1)","rgba(70,33,103,1)","rgba(102,195,100,1)","rgba(69,23,96,1)","rgba(37,163,133,1)","rgba(43,123,142,1)","rgba(43,145,137,1)","rgba(44,133,140,1)","rgba(45,117,142,1)","rgba(42,148,137,1)","rgba(50,111,141,1)","rgba(38,160,134,1)","rgba(44,136,139,1)","rgba(74,181,118,1)","rgba(35,166,132,1)","rgba(179,219,68,1)","rgba(79,183,115,1)","rgba(40,155,135,1)","rgba(70,34,104,1)","rgba(43,127,141,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(73,180,118,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(44,137,139,1)","rgba(42,122,142,1)","rgba(68,4,86,1)","rgba(35,166,133,1)","rgba(43,126,141,1)","rgba(50,111,141,1)","rgba(42,148,137,1)","rgba(41,169,130,1)","rgba(64,77,136,1)","rgba(36,165,133,1)","rgba(53,173,127,1)","rgba(35,168,132,1)","rgba(43,144,137,1)","rgba(83,185,113,1)","rgba(45,117,142,1)","rgba(41,153,135,1)","rgba(69,12,89,1)","rgba(58,97,139,1)","rgba(35,167,132,1)","rgba(74,180,118,1)","rgba(67,62,129,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(42,121,142,1)","rgba(68,178,121,1)","rgba(64,177,123,1)","rgba(203,223,60,1)","rgba(153,214,74,1)","rgba(105,197,97,1)","rgba(68,4,86,1)","rgba(42,147,137,1)","rgba(43,139,138,1)","rgba(43,144,137,1)","rgba(70,41,110,1)","rgba(41,154,135,1)","rgba(65,68,135,1)","rgba(50,111,141,1)","rgba(58,97,139,1)","rgba(98,193,103,1)","rgba(142,212,77,1)","rgba(69,21,94,1)","rgba(121,208,82,1)","rgba(144,213,76,1)","rgba(66,65,132,1)","rgba(164,216,72,1)","rgba(151,214,75,1)","rgba(36,165,133,1)","rgba(42,121,142,1)","rgba(34,168,132,1)","rgba(79,183,116,1)","rgba(45,117,142,1)","rgba(62,83,137,1)","rgba(48,114,141,1)","rgba(56,100,139,1)","rgba(65,68,135,1)","rgba(86,186,111,1)","rgba(43,126,141,1)","rgba(47,115,141,1)","rgba(70,26,98,1)","rgba(43,128,140,1)","rgba(66,63,131,1)","rgba(43,140,138,1)","rgba(42,121,142,1)","rgba(43,146,137,1)","rgba(49,172,128,1)","rgba(59,175,125,1)","rgba(51,172,128,1)","rgba(41,154,135,1)","rgba(42,121,142,1)","rgba(62,86,138,1)","rgba(44,132,140,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(42,121,142,1)","rgba(55,103,140,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(39,159,134,1)","rgba(64,75,136,1)","rgba(105,197,98,1)","rgba(70,40,109,1)","rgba(64,177,123,1)","rgba(61,87,138,1)","rgba(63,82,137,1)","rgba(44,132,140,1)","rgba(68,7,87,1)","rgba(65,71,135,1)","rgba(49,113,141,1)","rgba(70,28,99,1)","rgba(49,171,128,1)","rgba(56,101,140,1)","rgba(45,117,142,1)","rgba(58,95,139,1)","rgba(69,18,92,1)","rgba(99,194,102,1)","rgba(68,54,122,1)","rgba(48,113,141,1)","rgba(42,148,136,1)","rgba(69,43,111,1)","rgba(154,214,74,1)","rgba(41,153,136,1)","rgba(55,103,140,1)","rgba(40,155,135,1)","rgba(62,176,123,1)","rgba(60,91,138,1)","rgba(38,162,133,1)","rgba(43,146,137,1)","rgba(56,100,140,1)","rgba(42,150,136,1)","rgba(39,160,134,1)","rgba(42,151,136,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(62,84,137,1)","rgba(69,23,95,1)","rgba(42,151,136,1)","rgba(58,96,139,1)","rgba(72,180,119,1)","rgba(79,183,115,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(61,175,124,1)","rgba(38,161,134,1)","rgba(44,132,140,1)","rgba(43,119,142,1)","rgba(69,25,97,1)","rgba(44,136,139,1)","rgba(63,82,137,1)","rgba(66,177,122,1)","rgba(42,150,136,1)","rgba(39,158,134,1)","rgba(50,172,128,1)","rgba(76,181,117,1)","rgba(70,27,99,1)","rgba(56,174,126,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(38,161,134,1)","rgba(44,136,139,1)","rgba(43,141,138,1)","rgba(42,150,136,1)","rgba(44,136,139,1)","rgba(52,107,140,1)","rgba(35,167,132,1)","rgba(70,39,108,1)","rgba(44,133,140,1)","rgba(108,199,95,1)","rgba(38,162,134,1)","rgba(126,210,80,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(97,192,104,1)","rgba(44,132,140,1)","rgba(186,220,65,1)","rgba(70,39,108,1)","rgba(43,144,137,1)","rgba(63,80,137,1)","rgba(52,108,141,1)","rgba(43,147,137,1)","rgba(42,149,136,1)","rgba(70,26,98,1)","rgba(42,148,137,1)","rgba(44,137,139,1)","rgba(44,131,140,1)","rgba(42,151,136,1)","rgba(37,162,133,1)","rgba(42,148,137,1)","rgba(60,91,138,1)","rgba(70,41,109,1)","rgba(36,165,133,1)","rgba(70,32,102,1)","rgba(68,6,87,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(36,165,133,1)","rgba(49,172,128,1)","rgba(53,107,140,1)","rgba(44,138,139,1)","rgba(38,160,134,1)","rgba(43,145,137,1)","rgba(43,128,141,1)","rgba(54,173,127,1)","rgba(58,95,139,1)","rgba(43,126,141,1)","rgba(67,60,127,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(41,153,136,1)","rgba(42,148,137,1)","rgba(55,173,126,1)","rgba(35,166,133,1)","rgba(43,123,141,1)","rgba(65,72,136,1)","rgba(67,59,127,1)","rgba(44,133,140,1)","rgba(63,176,123,1)","rgba(67,59,126,1)","rgba(49,112,141,1)","rgba(43,146,137,1)","rgba(71,179,119,1)","rgba(62,84,137,1)","rgba(45,117,142,1)","rgba(43,141,138,1)","rgba(58,174,125,1)","rgba(208,224,58,1)","rgba(39,159,134,1)","rgba(42,149,136,1)","rgba(68,55,123,1)","rgba(44,131,140,1)","rgba(36,164,133,1)","rgba(43,139,138,1)","rgba(43,140,138,1)","rgba(44,139,138,1)","rgba(40,169,131,1)","rgba(38,162,133,1)","rgba(40,155,135,1)","rgba(43,143,138,1)","rgba(43,128,141,1)","rgba(58,97,139,1)","rgba(42,151,136,1)","rgba(52,108,140,1)","rgba(36,165,133,1)","rgba(61,175,124,1)","rgba(46,116,142,1)","rgba(44,131,140,1)","rgba(68,5,86,1)","rgba(50,111,141,1)","rgba(44,131,140,1)","rgba(55,173,126,1)","rgba(51,172,128,1)","rgba(59,94,139,1)","rgba(75,181,118,1)","rgba(69,13,90,1)","rgba(59,94,139,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(70,29,100,1)","rgba(43,144,137,1)","rgba(69,47,115,1)","rgba(50,172,128,1)","rgba(38,162,133,1)","rgba(42,151,136,1)","rgba(43,141,138,1)","rgba(158,215,73,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(121,209,82,1)","rgba(80,183,115,1)","rgba(41,153,135,1)","rgba(103,196,99,1)","rgba(44,137,139,1)","rgba(69,19,93,1)","rgba(69,15,91,1)","rgba(120,207,84,1)","rgba(118,206,85,1)","rgba(79,183,116,1)","rgba(52,108,140,1)","rgba(40,169,131,1)","rgba(42,149,136,1)","rgba(39,159,134,1)","rgba(40,156,135,1)","rgba(43,127,141,1)","rgba(65,67,134,1)","rgba(63,80,137,1)","rgba(60,91,138,1)","rgba(69,11,89,1)","rgba(51,109,141,1)","rgba(65,67,134,1)","rgba(43,141,138,1)","rgba(44,136,139,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(35,166,133,1)","rgba(52,107,140,1)","rgba(43,119,142,1)","rgba(65,177,122,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(44,132,140,1)","rgba(69,50,118,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(111,201,92,1)","rgba(168,217,71,1)","rgba(69,52,119,1)","rgba(67,178,121,1)","rgba(43,145,137,1)","rgba(70,29,100,1)","rgba(42,147,137,1)","rgba(55,173,126,1)","rgba(43,144,137,1)","rgba(65,177,122,1)","rgba(63,82,137,1)","rgba(68,7,87,1)","rgba(42,170,130,1)","rgba(76,182,117,1)","rgba(62,83,137,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(39,158,134,1)","rgba(45,170,129,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(44,133,140,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(39,160,134,1)","rgba(36,164,133,1)","rgba(42,150,136,1)","rgba(44,135,139,1)","rgba(37,162,133,1)","rgba(37,169,131,1)","rgba(43,170,130,1)","rgba(51,110,141,1)","rgba(82,184,114,1)","rgba(106,198,97,1)","rgba(56,174,126,1)","rgba(44,134,139,1)","rgba(157,215,74,1)","rgba(42,150,136,1)","rgba(68,6,86,1)","rgba(85,186,112,1)","rgba(43,140,138,1)","rgba(41,153,136,1)","rgba(38,162,133,1)","rgba(49,172,128,1)","rgba(43,123,141,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(42,151,136,1)","rgba(43,130,140,1)","rgba(70,34,104,1)","rgba(43,126,141,1)","rgba(87,187,111,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(40,169,131,1)","rgba(70,36,105,1)","rgba(34,168,132,1)","rgba(40,156,135,1)","rgba(56,174,126,1)","rgba(65,68,135,1)","rgba(51,109,141,1)","rgba(48,171,128,1)","rgba(40,155,135,1)","rgba(104,197,98,1)","rgba(70,26,98,1)","rgba(42,121,142,1)","rgba(44,136,139,1)","rgba(42,147,137,1)","rgba(43,147,137,1)","rgba(44,133,140,1)","rgba(71,179,120,1)","rgba(42,150,136,1)","rgba(42,147,137,1)","rgba(65,68,135,1)","rgba(63,176,123,1)","rgba(42,147,137,1)","rgba(59,94,139,1)","rgba(40,157,135,1)","rgba(39,159,134,1)","rgba(39,158,134,1)","rgba(87,187,111,1)","rgba(80,183,115,1)","rgba(37,162,133,1)","rgba(36,164,133,1)","rgba(56,101,140,1)","rgba(44,134,139,1)","rgba(81,184,114,1)","rgba(50,110,141,1)","rgba(42,151,136,1)","rgba(44,118,142,1)","rgba(94,191,106,1)","rgba(69,18,93,1)","rgba(59,93,138,1)","rgba(42,120,142,1)","rgba(42,148,137,1)","rgba(67,62,129,1)","rgba(54,105,140,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(65,70,135,1)","rgba(157,215,74,1)","rgba(40,169,131,1)","rgba(62,85,137,1)","rgba(43,141,138,1)","rgba(52,172,127,1)","rgba(45,171,129,1)","rgba(43,145,137,1)","rgba(65,68,135,1)","rgba(69,8,87,1)","rgba(37,162,133,1)","rgba(41,155,135,1)","rgba(43,125,141,1)","rgba(68,7,87,1)","rgba(44,136,139,1)","rgba(44,135,139,1)","rgba(83,185,113,1)","rgba(58,95,139,1)","rgba(41,153,135,1)","rgba(43,145,137,1)","rgba(43,128,141,1)","rgba(62,86,137,1)","rgba(39,158,134,1)","rgba(170,217,70,1)","rgba(43,170,130,1)","rgba(52,172,127,1)","rgba(51,172,127,1)","rgba(48,114,141,1)","rgba(43,139,138,1)","rgba(37,162,133,1)","rgba(43,140,138,1)","rgba(44,134,139,1)","rgba(44,137,139,1)","rgba(40,156,135,1)","rgba(42,147,137,1)","rgba(42,121,142,1)","rgba(109,200,94,1)","rgba(53,173,127,1)","rgba(70,40,109,1)","rgba(68,4,86,1)","rgba(43,125,141,1)","rgba(118,206,86,1)","rgba(66,66,133,1)","rgba(145,213,76,1)","rgba(43,142,138,1)","rgba(44,135,139,1)","rgba(69,10,89,1)","rgba(64,75,136,1)","rgba(43,140,138,1)","rgba(42,148,136,1)","rgba(70,39,108,1)","rgba(43,142,138,1)","rgba(61,88,138,1)","rgba(99,194,102,1)","rgba(40,155,135,1)","rgba(88,187,110,1)","rgba(117,205,87,1)","rgba(42,121,142,1)","rgba(41,152,136,1)","rgba(43,141,138,1)","rgba(42,147,137,1)","rgba(37,163,133,1)","rgba(37,163,133,1)","rgba(58,96,139,1)","rgba(46,116,141,1)","rgba(61,88,138,1)","rgba(60,175,124,1)","rgba(61,175,124,1)","rgba(63,176,123,1)","rgba(47,114,141,1)","rgba(44,138,139,1)","rgba(51,109,141,1)","rgba(44,134,139,1)","rgba(70,40,109,1)","rgba(42,149,136,1)","rgba(75,181,117,1)","rgba(48,113,141,1)","rgba(43,146,137,1)","rgba(67,61,128,1)","rgba(65,72,136,1)","rgba(44,135,139,1)","rgba(69,17,92,1)","rgba(62,84,137,1)","rgba(61,87,138,1)","rgba(37,164,133,1)","rgba(69,45,113,1)","rgba(56,101,140,1)","rgba(128,210,80,1)","rgba(44,137,139,1)","rgba(61,88,138,1)","rgba(71,179,120,1)","rgba(52,172,127,1)","rgba(60,175,124,1)","rgba(34,168,132,1)","rgba(43,145,137,1)","rgba(158,215,73,1)","rgba(43,144,137,1)","rgba(41,153,135,1)","rgba(81,184,114,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(64,75,136,1)","rgba(103,196,100,1)","rgba(57,99,139,1)","rgba(42,148,137,1)","rgba(37,163,133,1)","rgba(39,158,134,1)","rgba(61,87,138,1)","rgba(69,8,88,1)","rgba(81,184,114,1)","rgba(43,170,130,1)","rgba(69,15,91,1)","rgba(70,32,102,1)","rgba(132,211,79,1)","rgba(45,117,142,1)","rgba(54,173,127,1)","rgba(50,172,128,1)","rgba(42,148,136,1)","rgba(56,101,140,1)","rgba(42,148,136,1)","rgba(43,129,140,1)","rgba(65,177,122,1)","rgba(67,178,121,1)","rgba(43,119,142,1)","rgba(65,68,135,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(43,124,141,1)","rgba(80,183,115,1)","rgba(66,66,133,1)","rgba(34,168,132,1)","rgba(44,135,139,1)","rgba(60,90,138,1)","rgba(72,179,119,1)","rgba(43,139,138,1)","rgba(51,172,128,1)","rgba(88,187,110,1)","rgba(44,132,140,1)","rgba(45,170,129,1)","rgba(39,158,134,1)","rgba(43,124,141,1)","rgba(39,158,134,1)","rgba(46,171,129,1)","rgba(69,24,96,1)","rgba(43,145,137,1)","rgba(68,53,120,1)","rgba(65,72,136,1)","rgba(69,18,93,1)","rgba(37,162,133,1)","rgba(43,142,138,1)","rgba(43,130,140,1)","rgba(60,92,138,1)","rgba(43,141,138,1)","rgba(66,66,133,1)","rgba(118,206,86,1)","rgba(126,210,80,1)","rgba(139,212,78,1)","rgba(124,209,81,1)","rgba(164,216,72,1)","rgba(41,155,135,1)","rgba(42,148,137,1)","rgba(43,125,141,1)","rgba(50,111,141,1)","rgba(43,140,138,1)","rgba(43,125,141,1)","rgba(112,202,91,1)","rgba(64,74,136,1)","rgba(44,135,139,1)","rgba(44,136,139,1)","rgba(64,176,123,1)","rgba(68,53,121,1)","rgba(69,20,94,1)","rgba(103,196,100,1)","rgba(58,96,139,1)","rgba(54,105,140,1)","rgba(44,131,140,1)","rgba(40,157,135,1)","rgba(59,94,139,1)","rgba(56,101,140,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(37,169,131,1)","rgba(48,171,128,1)","rgba(41,152,136,1)","rgba(43,145,137,1)","rgba(41,153,135,1)","rgba(40,155,135,1)","rgba(55,103,140,1)","rgba(58,174,125,1)","rgba(38,160,134,1)","rgba(42,148,137,1)","rgba(69,47,115,1)","rgba(93,190,107,1)","rgba(53,106,140,1)","rgba(42,149,136,1)","rgba(44,134,139,1)","rgba(38,161,134,1)","rgba(40,157,135,1)","rgba(54,105,140,1)","rgba(43,143,138,1)","rgba(40,155,135,1)","rgba(43,139,138,1)","rgba(36,164,133,1)","rgba(45,117,142,1)","rgba(43,126,141,1)","rgba(43,143,138,1)","rgba(41,153,136,1)","rgba(46,171,129,1)","rgba(42,151,136,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(39,158,134,1)","rgba(43,125,141,1)","rgba(35,166,132,1)","rgba(56,101,140,1)","rgba(39,160,134,1)","rgba(104,197,98,1)","rgba(44,131,140,1)","rgba(41,154,135,1)","rgba(43,119,142,1)","rgba(41,152,136,1)","rgba(42,151,136,1)","rgba(48,113,141,1)","rgba(50,111,141,1)","rgba(44,136,139,1)","rgba(101,194,101,1)","rgba(68,178,121,1)","rgba(84,185,112,1)","rgba(43,145,137,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(112,202,92,1)","rgba(43,130,140,1)","rgba(39,158,134,1)","rgba(60,92,138,1)","rgba(50,110,141,1)","rgba(54,105,140,1)","rgba(142,212,77,1)","rgba(44,136,139,1)","rgba(70,27,99,1)","rgba(69,18,92,1)","rgba(162,216,72,1)","rgba(43,130,140,1)","rgba(69,42,111,1)","rgba(69,50,118,1)","rgba(40,157,135,1)","rgba(43,130,140,1)","rgba(54,104,140,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(43,125,141,1)","rgba(47,114,141,1)","rgba(50,172,128,1)","rgba(41,153,135,1)","rgba(43,142,138,1)","rgba(43,143,138,1)","rgba(65,69,135,1)","rgba(43,144,137,1)","rgba(44,134,139,1)","rgba(44,118,142,1)","rgba(38,161,134,1)","rgba(43,170,130,1)","rgba(39,158,134,1)","rgba(62,85,137,1)","rgba(36,165,133,1)","rgba(40,156,135,1)","rgba(36,165,133,1)","rgba(70,30,101,1)","rgba(64,77,136,1)","rgba(43,126,141,1)","rgba(64,76,136,1)","rgba(37,169,131,1)","rgba(40,155,135,1)","rgba(48,171,128,1)","rgba(40,156,135,1)","rgba(42,122,142,1)","rgba(43,144,137,1)","rgba(69,9,88,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(123,209,81,1)","rgba(42,149,136,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(57,99,139,1)","rgba(40,157,135,1)","rgba(69,21,95,1)","rgba(36,164,133,1)","rgba(37,164,133,1)","rgba(41,153,135,1)","rgba(68,57,124,1)","rgba(42,151,136,1)","rgba(37,164,133,1)","rgba(43,143,138,1)","rgba(104,196,99,1)","rgba(44,133,140,1)","rgba(88,187,110,1)","rgba(44,132,140,1)","rgba(38,160,134,1)","rgba(44,134,139,1)","rgba(44,138,139,1)","rgba(39,160,134,1)","rgba(35,168,132,1)","rgba(41,153,136,1)","rgba(69,17,92,1)","rgba(61,175,124,1)","rgba(44,132,140,1)","rgba(70,179,120,1)","rgba(54,104,140,1)","rgba(39,158,134,1)","rgba(41,152,136,1)","rgba(71,179,119,1)","rgba(40,157,135,1)","rgba(59,95,139,1)","rgba(49,111,141,1)","rgba(41,153,135,1)","rgba(43,130,140,1)","rgba(37,163,133,1)","rgba(44,135,139,1)","rgba(48,113,141,1)","rgba(43,125,141,1)","rgba(43,131,140,1)","rgba(72,180,119,1)","rgba(43,143,138,1)","rgba(43,146,137,1)","rgba(168,217,71,1)","rgba(61,87,138,1)","rgba(43,131,140,1)","rgba(36,165,133,1)","rgba(40,155,135,1)","rgba(70,179,120,1)","rgba(44,137,139,1)","rgba(70,39,108,1)","rgba(35,167,132,1)","rgba(41,153,136,1)","rgba(43,145,137,1)","rgba(43,130,140,1)","rgba(62,84,137,1)","rgba(36,164,133,1)","rgba(106,198,97,1)","rgba(60,91,138,1)","rgba(38,169,131,1)","rgba(122,209,81,1)","rgba(43,145,137,1)","rgba(37,164,133,1)","rgba(43,130,140,1)","rgba(42,120,142,1)","rgba(61,89,138,1)","rgba(69,25,97,1)","rgba(69,22,95,1)","rgba(49,112,141,1)","rgba(44,137,139,1)","rgba(51,109,141,1)","rgba(37,163,133,1)","rgba(70,179,120,1)","rgba(44,118,142,1)","rgba(48,113,141,1)","rgba(43,119,142,1)","rgba(38,162,134,1)","rgba(59,94,139,1)","rgba(135,211,79,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(40,157,134,1)","rgba(43,129,140,1)","rgba(68,178,121,1)","rgba(34,168,132,1)","rgba(46,116,142,1)","rgba(132,211,79,1)","rgba(43,142,138,1)","rgba(36,165,133,1)","rgba(138,212,78,1)","rgba(63,78,136,1)","rgba(69,10,88,1)","rgba(58,96,139,1)","rgba(44,137,139,1)","rgba(70,37,107,1)","rgba(43,125,141,1)","rgba(44,118,142,1)","rgba(68,5,86,1)","rgba(69,45,114,1)","rgba(42,122,142,1)","rgba(66,63,130,1)","rgba(63,176,123,1)","rgba(39,158,134,1)","rgba(69,23,96,1)","rgba(51,172,127,1)","rgba(43,129,140,1)","rgba(63,176,123,1)","rgba(36,164,133,1)","rgba(36,164,133,1)","rgba(69,11,89,1)","rgba(40,157,135,1)","rgba(43,146,137,1)","rgba(39,159,134,1)","rgba(111,201,92,1)","rgba(42,151,136,1)","rgba(42,151,136,1)","rgba(40,156,135,1)","rgba(36,165,133,1)","rgba(43,143,138,1)","rgba(36,165,133,1)","rgba(97,192,104,1)","rgba(50,110,141,1)","rgba(43,128,141,1)","rgba(42,149,136,1)","rgba(159,215,73,1)","rgba(38,161,134,1)","rgba(43,170,130,1)","rgba(69,50,118,1)","rgba(55,103,140,1)","rgba(78,182,116,1)","rgba(66,177,122,1)","rgba(37,163,133,1)","rgba(51,110,141,1)","rgba(43,142,138,1)","rgba(42,170,130,1)","rgba(60,92,138,1)","rgba(120,208,83,1)","rgba(50,172,128,1)","rgba(76,181,117,1)","rgba(54,173,127,1)","rgba(44,134,139,1)","rgba(44,131,140,1)","rgba(55,103,140,1)","rgba(70,34,104,1)","rgba(43,141,138,1)","rgba(43,129,140,1)","rgba(43,146,137,1)","rgba(121,209,82,1)","rgba(44,135,139,1)","rgba(68,56,124,1)","rgba(39,158,134,1)","rgba(42,149,136,1)","rgba(59,93,139,1)","rgba(44,170,130,1)","rgba(44,134,139,1)","rgba(44,138,139,1)","rgba(41,153,136,1)","rgba(70,29,100,1)","rgba(41,153,135,1)","rgba(69,18,93,1)","rgba(51,172,127,1)","rgba(41,152,136,1)","rgba(43,140,138,1)","rgba(42,120,142,1)","rgba(43,144,137,1)","rgba(48,171,129,1)","rgba(49,113,141,1)","rgba(50,111,141,1)","rgba(38,169,131,1)","rgba(43,140,138,1)","rgba(43,141,138,1)","rgba(84,185,113,1)","rgba(37,163,133,1)","rgba(63,81,137,1)","rgba(43,145,137,1)","rgba(70,31,101,1)","rgba(63,80,137,1)","rgba(58,96,139,1)","rgba(54,105,140,1)","rgba(43,128,141,1)","rgba(37,164,133,1)","rgba(65,68,135,1)","rgba(94,191,106,1)","rgba(43,128,141,1)","rgba(43,127,141,1)","rgba(43,127,141,1)","rgba(93,190,106,1)","rgba(40,155,135,1)","rgba(82,184,114,1)","rgba(43,146,137,1)","rgba(43,119,142,1)","rgba(54,104,140,1)","rgba(44,135,139,1)","rgba(40,155,135,1)","rgba(37,163,133,1)","rgba(39,158,134,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(82,184,114,1)","rgba(43,140,138,1)","rgba(42,149,136,1)","rgba(43,123,142,1)","rgba(44,136,139,1)","rgba(69,46,114,1)","rgba(43,145,137,1)","rgba(44,131,140,1)","rgba(55,173,126,1)","rgba(44,138,139,1)","rgba(69,16,91,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(44,139,138,1)","rgba(43,119,142,1)","rgba(42,120,142,1)","rgba(44,133,140,1)","rgba(45,116,142,1)","rgba(44,133,140,1)","rgba(35,166,132,1)","rgba(43,143,138,1)","rgba(42,148,137,1)","rgba(67,178,121,1)","rgba(44,133,140,1)","rgba(47,171,129,1)","rgba(35,166,132,1)","rgba(47,171,129,1)","rgba(53,106,140,1)","rgba(108,200,95,1)","rgba(59,94,139,1)","rgba(39,158,134,1)","rgba(40,155,135,1)","rgba(37,164,133,1)","rgba(70,27,99,1)","rgba(43,119,142,1)","rgba(44,132,140,1)","rgba(54,104,140,1)","rgba(68,55,123,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(40,157,135,1)","rgba(54,105,140,1)","rgba(42,121,142,1)","rgba(59,94,139,1)","rgba(69,24,97,1)","rgba(44,134,139,1)","rgba(56,101,140,1)","rgba(42,120,142,1)","rgba(51,109,141,1)","rgba(72,180,119,1)","rgba(43,144,137,1)","rgba(43,125,141,1)","rgba(70,30,101,1)","rgba(38,161,134,1)","rgba(43,141,138,1)","rgba(41,153,135,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(43,128,140,1)","rgba(84,185,113,1)","rgba(64,176,123,1)","rgba(80,183,115,1)","rgba(49,112,141,1)","rgba(39,159,134,1)","rgba(57,99,139,1)","rgba(42,150,136,1)","rgba(43,144,137,1)","rgba(64,177,123,1)","rgba(41,153,135,1)","rgba(130,210,79,1)","rgba(50,111,141,1)","rgba(70,31,102,1)","rgba(44,135,139,1)","rgba(68,55,122,1)","rgba(59,94,139,1)","rgba(54,173,127,1)","rgba(43,143,138,1)","rgba(92,189,108,1)","rgba(51,109,141,1)","rgba(41,154,135,1)","rgba(69,178,120,1)","rgba(42,148,137,1)","rgba(56,101,140,1)","rgba(44,131,140,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(40,156,135,1)","rgba(44,138,139,1)","rgba(44,137,139,1)","rgba(40,156,135,1)","rgba(40,156,135,1)","rgba(66,177,122,1)","rgba(44,133,140,1)","rgba(43,140,138,1)","rgba(43,142,138,1)","rgba(43,146,137,1)","rgba(55,103,140,1)","rgba(67,178,121,1)","rgba(41,152,136,1)","rgba(44,131,140,1)","rgba(70,36,105,1)","rgba(44,131,140,1)","rgba(44,133,140,1)","rgba(39,159,134,1)","rgba(41,154,135,1)","rgba(44,132,140,1)","rgba(52,108,141,1)","rgba(40,155,135,1)","rgba(37,169,131,1)","rgba(43,146,137,1)","rgba(42,122,142,1)","rgba(43,123,142,1)","rgba(51,109,141,1)","rgba(43,144,137,1)","rgba(47,115,141,1)","rgba(41,170,130,1)","rgba(44,134,139,1)","rgba(149,213,75,1)","rgba(43,127,141,1)","rgba(42,120,142,1)","rgba(62,86,137,1)","rgba(65,177,122,1)","rgba(39,158,134,1)","rgba(38,161,134,1)","rgba(46,115,141,1)","rgba(57,98,139,1)","rgba(46,115,141,1)","rgba(42,120,142,1)","rgba(44,132,140,1)","rgba(42,121,142,1)","rgba(69,49,117,1)","rgba(36,165,133,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(76,181,117,1)","rgba(43,146,137,1)","rgba(43,144,137,1)","rgba(43,143,138,1)","rgba(43,123,141,1)","rgba(53,173,127,1)","rgba(44,133,140,1)","rgba(58,97,139,1)","rgba(88,187,110,1)","rgba(43,140,138,1)","rgba(43,129,140,1)","rgba(51,109,141,1)","rgba(43,140,138,1)","rgba(36,164,133,1)","rgba(64,77,136,1)","rgba(69,178,121,1)","rgba(63,79,137,1)","rgba(43,140,138,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(61,89,138,1)","rgba(70,38,107,1)","rgba(97,192,104,1)","rgba(43,143,138,1)","rgba(38,161,134,1)","rgba(37,164,133,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(43,140,138,1)","rgba(42,122,142,1)","rgba(56,100,139,1)","rgba(40,157,134,1)","rgba(69,46,115,1)","rgba(44,134,139,1)","rgba(62,85,137,1)","rgba(131,210,79,1)","rgba(44,136,139,1)","rgba(39,159,134,1)","rgba(49,113,141,1)","rgba(44,131,140,1)","rgba(35,167,132,1)","rgba(43,143,138,1)","rgba(42,151,136,1)","rgba(42,149,136,1)","rgba(57,99,139,1)","rgba(121,208,82,1)","rgba(121,209,82,1)","rgba(37,164,133,1)","rgba(43,146,137,1)","rgba(70,41,110,1)","rgba(39,159,134,1)","rgba(69,9,88,1)","rgba(51,109,141,1)","rgba(59,94,139,1)","rgba(43,147,137,1)","rgba(44,136,139,1)","rgba(44,131,140,1)","rgba(39,159,134,1)","rgba(38,161,134,1)","rgba(43,147,137,1)","rgba(48,113,141,1)","rgba(104,197,98,1)","rgba(40,155,135,1)","rgba(41,151,136,1)","rgba(38,161,134,1)","rgba(46,116,141,1)","rgba(42,150,136,1)","rgba(43,145,137,1)","rgba(41,154,135,1)","rgba(39,159,134,1)","rgba(43,142,138,1)","rgba(43,129,140,1)","rgba(60,90,138,1)","rgba(39,158,134,1)","rgba(51,109,141,1)","rgba(43,141,138,1)","rgba(88,187,110,1)","rgba(67,178,121,1)","rgba(105,197,97,1)","rgba(37,162,133,1)","rgba(41,154,135,1)","rgba(67,60,127,1)","rgba(43,141,138,1)","rgba(46,116,142,1)","rgba(78,182,116,1)","rgba(43,143,138,1)","rgba(44,135,139,1)","rgba(68,2,85,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(41,154,135,1)","rgba(97,193,104,1)","rgba(70,27,99,1)","rgba(55,103,140,1)","rgba(60,175,124,1)","rgba(43,128,140,1)","rgba(80,183,115,1)","rgba(69,11,89,1)","rgba(92,189,108,1)","rgba(41,152,136,1)","rgba(51,172,127,1)","rgba(58,97,139,1)","rgba(37,164,133,1)","rgba(37,164,133,1)","rgba(67,62,129,1)","rgba(66,63,130,1)","rgba(43,126,141,1)","rgba(58,97,139,1)","rgba(53,106,140,1)","rgba(35,167,132,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(53,106,140,1)","rgba(44,135,139,1)","rgba(47,115,141,1)","rgba(41,153,135,1)","rgba(92,189,108,1)","rgba(104,196,99,1)","rgba(90,188,109,1)","rgba(44,136,139,1)","rgba(68,178,121,1)","rgba(70,37,107,1)","rgba(46,116,142,1)","rgba(38,161,134,1)","rgba(71,179,119,1)","rgba(42,148,137,1)","rgba(41,154,135,1)","rgba(72,180,119,1)","rgba(68,3,85,1)","rgba(42,148,136,1)","rgba(43,146,137,1)","rgba(43,146,137,1)","rgba(132,211,79,1)","rgba(37,163,133,1)","rgba(41,154,135,1)","rgba(44,139,138,1)","rgba(70,35,105,1)","rgba(41,151,136,1)","rgba(40,156,135,1)","rgba(54,173,127,1)","rgba(67,177,122,1)","rgba(43,143,138,1)","rgba(39,159,134,1)","rgba(43,144,137,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(43,147,137,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(147,213,76,1)","rgba(44,131,140,1)","rgba(48,114,141,1)","rgba(36,165,133,1)","rgba(39,160,134,1)","rgba(44,131,140,1)","rgba(42,151,136,1)","rgba(69,25,97,1)","rgba(44,137,139,1)","rgba(69,14,91,1)","rgba(39,160,134,1)","rgba(38,162,133,1)","rgba(43,123,141,1)","rgba(44,133,140,1)","rgba(41,154,135,1)","rgba(41,153,135,1)","rgba(43,126,141,1)","rgba(44,131,140,1)","rgba(108,199,95,1)","rgba(40,155,135,1)","rgba(43,126,141,1)","rgba(47,114,141,1)","rgba(38,162,134,1)","rgba(39,160,134,1)","rgba(42,147,137,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(43,145,137,1)","rgba(79,183,115,1)","rgba(45,116,142,1)","rgba(43,143,138,1)","rgba(41,154,135,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(63,176,123,1)","rgba(43,142,138,1)","rgba(43,144,137,1)","rgba(36,165,133,1)","rgba(42,148,137,1)","rgba(35,166,132,1)","rgba(34,168,132,1)","rgba(60,92,138,1)","rgba(44,131,140,1)","rgba(64,76,136,1)","rgba(34,168,132,1)","rgba(44,132,140,1)","rgba(43,143,138,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(37,163,133,1)","rgba(69,47,115,1)","rgba(52,108,141,1)","rgba(62,84,137,1)","rgba(43,130,140,1)","rgba(43,125,141,1)","rgba(46,116,142,1)","rgba(40,155,135,1)","rgba(61,88,138,1)","rgba(61,87,138,1)","rgba(40,156,135,1)","rgba(50,111,141,1)","rgba(70,179,120,1)","rgba(38,161,134,1)","rgba(70,37,107,1)","rgba(43,123,142,1)","rgba(44,139,138,1)","rgba(43,130,140,1)","rgba(63,176,123,1)","rgba(65,71,135,1)","rgba(107,199,96,1)","rgba(44,132,140,1)","rgba(50,111,141,1)","rgba(42,151,136,1)","rgba(49,113,141,1)","rgba(52,172,127,1)","rgba(42,149,136,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(46,116,141,1)","rgba(44,136,139,1)","rgba(42,121,142,1)","rgba(45,116,142,1)","rgba(41,152,136,1)","rgba(43,145,137,1)","rgba(69,16,91,1)","rgba(70,26,98,1)","rgba(52,108,141,1)","rgba(62,83,137,1)","rgba(68,57,124,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(42,121,142,1)","rgba(90,189,109,1)","rgba(44,137,139,1)","rgba(44,138,139,1)","rgba(89,188,110,1)","rgba(42,121,142,1)","rgba(58,174,125,1)","rgba(41,154,135,1)","rgba(42,147,137,1)","rgba(41,154,135,1)","rgba(55,102,140,1)","rgba(42,121,142,1)","rgba(44,131,140,1)","rgba(42,148,137,1)","rgba(90,188,109,1)","rgba(43,129,140,1)","rgba(43,141,138,1)","rgba(43,119,142,1)","rgba(42,150,136,1)","rgba(42,120,142,1)","rgba(41,155,135,1)","rgba(43,124,141,1)","rgba(41,153,135,1)","rgba(68,58,126,1)","rgba(41,153,136,1)","rgba(76,182,117,1)","rgba(40,155,135,1)","rgba(34,168,132,1)","rgba(54,104,140,1)","rgba(43,139,138,1)","rgba(39,158,134,1)","rgba(43,146,137,1)","rgba(39,160,134,1)","rgba(41,153,135,1)","rgba(42,151,136,1)","rgba(44,135,139,1)","rgba(43,141,138,1)","rgba(43,130,140,1)","rgba(43,126,141,1)","rgba(44,133,140,1)","rgba(36,164,133,1)","rgba(43,129,140,1)","rgba(57,98,139,1)","rgba(58,95,139,1)","rgba(44,138,139,1)","rgba(69,23,96,1)","rgba(69,45,113,1)","rgba(63,78,136,1)","rgba(44,118,142,1)","rgba(43,127,141,1)","rgba(43,147,137,1)","rgba(75,181,118,1)","rgba(43,123,141,1)","rgba(44,134,139,1)","rgba(57,100,139,1)","rgba(63,82,137,1)","rgba(40,155,135,1)","rgba(56,100,139,1)","rgba(69,49,117,1)","rgba(44,136,139,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(39,160,134,1)","rgba(43,124,141,1)","rgba(47,171,129,1)","rgba(43,142,138,1)","rgba(43,127,141,1)","rgba(49,112,141,1)","rgba(43,128,141,1)","rgba(43,140,138,1)","rgba(44,132,140,1)","rgba(44,137,139,1)","rgba(51,110,141,1)","rgba(39,158,134,1)","rgba(53,173,127,1)","rgba(51,172,128,1)","rgba(51,109,141,1)","rgba(44,139,138,1)","rgba(42,120,142,1)","rgba(43,147,137,1)","rgba(41,152,136,1)","rgba(78,182,116,1)","rgba(39,159,134,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(122,209,81,1)","rgba(42,151,136,1)","rgba(62,84,137,1)","rgba(39,158,134,1)","rgba(43,139,138,1)","rgba(42,147,137,1)","rgba(35,167,132,1)","rgba(46,115,141,1)","rgba(42,148,137,1)","rgba(50,172,128,1)","rgba(53,106,140,1)","rgba(69,46,115,1)","rgba(43,129,140,1)","rgba(43,128,140,1)","rgba(43,143,138,1)","rgba(67,59,126,1)","rgba(44,133,140,1)","rgba(41,154,135,1)","rgba(43,127,141,1)","rgba(36,165,133,1)","rgba(53,107,140,1)","rgba(57,100,139,1)","rgba(35,166,133,1)","rgba(39,158,134,1)","rgba(44,138,139,1)","rgba(44,137,139,1)","rgba(41,155,135,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(108,199,95,1)","rgba(36,164,133,1)","rgba(44,132,140,1)","rgba(40,156,135,1)","rgba(42,150,136,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(42,149,136,1)","rgba(70,38,108,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(50,111,141,1)","rgba(44,132,140,1)","rgba(42,151,136,1)","rgba(60,90,138,1)","rgba(49,113,141,1)","rgba(61,89,138,1)","rgba(41,153,135,1)","rgba(39,158,134,1)","rgba(171,217,70,1)","rgba(69,43,111,1)","rgba(43,124,141,1)","rgba(42,170,130,1)","rgba(43,124,141,1)","rgba(43,147,137,1)","rgba(43,140,138,1)","rgba(44,137,139,1)","rgba(54,104,140,1)","rgba(68,54,121,1)","rgba(43,129,140,1)","rgba(54,105,140,1)","rgba(60,91,138,1)","rgba(44,135,139,1)","rgba(51,109,141,1)","rgba(43,146,137,1)","rgba(43,124,141,1)","rgba(40,156,135,1)","rgba(58,95,139,1)","rgba(43,130,140,1)","rgba(44,132,140,1)","rgba(42,120,142,1)","rgba(49,111,141,1)","rgba(44,118,142,1)","rgba(38,160,134,1)","rgba(44,139,138,1)","rgba(49,112,141,1)","rgba(42,122,142,1)","rgba(117,205,87,1)","rgba(43,142,138,1)","rgba(43,128,141,1)","rgba(38,162,133,1)","rgba(49,112,141,1)","rgba(79,183,115,1)","rgba(44,133,140,1)","rgba(80,183,115,1)","rgba(55,103,140,1)","rgba(63,82,137,1)","rgba(70,27,99,1)","rgba(42,122,142,1)","rgba(57,98,139,1)","rgba(41,155,135,1)","rgba(35,166,132,1)","rgba(67,60,127,1)","rgba(43,147,137,1)","rgba(42,147,137,1)","rgba(38,162,133,1)","rgba(50,110,141,1)","rgba(43,128,141,1)","rgba(64,73,136,1)","rgba(42,120,142,1)","rgba(52,107,140,1)","rgba(40,156,135,1)","rgba(42,121,142,1)","rgba(62,176,123,1)","rgba(44,133,140,1)","rgba(70,25,97,1)","rgba(41,152,136,1)","rgba(42,147,137,1)","rgba(70,37,106,1)","rgba(43,127,141,1)","rgba(43,139,138,1)","rgba(45,116,142,1)","rgba(41,154,135,1)","rgba(69,45,114,1)","rgba(45,171,129,1)","rgba(44,132,140,1)","rgba(45,117,142,1)","rgba(44,138,139,1)","rgba(62,85,137,1)","rgba(44,138,139,1)","rgba(54,105,140,1)","rgba(60,92,138,1)","rgba(43,146,137,1)","rgba(43,129,140,1)","rgba(43,143,138,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(63,83,137,1)","rgba(70,36,105,1)","rgba(39,159,134,1)","rgba(44,118,142,1)","rgba(95,191,105,1)","rgba(44,170,130,1)","rgba(90,188,109,1)","rgba(59,175,125,1)","rgba(44,137,139,1)","rgba(65,177,122,1)","rgba(64,75,136,1)","rgba(44,138,139,1)","rgba(63,78,136,1)","rgba(56,174,126,1)","rgba(43,144,137,1)","rgba(52,173,127,1)","rgba(61,88,138,1)","rgba(70,38,108,1)","rgba(43,139,138,1)","rgba(44,133,140,1)","rgba(56,174,126,1)","rgba(74,181,118,1)","rgba(44,170,130,1)","rgba(43,126,141,1)","rgba(44,138,139,1)","rgba(36,164,133,1)","rgba(43,145,137,1)","rgba(43,140,138,1)","rgba(44,133,140,1)","rgba(45,117,142,1)","rgba(43,140,138,1)","rgba(41,154,135,1)","rgba(42,150,136,1)","rgba(35,166,132,1)","rgba(43,123,141,1)","rgba(60,92,138,1)","rgba(58,96,139,1)","rgba(56,102,140,1)","rgba(43,142,138,1)","rgba(39,159,134,1)","rgba(55,103,140,1)","rgba(44,135,139,1)","rgba(43,123,141,1)","rgba(43,123,141,1)","rgba(62,85,137,1)","rgba(69,13,90,1)","rgba(43,123,142,1)","rgba(59,175,125,1)","rgba(66,177,122,1)","rgba(43,145,137,1)","rgba(49,113,141,1)","rgba(68,7,87,1)","rgba(114,203,90,1)","rgba(43,143,138,1)","rgba(42,151,136,1)","rgba(46,115,141,1)","rgba(43,144,137,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(43,125,141,1)","rgba(59,175,125,1)","rgba(44,138,139,1)","rgba(57,100,139,1)","rgba(44,135,139,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(44,134,139,1)","rgba(47,115,141,1)","rgba(70,29,100,1)","rgba(44,134,139,1)","rgba(65,71,135,1)","rgba(50,111,141,1)","rgba(43,141,138,1)","rgba(41,154,135,1)","rgba(57,174,125,1)","rgba(69,46,115,1)","rgba(51,109,141,1)","rgba(70,31,101,1)","rgba(43,124,141,1)","rgba(49,172,128,1)","rgba(99,194,102,1)","rgba(42,150,136,1)","rgba(40,157,135,1)","rgba(64,73,136,1)","rgba(42,148,137,1)","rgba(39,159,134,1)","rgba(43,128,141,1)","rgba(41,154,135,1)","rgba(44,132,140,1)","rgba(64,78,136,1)","rgba(49,172,128,1)","rgba(88,187,110,1)","rgba(70,30,101,1)","rgba(60,91,138,1)","rgba(79,183,115,1)","rgba(45,117,142,1)","rgba(43,143,138,1)","rgba(56,101,140,1)","rgba(42,121,142,1)","rgba(38,160,134,1)","rgba(43,145,137,1)","rgba(43,127,141,1)","rgba(44,137,139,1)","rgba(56,101,140,1)","rgba(43,128,140,1)","rgba(42,151,136,1)","rgba(52,107,140,1)","rgba(43,127,141,1)","rgba(49,113,141,1)","rgba(42,150,136,1)","rgba(36,164,133,1)","rgba(45,117,142,1)","rgba(186,220,66,1)","rgba(43,124,141,1)","rgba(49,112,141,1)","rgba(38,169,131,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(48,114,141,1)","rgba(43,145,137,1)","rgba(59,95,139,1)","rgba(44,132,140,1)","rgba(36,164,133,1)","rgba(50,111,141,1)","rgba(43,127,141,1)","rgba(90,188,109,1)","rgba(65,67,134,1)","rgba(53,106,140,1)","rgba(90,188,109,1)","rgba(37,162,133,1)","rgba(44,131,140,1)","rgba(43,142,138,1)","rgba(51,109,141,1)","rgba(43,123,141,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(43,142,138,1)","rgba(40,157,135,1)","rgba(61,175,124,1)","rgba(163,216,72,1)","rgba(44,135,139,1)","rgba(42,147,137,1)","rgba(43,129,140,1)","rgba(43,142,138,1)","rgba(78,182,116,1)","rgba(44,136,139,1)","rgba(57,174,125,1)","rgba(46,115,141,1)","rgba(39,158,134,1)","rgba(70,41,109,1)","rgba(61,86,138,1)","rgba(41,153,136,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(64,176,123,1)","rgba(46,116,141,1)","rgba(48,113,141,1)","rgba(34,167,132,1)","rgba(43,143,138,1)","rgba(46,116,142,1)","rgba(55,102,140,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(39,159,134,1)","rgba(64,77,136,1)","rgba(51,172,127,1)","rgba(43,129,140,1)","rgba(52,107,140,1)","rgba(49,112,141,1)","rgba(53,105,140,1)","rgba(46,116,141,1)","rgba(44,136,139,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(69,44,113,1)","rgba(43,146,137,1)","rgba(44,136,139,1)","rgba(51,110,141,1)","rgba(37,164,133,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(67,62,129,1)","rgba(51,110,141,1)","rgba(69,11,89,1)","rgba(48,114,141,1)","rgba(51,110,141,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(44,137,139,1)","rgba(43,128,141,1)","rgba(54,173,127,1)","rgba(68,56,123,1)","rgba(37,169,131,1)","rgba(42,151,136,1)","rgba(66,177,122,1)","rgba(54,105,140,1)","rgba(53,106,140,1)","rgba(40,157,134,1)","rgba(42,122,142,1)","rgba(34,167,132,1)","rgba(36,164,133,1)","rgba(36,164,133,1)","rgba(43,147,137,1)","rgba(43,146,137,1)","rgba(43,144,137,1)","rgba(76,182,117,1)","rgba(40,156,135,1)","rgba(43,128,141,1)","rgba(49,112,141,1)","rgba(43,124,141,1)","rgba(69,46,114,1)","rgba(46,116,142,1)","rgba(35,167,132,1)","rgba(71,179,119,1)","rgba(42,149,136,1)","rgba(39,160,134,1)","rgba(53,106,140,1)","rgba(77,182,116,1)","rgba(44,135,139,1)","rgba(43,125,141,1)","rgba(96,192,105,1)","rgba(43,170,130,1)","rgba(42,149,136,1)","rgba(46,116,141,1)","rgba(56,100,139,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(40,156,135,1)","rgba(43,141,138,1)","rgba(42,122,142,1)","rgba(44,138,139,1)","rgba(43,126,141,1)","rgba(69,43,112,1)","rgba(69,178,120,1)","rgba(73,180,119,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(41,152,136,1)","rgba(53,106,140,1)","rgba(43,139,138,1)","rgba(44,134,139,1)","rgba(44,137,139,1)","rgba(41,152,136,1)","rgba(42,121,142,1)","rgba(43,126,141,1)","rgba(44,133,140,1)","rgba(44,135,139,1)","rgba(41,154,135,1)","rgba(44,134,139,1)","rgba(50,110,141,1)","rgba(49,112,141,1)","rgba(43,123,141,1)","rgba(44,139,138,1)","rgba(68,55,123,1)","rgba(51,110,141,1)","rgba(40,169,131,1)","rgba(47,114,141,1)","rgba(43,146,137,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(44,139,138,1)","rgba(50,172,128,1)","rgba(56,100,140,1)","rgba(43,125,141,1)","rgba(69,25,97,1)","rgba(43,141,138,1)","rgba(42,149,136,1)","rgba(43,145,137,1)","rgba(53,106,140,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(43,143,138,1)","rgba(42,121,142,1)","rgba(43,145,137,1)","rgba(44,118,142,1)","rgba(69,11,89,1)","rgba(39,159,134,1)","rgba(70,33,103,1)","rgba(43,119,142,1)","rgba(45,117,142,1)","rgba(35,166,133,1)","rgba(40,156,135,1)","rgba(40,157,135,1)","rgba(69,51,118,1)","rgba(192,221,64,1)","rgba(44,137,139,1)","rgba(42,122,142,1)","rgba(68,55,123,1)","rgba(42,149,136,1)","rgba(43,146,137,1)","rgba(42,148,137,1)","rgba(42,150,136,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(65,68,135,1)","rgba(43,143,138,1)","rgba(67,177,122,1)","rgba(43,124,141,1)","rgba(42,147,137,1)","rgba(81,184,114,1)","rgba(44,135,139,1)","rgba(42,122,142,1)","rgba(82,184,114,1)","rgba(42,120,142,1)","rgba(43,124,141,1)","rgba(43,129,140,1)","rgba(40,156,135,1)","rgba(49,112,141,1)","rgba(47,171,129,1)","rgba(39,159,134,1)","rgba(44,133,140,1)","rgba(41,153,135,1)","rgba(35,166,132,1)","rgba(51,110,141,1)","rgba(40,155,135,1)","rgba(53,106,140,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(67,59,126,1)","rgba(35,167,132,1)","rgba(42,120,142,1)","rgba(62,86,138,1)","rgba(39,159,134,1)","rgba(42,120,142,1)","rgba(41,152,136,1)","rgba(58,95,139,1)","rgba(43,144,137,1)","rgba(43,145,137,1)","rgba(44,136,139,1)","rgba(69,52,120,1)","rgba(44,136,139,1)","rgba(63,78,136,1)","rgba(92,190,107,1)","rgba(40,169,131,1)","rgba(52,107,140,1)","rgba(43,145,137,1)","rgba(44,135,139,1)","rgba(61,87,138,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(40,155,135,1)","rgba(58,96,139,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(105,198,97,1)","rgba(44,133,140,1)","rgba(54,104,140,1)","rgba(86,186,111,1)","rgba(42,150,136,1)","rgba(42,121,142,1)","rgba(38,160,134,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(36,165,133,1)","rgba(50,172,128,1)","rgba(60,175,124,1)","rgba(41,152,136,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(40,157,135,1)","rgba(44,132,140,1)","rgba(50,111,141,1)","rgba(66,64,131,1)","rgba(44,136,139,1)","rgba(69,19,93,1)","rgba(43,125,141,1)","rgba(51,110,141,1)","rgba(77,182,116,1)","rgba(42,120,142,1)","rgba(58,96,139,1)","rgba(44,118,142,1)","rgba(53,106,140,1)","rgba(34,167,132,1)","rgba(44,137,139,1)","rgba(46,116,142,1)","rgba(54,104,140,1)","rgba(67,62,129,1)","rgba(43,123,141,1)","rgba(69,17,92,1)","rgba(40,156,135,1)","rgba(42,150,136,1)","rgba(44,133,140,1)","rgba(43,144,137,1)","rgba(46,171,129,1)","rgba(43,130,140,1)","rgba(44,139,139,1)","rgba(62,85,137,1)","rgba(43,129,140,1)","rgba(41,154,135,1)","rgba(62,84,137,1)","rgba(63,82,137,1)","rgba(54,105,140,1)","rgba(43,144,137,1)","rgba(43,142,138,1)","rgba(42,149,136,1)","rgba(44,135,139,1)","rgba(119,207,84,1)","rgba(41,153,135,1)","rgba(42,151,136,1)","rgba(43,142,138,1)","rgba(35,167,132,1)","rgba(40,156,135,1)","rgba(63,79,137,1)","rgba(42,148,137,1)","rgba(43,141,138,1)","rgba(42,120,142,1)","rgba(87,187,110,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(42,151,136,1)","rgba(54,104,140,1)","rgba(42,148,137,1)","rgba(57,98,139,1)","rgba(43,124,141,1)","rgba(43,129,140,1)","rgba(43,144,137,1)","rgba(65,69,135,1)","rgba(43,130,140,1)","rgba(42,150,136,1)","rgba(70,28,99,1)","rgba(44,131,140,1)","rgba(51,109,141,1)","rgba(50,111,141,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(60,90,138,1)","rgba(41,154,135,1)","rgba(43,145,137,1)","rgba(44,133,140,1)","rgba(44,136,139,1)","rgba(57,98,139,1)","rgba(43,125,141,1)","rgba(42,148,137,1)","rgba(69,11,89,1)","rgba(43,143,138,1)","rgba(44,118,142,1)","rgba(62,86,138,1)","rgba(36,164,133,1)","rgba(43,130,140,1)","rgba(39,159,134,1)","rgba(69,11,89,1)","rgba(44,133,140,1)","rgba(53,106,140,1)","rgba(49,112,141,1)","rgba(68,58,125,1)","rgba(38,161,134,1)","rgba(44,139,138,1)","rgba(59,93,138,1)","rgba(38,162,134,1)","rgba(51,172,128,1)","rgba(36,164,133,1)","rgba(55,103,140,1)","rgba(56,100,139,1)","rgba(43,142,138,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(35,166,133,1)","rgba(42,150,136,1)","rgba(38,161,134,1)","rgba(41,155,135,1)","rgba(43,141,138,1)","rgba(51,172,127,1)","rgba(43,140,138,1)","rgba(41,153,136,1)","rgba(35,166,132,1)","rgba(108,199,95,1)","rgba(46,116,142,1)","rgba(43,128,141,1)","rgba(68,54,121,1)","rgba(65,69,135,1)","rgba(52,107,140,1)","rgba(42,150,136,1)","rgba(50,172,128,1)","rgba(89,188,109,1)","rgba(63,80,137,1)","rgba(63,80,137,1)","rgba(69,50,118,1)","rgba(43,145,137,1)","rgba(61,88,138,1)","rgba(43,124,141,1)","rgba(43,124,141,1)","rgba(70,34,104,1)","rgba(58,96,139,1)","rgba(60,91,138,1)","rgba(44,132,140,1)","rgba(64,77,136,1)","rgba(61,87,138,1)","rgba(70,40,109,1)","rgba(68,7,87,1)","rgba(69,12,90,1)","rgba(67,59,126,1)","rgba(44,138,139,1)","rgba(55,103,140,1)","rgba(58,96,139,1)","rgba(48,114,141,1)","rgba(61,89,138,1)","rgba(63,82,137,1)","rgba(43,142,138,1)","rgba(69,44,113,1)","rgba(43,123,141,1)","rgba(70,30,101,1)","rgba(63,80,137,1)","rgba(41,154,135,1)","rgba(41,154,135,1)","rgba(65,69,135,1)","rgba(42,121,142,1)","rgba(43,124,141,1)","rgba(43,130,140,1)","rgba(41,170,130,1)","rgba(61,176,124,1)","rgba(43,124,141,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(53,107,140,1)","rgba(49,172,128,1)","rgba(45,117,142,1)","rgba(76,181,117,1)","rgba(44,137,139,1)","rgba(70,41,110,1)","rgba(42,149,136,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(66,64,131,1)","rgba(53,105,140,1)","rgba(41,152,136,1)","rgba(39,158,134,1)","rgba(43,145,137,1)","rgba(43,124,141,1)","rgba(43,145,137,1)","rgba(66,65,132,1)","rgba(86,186,112,1)","rgba(103,196,100,1)","rgba(44,133,140,1)","rgba(56,174,126,1)","rgba(38,162,133,1)","rgba(42,149,136,1)","rgba(57,98,139,1)","rgba(43,128,141,1)","rgba(55,102,140,1)","rgba(68,5,86,1)","rgba(42,148,136,1)","rgba(36,164,133,1)","rgba(40,155,135,1)","rgba(68,54,121,1)","rgba(43,141,138,1)","rgba(43,130,140,1)","rgba(44,132,140,1)","rgba(52,172,127,1)","rgba(43,145,137,1)","rgba(54,105,140,1)","rgba(50,111,141,1)","rgba(39,159,134,1)","rgba(129,210,80,1)","rgba(42,122,142,1)","rgba(46,116,141,1)","rgba(43,143,138,1)","rgba(100,194,102,1)","rgba(41,152,136,1)","rgba(62,84,137,1)","rgba(43,142,138,1)","rgba(55,103,140,1)","rgba(62,86,138,1)","rgba(67,62,129,1)","rgba(70,42,110,1)","rgba(70,35,105,1)","rgba(45,117,142,1)","rgba(65,68,135,1)","rgba(62,85,137,1)","rgba(64,72,136,1)","rgba(70,30,101,1)","rgba(69,43,111,1)","rgba(70,42,111,1)","rgba(69,20,94,1)","rgba(69,53,120,1)","rgba(42,121,142,1)","rgba(65,70,135,1)","rgba(44,131,140,1)","rgba(75,181,117,1)","rgba(68,6,86,1)","rgba(66,63,130,1)","rgba(70,38,107,1)","rgba(65,72,136,1)","rgba(63,80,137,1)","rgba(64,76,136,1)","rgba(42,122,142,1)","rgba(64,73,136,1)","rgba(43,130,140,1)","rgba(59,93,139,1)","rgba(43,124,141,1)","rgba(69,48,116,1)","rgba(70,40,109,1)","rgba(47,115,141,1)","rgba(54,104,140,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(38,160,134,1)","rgba(46,171,129,1)","rgba(46,116,141,1)","rgba(44,131,140,1)","rgba(69,16,91,1)","rgba(43,129,140,1)","rgba(43,142,138,1)","rgba(41,154,135,1)","rgba(68,55,122,1)","rgba(56,100,140,1)","rgba(69,48,116,1)","rgba(43,127,141,1)","rgba(53,107,140,1)","rgba(44,134,139,1)","rgba(50,110,141,1)","rgba(69,50,118,1)","rgba(70,33,103,1)","rgba(43,143,138,1)","rgba(43,147,137,1)","rgba(35,166,132,1)","rgba(69,25,97,1)","rgba(70,29,100,1)","rgba(43,123,141,1)","rgba(58,97,139,1)","rgba(69,44,113,1)","rgba(69,48,116,1)","rgba(43,145,137,1)","rgba(54,173,126,1)","rgba(70,32,103,1)","rgba(115,204,88,1)","rgba(43,126,141,1)","rgba(43,128,141,1)","rgba(62,85,137,1)","rgba(121,208,82,1)","rgba(43,119,142,1)","rgba(58,96,139,1)","rgba(70,35,105,1)","rgba(43,130,140,1)","rgba(64,77,136,1)","rgba(62,85,137,1)","rgba(42,151,136,1)","rgba(63,80,137,1)","rgba(68,7,87,1)","rgba(68,2,85,1)","rgba(56,101,140,1)","rgba(64,74,136,1)","rgba(42,147,137,1)","rgba(44,132,140,1)","rgba(43,147,137,1)","rgba(62,83,137,1)","rgba(44,131,140,1)","rgba(38,161,134,1)","rgba(49,113,141,1)","rgba(62,83,137,1)","rgba(69,22,95,1)","rgba(69,13,90,1)","rgba(43,126,141,1)","rgba(69,43,112,1)","rgba(43,143,138,1)","rgba(68,2,84,1)","rgba(70,32,103,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(68,54,122,1)","rgba(44,136,139,1)","rgba(42,120,142,1)","rgba(41,153,136,1)","rgba(43,130,140,1)","rgba(55,102,140,1)","rgba(44,132,140,1)","rgba(69,43,111,1)","rgba(41,152,136,1)","rgba(43,144,137,1)","rgba(38,162,133,1)","rgba(43,140,138,1)","rgba(44,118,142,1)","rgba(70,38,108,1)","rgba(40,157,135,1)","rgba(69,48,116,1)","rgba(70,35,105,1)","rgba(57,174,125,1)","rgba(43,143,138,1)","rgba(43,119,142,1)","rgba(42,149,136,1)","rgba(43,130,140,1)","rgba(63,80,137,1)","rgba(43,146,137,1)","rgba(63,82,137,1)","rgba(39,160,134,1)","rgba(48,113,141,1)","rgba(60,91,138,1)","rgba(43,125,141,1)","rgba(147,213,76,1)","rgba(68,4,85,1)","rgba(40,169,131,1)","rgba(54,173,126,1)","rgba(44,133,140,1)","rgba(69,12,89,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(61,87,138,1)","rgba(45,117,142,1)","rgba(44,118,142,1)","rgba(59,93,139,1)","rgba(52,108,140,1)","rgba(68,56,123,1)","rgba(45,117,142,1)","rgba(56,101,140,1)","rgba(53,105,140,1)","rgba(44,136,139,1)","rgba(69,10,88,1)","rgba(36,165,133,1)","rgba(55,103,140,1)","rgba(47,115,141,1)","rgba(43,128,141,1)","rgba(70,41,110,1)","rgba(69,43,112,1)","rgba(70,42,110,1)","rgba(44,135,139,1)","rgba(45,116,142,1)","rgba(45,117,142,1)","rgba(62,84,137,1)","rgba(43,124,141,1)","rgba(59,94,139,1)","rgba(44,138,139,1)","rgba(68,58,125,1)","rgba(36,165,133,1)","rgba(38,161,134,1)","rgba(66,63,130,1)","rgba(37,162,133,1)","rgba(47,114,141,1)","rgba(69,44,113,1)","rgba(69,16,91,1)","rgba(70,42,111,1)","rgba(43,140,138,1)","rgba(70,37,107,1)","rgba(42,148,137,1)","rgba(54,105,140,1)","rgba(43,129,140,1)","rgba(42,120,142,1)","rgba(66,65,132,1)","rgba(43,129,140,1)","rgba(63,81,137,1)","rgba(60,90,138,1)","rgba(44,136,139,1)","rgba(54,105,140,1)","rgba(63,78,136,1)","rgba(69,49,117,1)","rgba(52,107,140,1)","rgba(42,122,142,1)","rgba(61,87,138,1)","rgba(42,150,136,1)","rgba(49,112,141,1)","rgba(54,105,140,1)","rgba(66,63,130,1)","rgba(68,2,84,1)","rgba(58,96,139,1)","rgba(42,148,137,1)","rgba(42,150,136,1)","rgba(44,134,139,1)","rgba(40,157,135,1)","rgba(55,103,140,1)","rgba(39,159,134,1)","rgba(62,83,137,1)","rgba(44,132,140,1)","rgba(37,163,133,1)","rgba(68,58,125,1)","rgba(81,184,114,1)","rgba(43,126,141,1)","rgba(37,163,133,1)","rgba(46,116,142,1)","rgba(42,150,136,1)","rgba(45,117,142,1)","rgba(44,131,140,1)","rgba(44,133,140,1)","rgba(52,108,140,1)","rgba(41,153,136,1)","rgba(53,107,140,1)","rgba(40,157,135,1)","rgba(66,66,133,1)","rgba(43,145,137,1)","rgba(42,120,142,1)","rgba(64,77,136,1)","rgba(37,163,133,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(80,183,115,1)","rgba(37,163,133,1)","rgba(57,98,139,1)","rgba(38,162,134,1)","rgba(57,98,139,1)","rgba(43,141,138,1)","rgba(57,99,139,1)","rgba(40,156,135,1)","rgba(43,126,141,1)","rgba(45,116,142,1)","rgba(65,70,135,1)","rgba(64,76,136,1)","rgba(37,162,133,1)","rgba(43,142,138,1)","rgba(69,43,112,1)","rgba(42,122,142,1)","rgba(42,121,142,1)","rgba(114,203,89,1)","rgba(63,80,137,1)","rgba(44,134,139,1)","rgba(43,127,141,1)","rgba(44,131,140,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(69,50,118,1)","rgba(52,108,140,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(62,85,137,1)","rgba(62,85,137,1)","rgba(44,118,142,1)","rgba(70,37,107,1)","rgba(51,109,141,1)","rgba(103,196,99,1)","rgba(44,118,142,1)","rgba(43,143,138,1)","rgba(61,89,138,1)","rgba(60,91,138,1)","rgba(42,120,142,1)","rgba(44,136,139,1)","rgba(57,99,139,1)","rgba(40,169,131,1)","rgba(39,158,134,1)","rgba(48,113,141,1)","rgba(44,135,139,1)","rgba(43,146,137,1)","rgba(56,101,140,1)","rgba(40,155,135,1)","rgba(68,6,86,1)","rgba(38,160,134,1)","rgba(42,150,136,1)","rgba(69,15,91,1)","rgba(69,49,117,1)","rgba(70,39,108,1)","rgba(41,154,135,1)","rgba(38,161,134,1)","rgba(45,170,129,1)","rgba(43,128,141,1)","rgba(69,13,90,1)","rgba(68,56,124,1)","rgba(69,11,89,1)","rgba(43,127,141,1)","rgba(41,153,135,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(61,87,138,1)","rgba(53,105,140,1)","rgba(41,152,136,1)","rgba(50,110,141,1)","rgba(44,138,139,1)","rgba(61,89,138,1)","rgba(43,140,138,1)","rgba(43,127,141,1)","rgba(43,143,138,1)","rgba(83,185,113,1)","rgba(39,159,134,1)","rgba(44,134,139,1)","rgba(43,129,140,1)","rgba(43,140,138,1)","rgba(65,70,135,1)","rgba(38,161,134,1)","rgba(69,21,94,1)","rgba(44,135,139,1)","rgba(35,167,132,1)","rgba(42,122,142,1)","rgba(58,97,139,1)","rgba(59,95,139,1)","rgba(42,121,142,1)","rgba(39,158,134,1)","rgba(44,138,139,1)","rgba(44,118,142,1)","rgba(43,143,138,1)","rgba(43,127,141,1)","rgba(93,190,107,1)","rgba(42,149,136,1)","rgba(43,143,138,1)","rgba(43,145,137,1)","rgba(43,129,140,1)","rgba(41,152,136,1)","rgba(42,150,136,1)","rgba(51,109,141,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(69,21,94,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(59,94,139,1)","rgba(51,110,141,1)","rgba(40,157,135,1)","rgba(62,83,137,1)","rgba(53,106,140,1)","rgba(69,51,119,1)","rgba(59,93,139,1)","rgba(79,183,116,1)","rgba(43,128,141,1)","rgba(44,136,139,1)","rgba(54,173,126,1)","rgba(64,76,136,1)","rgba(64,72,136,1)","rgba(50,111,141,1)","rgba(60,92,138,1)","rgba(44,118,142,1)","rgba(69,10,88,1)","rgba(68,57,124,1)","rgba(69,15,91,1)","rgba(42,149,136,1)","rgba(43,143,138,1)","rgba(54,104,140,1)","rgba(65,68,135,1)","rgba(61,88,138,1)","rgba(61,88,138,1)","rgba(42,148,137,1)","rgba(61,88,138,1)","rgba(45,117,142,1)","rgba(43,143,138,1)","rgba(56,101,140,1)","rgba(68,2,84,1)","rgba(43,126,141,1)","rgba(68,58,125,1)","rgba(43,125,141,1)","rgba(44,137,139,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(43,127,141,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(44,133,140,1)","rgba(43,126,141,1)","rgba(69,14,90,1)","rgba(42,122,142,1)","rgba(44,131,140,1)","rgba(36,165,133,1)","rgba(44,137,139,1)","rgba(41,154,135,1)","rgba(39,160,134,1)","rgba(43,141,138,1)","rgba(42,150,136,1)","rgba(51,109,141,1)","rgba(54,105,140,1)","rgba(43,123,142,1)","rgba(57,174,125,1)","rgba(43,146,137,1)","rgba(47,115,141,1)","rgba(38,160,134,1)","rgba(44,136,139,1)","rgba(44,131,140,1)","rgba(43,147,137,1)","rgba(36,164,133,1)","rgba(57,98,139,1)","rgba(35,167,132,1)","rgba(41,154,135,1)","rgba(42,120,142,1)","rgba(44,118,142,1)","rgba(69,15,91,1)","rgba(43,123,141,1)","rgba(55,102,140,1)","rgba(42,148,137,1)","rgba(41,153,135,1)","rgba(49,111,141,1)","rgba(43,143,138,1)","rgba(66,177,122,1)","rgba(66,177,122,1)","rgba(50,110,141,1)","rgba(69,18,92,1)","rgba(59,94,139,1)","rgba(90,188,109,1)","rgba(41,154,135,1)","rgba(39,158,134,1)","rgba(47,115,141,1)","rgba(43,126,141,1)","rgba(49,112,141,1)","rgba(53,106,140,1)","rgba(42,150,136,1)","rgba(67,62,129,1)","rgba(52,108,141,1)","rgba(46,116,141,1)","rgba(67,60,127,1)","rgba(70,37,106,1)","rgba(65,71,135,1)","rgba(49,112,141,1)","rgba(43,126,141,1)","rgba(43,142,138,1)","rgba(66,64,131,1)","rgba(48,113,141,1)","rgba(69,11,89,1)","rgba(55,102,140,1)","rgba(68,58,125,1)","rgba(70,30,101,1)","rgba(70,30,101,1)","rgba(70,29,100,1)","rgba(69,46,114,1)","rgba(69,24,96,1)","rgba(67,61,129,1)","rgba(37,163,133,1)","rgba(44,132,140,1)","rgba(44,132,140,1)","rgba(43,127,141,1)","rgba(69,49,117,1)","rgba(50,110,141,1)","rgba(60,92,138,1)","rgba(42,120,142,1)","rgba(69,25,97,1)","rgba(63,80,137,1)","rgba(121,209,82,1)","rgba(50,111,141,1)","rgba(43,123,141,1)","rgba(43,140,138,1)","rgba(43,127,141,1)","rgba(59,94,139,1)","rgba(43,129,140,1)","rgba(42,147,137,1)","rgba(69,14,90,1)","rgba(70,40,109,1)","rgba(43,141,138,1)","rgba(69,47,115,1)","rgba(69,21,94,1)","rgba(62,84,137,1)","rgba(53,106,140,1)","rgba(70,29,100,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(61,89,138,1)","rgba(48,113,141,1)","rgba(63,82,137,1)","rgba(43,144,137,1)","rgba(43,123,141,1)","rgba(57,99,139,1)","rgba(69,51,119,1)","rgba(44,134,139,1)","rgba(70,30,101,1)","rgba(68,3,85,1)","rgba(70,33,103,1)","rgba(60,92,138,1)","rgba(52,108,140,1)","rgba(53,106,140,1)","rgba(44,131,140,1)","rgba(48,113,141,1)","rgba(75,181,118,1)","rgba(62,84,137,1)","rgba(93,190,107,1)","rgba(63,78,136,1)","rgba(69,24,97,1)","rgba(53,106,140,1)","rgba(43,123,142,1)","rgba(43,129,140,1)","rgba(51,109,141,1)","rgba(39,159,134,1)","rgba(43,124,141,1)","rgba(44,136,139,1)","rgba(69,51,118,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(53,106,140,1)","rgba(54,104,140,1)","rgba(69,48,116,1)","rgba(44,134,139,1)","rgba(65,69,135,1)","rgba(42,148,137,1)","rgba(40,155,135,1)","rgba(56,101,140,1)","rgba(70,38,107,1)","rgba(54,105,140,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(46,115,141,1)","rgba(43,142,138,1)","rgba(36,164,133,1)","rgba(42,148,137,1)","rgba(63,81,137,1)","rgba(69,51,118,1)","rgba(67,59,126,1)","rgba(42,122,142,1)","rgba(70,28,99,1)","rgba(42,149,136,1)","rgba(40,169,131,1)","rgba(43,146,137,1)","rgba(60,90,138,1)","rgba(43,130,140,1)","rgba(45,117,142,1)","rgba(46,171,129,1)","rgba(69,42,111,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(44,135,139,1)","rgba(67,61,128,1)","rgba(42,149,136,1)","rgba(43,140,138,1)","rgba(43,142,138,1)","rgba(44,170,130,1)","rgba(40,156,135,1)","rgba(60,92,138,1)","rgba(56,101,140,1)","rgba(42,150,136,1)","rgba(43,124,141,1)","rgba(42,122,142,1)","rgba(70,38,107,1)","rgba(44,138,139,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(44,132,140,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(52,108,141,1)","rgba(44,132,140,1)","rgba(44,133,140,1)","rgba(44,137,139,1)","rgba(43,126,141,1)","rgba(64,77,136,1)","rgba(59,94,139,1)","rgba(94,190,106,1)","rgba(42,122,142,1)","rgba(59,94,139,1)","rgba(43,119,142,1)","rgba(60,92,138,1)","rgba(52,108,141,1)","rgba(42,120,142,1)","rgba(45,117,142,1)","rgba(50,111,141,1)","rgba(70,25,98,1)","rgba(36,164,133,1)","rgba(43,170,130,1)","rgba(39,158,134,1)","rgba(64,75,136,1)","rgba(61,87,138,1)","rgba(69,12,89,1)","rgba(69,43,112,1)","rgba(57,98,139,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(57,98,139,1)","rgba(43,147,137,1)","rgba(69,49,117,1)","rgba(43,142,138,1)","rgba(58,95,139,1)","rgba(68,54,122,1)","rgba(43,140,138,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(43,145,137,1)","rgba(69,23,96,1)","rgba(41,153,136,1)","rgba(43,127,141,1)","rgba(40,157,135,1)","rgba(44,133,140,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(55,102,140,1)","rgba(61,87,138,1)","rgba(44,135,139,1)","rgba(43,142,138,1)","rgba(56,100,139,1)","rgba(63,81,137,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(43,119,142,1)","rgba(40,157,135,1)","rgba(43,142,138,1)","rgba(70,30,101,1)","rgba(53,106,140,1)","rgba(69,25,97,1)","rgba(68,58,125,1)","rgba(70,41,110,1)","rgba(68,55,122,1)","rgba(69,9,88,1)","rgba(40,155,135,1)","rgba(69,49,117,1)","rgba(69,23,96,1)","rgba(68,4,86,1)","rgba(69,50,118,1)","rgba(42,149,136,1)","rgba(44,133,140,1)","rgba(43,128,141,1)","rgba(45,117,142,1)","rgba(37,162,133,1)","rgba(70,39,108,1)","rgba(56,174,126,1)","rgba(59,94,139,1)","rgba(69,51,119,1)","rgba(51,110,141,1)","rgba(51,109,141,1)","rgba(70,41,110,1)","rgba(61,87,138,1)","rgba(51,110,141,1)","rgba(61,88,138,1)","rgba(43,128,140,1)","rgba(43,123,141,1)","rgba(44,137,139,1)","rgba(42,150,136,1)","rgba(68,55,122,1)","rgba(69,46,114,1)","rgba(38,161,134,1)","rgba(47,171,129,1)","rgba(57,99,139,1)","rgba(47,171,129,1)","rgba(64,77,136,1)","rgba(61,87,138,1)","rgba(43,142,138,1)","rgba(69,24,96,1)","rgba(43,127,141,1)","rgba(47,115,141,1)","rgba(41,154,135,1)","rgba(44,133,140,1)","rgba(68,6,86,1)","rgba(59,175,125,1)","rgba(43,140,138,1)","rgba(60,91,138,1)","rgba(66,63,131,1)","rgba(39,160,134,1)","rgba(42,120,142,1)","rgba(52,108,140,1)","rgba(39,159,134,1)","rgba(60,175,124,1)","rgba(68,58,126,1)","rgba(65,68,135,1)","rgba(68,56,123,1)","rgba(63,81,137,1)","rgba(132,211,79,1)","rgba(44,118,142,1)","rgba(43,124,141,1)","rgba(69,47,115,1)","rgba(59,93,138,1)","rgba(64,73,136,1)","rgba(43,126,141,1)","rgba(43,123,142,1)","rgba(64,75,136,1)","rgba(41,152,136,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(40,156,135,1)","rgba(40,155,135,1)","rgba(43,119,142,1)","rgba(43,127,141,1)","rgba(35,167,132,1)","rgba(50,172,128,1)","rgba(62,85,137,1)","rgba(59,94,139,1)","rgba(46,116,141,1)","rgba(43,128,140,1)","rgba(43,124,141,1)","rgba(36,165,133,1)","rgba(42,122,142,1)","rgba(36,165,133,1)","rgba(44,136,139,1)","rgba(70,30,101,1)","rgba(42,148,136,1)","rgba(70,179,120,1)","rgba(51,172,127,1)","rgba(43,141,138,1)","rgba(70,32,102,1)","rgba(68,1,84,1)","rgba(53,106,140,1)","rgba(40,157,135,1)","rgba(43,128,141,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(54,104,140,1)","rgba(62,84,137,1)","rgba(43,123,141,1)","rgba(65,69,135,1)","rgba(43,124,141,1)","rgba(35,166,132,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(69,46,115,1)","rgba(46,116,141,1)","rgba(37,163,133,1)","rgba(51,110,141,1)","rgba(44,118,142,1)","rgba(69,22,95,1)","rgba(69,17,92,1)","rgba(66,64,131,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(40,156,135,1)","rgba(62,84,137,1)","rgba(36,165,133,1)","rgba(64,177,123,1)","rgba(44,136,139,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(44,131,140,1)","rgba(70,32,102,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(40,157,135,1)","rgba(70,36,105,1)","rgba(42,122,142,1)","rgba(63,82,137,1)","rgba(42,151,136,1)","rgba(36,164,133,1)","rgba(42,120,142,1)","rgba(58,96,139,1)","rgba(42,151,136,1)","rgba(42,121,142,1)","rgba(44,136,139,1)","rgba(56,100,139,1)","rgba(69,48,116,1)","rgba(65,70,135,1)","rgba(44,135,139,1)","rgba(69,15,91,1)","rgba(44,133,140,1)","rgba(44,137,139,1)","rgba(68,57,124,1)","rgba(36,164,133,1)","rgba(46,116,142,1)","rgba(61,88,138,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(55,103,140,1)","rgba(49,112,141,1)","rgba(52,108,141,1)","rgba(70,27,99,1)","rgba(68,4,85,1)","rgba(55,102,140,1)","rgba(55,103,140,1)","rgba(59,95,139,1)","rgba(43,124,141,1)","rgba(42,121,142,1)","rgba(56,101,140,1)","rgba(63,80,137,1)","rgba(68,57,125,1)","rgba(42,150,136,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(38,160,134,1)","rgba(36,165,133,1)","rgba(42,151,136,1)","rgba(68,5,86,1)","rgba(43,124,141,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(60,92,138,1)","rgba(67,59,126,1)","rgba(55,102,140,1)","rgba(59,93,138,1)","rgba(69,15,91,1)","rgba(56,100,139,1)","rgba(39,169,131,1)","rgba(47,115,141,1)","rgba(52,107,140,1)","rgba(70,37,106,1)","rgba(69,42,111,1)","rgba(43,140,138,1)","rgba(63,81,137,1)","rgba(43,142,138,1)","rgba(64,76,136,1)","rgba(41,153,136,1)","rgba(40,155,135,1)","rgba(52,107,140,1)","rgba(42,122,142,1)","rgba(44,138,139,1)","rgba(42,149,136,1)","rgba(42,150,136,1)","rgba(42,120,142,1)","rgba(54,105,140,1)","rgba(44,134,139,1)","rgba(42,150,136,1)","rgba(44,132,140,1)","rgba(55,103,140,1)","rgba(50,111,141,1)","rgba(42,120,142,1)","rgba(44,135,139,1)","rgba(55,103,140,1)","rgba(63,82,137,1)","rgba(42,147,137,1)","rgba(51,110,141,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(61,88,138,1)","rgba(50,111,141,1)","rgba(39,158,134,1)","rgba(43,146,137,1)","rgba(43,130,140,1)","rgba(61,89,138,1)","rgba(43,144,137,1)","rgba(50,172,128,1)","rgba(62,83,137,1)","rgba(39,158,134,1)","rgba(37,162,133,1)","rgba(52,107,140,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(69,47,115,1)","rgba(62,86,138,1)","rgba(44,135,139,1)","rgba(44,138,139,1)","rgba(60,92,138,1)","rgba(56,100,139,1)","rgba(49,172,128,1)","rgba(61,87,138,1)","rgba(67,59,127,1)","rgba(42,149,136,1)","rgba(68,56,124,1)","rgba(43,130,140,1)","rgba(49,112,141,1)","rgba(64,73,136,1)","rgba(63,81,137,1)","rgba(69,16,91,1)","rgba(67,61,128,1)","rgba(67,61,128,1)","rgba(44,132,140,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(70,37,106,1)","rgba(43,126,141,1)","rgba(40,155,135,1)","rgba(43,125,141,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(42,148,137,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(68,54,122,1)","rgba(57,100,139,1)","rgba(67,59,126,1)","rgba(34,168,132,1)","rgba(68,4,85,1)","rgba(43,125,141,1)","rgba(48,114,141,1)","rgba(44,132,140,1)","rgba(43,128,141,1)","rgba(52,107,140,1)","rgba(54,105,140,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(43,130,140,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(62,85,137,1)","rgba(43,143,138,1)","rgba(69,42,111,1)","rgba(43,170,130,1)","rgba(59,94,139,1)","rgba(61,87,138,1)","rgba(44,132,140,1)","rgba(69,23,96,1)","rgba(69,19,93,1)","rgba(44,138,139,1)","rgba(68,8,87,1)","rgba(70,39,108,1)","rgba(64,72,136,1)","rgba(43,123,142,1)","rgba(46,116,142,1)","rgba(43,145,137,1)","rgba(45,117,142,1)","rgba(43,128,141,1)","rgba(43,130,140,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(50,111,141,1)","rgba(65,70,135,1)","rgba(43,128,141,1)","rgba(43,126,141,1)","rgba(67,61,128,1)","rgba(66,63,130,1)","rgba(60,90,138,1)","rgba(51,109,141,1)","rgba(45,117,142,1)","rgba(70,26,98,1)","rgba(68,2,85,1)","rgba(41,153,136,1)","rgba(38,162,134,1)","rgba(60,91,138,1)","rgba(43,123,141,1)","rgba(59,93,139,1)","rgba(43,123,142,1)","rgba(57,99,139,1)","rgba(57,100,139,1)","rgba(43,144,137,1)","rgba(44,138,139,1)","rgba(44,133,140,1)","rgba(57,99,139,1)","rgba(53,106,140,1)","rgba(63,80,137,1)","rgba(51,110,141,1)","rgba(69,23,96,1)","rgba(62,83,137,1)","rgba(69,23,96,1)","rgba(61,89,138,1)","rgba(43,140,138,1)","rgba(68,4,86,1)","rgba(44,137,139,1)","rgba(43,146,137,1)","rgba(44,118,142,1)","rgba(50,111,141,1)","rgba(42,121,142,1)","rgba(39,158,134,1)","rgba(55,103,140,1)","rgba(45,116,142,1)","rgba(68,3,85,1)","rgba(42,120,142,1)","rgba(63,83,137,1)","rgba(53,107,140,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(43,130,140,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(44,136,139,1)","rgba(43,130,140,1)","rgba(43,123,141,1)","rgba(42,149,136,1)","rgba(41,154,135,1)","rgba(65,67,134,1)","rgba(50,111,141,1)","rgba(66,64,131,1)","rgba(40,156,135,1)","rgba(38,161,134,1)","rgba(45,117,142,1)","rgba(52,108,140,1)","rgba(42,122,142,1)","rgba(68,56,123,1)","rgba(50,111,141,1)","rgba(69,20,94,1)","rgba(53,106,140,1)","rgba(70,35,104,1)","rgba(70,29,100,1)","rgba(43,142,138,1)","rgba(51,172,127,1)","rgba(62,84,137,1)","rgba(68,5,86,1)","rgba(52,108,141,1)","rgba(45,117,142,1)","rgba(44,137,139,1)","rgba(44,131,140,1)","rgba(64,73,136,1)","rgba(68,4,85,1)","rgba(44,136,139,1)","rgba(69,15,91,1)","rgba(43,142,138,1)","rgba(54,105,140,1)","rgba(44,133,140,1)","rgba(69,13,90,1)","rgba(43,145,137,1)","rgba(70,41,110,1)","rgba(68,7,87,1)","rgba(68,58,125,1)","rgba(59,175,125,1)","rgba(43,124,141,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(43,119,142,1)","rgba(42,147,137,1)","rgba(40,155,135,1)","rgba(55,103,140,1)","rgba(57,99,139,1)","rgba(55,103,140,1)","rgba(69,20,94,1)","rgba(44,137,139,1)","rgba(63,82,137,1)","rgba(44,133,140,1)","rgba(43,147,137,1)","rgba(44,131,140,1)","rgba(44,134,139,1)","rgba(34,168,132,1)","rgba(62,83,137,1)","rgba(46,116,141,1)","rgba(42,121,142,1)","rgba(44,135,139,1)","rgba(44,131,140,1)","rgba(58,96,139,1)","rgba(58,97,139,1)","rgba(43,139,138,1)","rgba(52,107,140,1)","rgba(52,108,141,1)","rgba(69,9,88,1)","rgba(42,148,137,1)","rgba(70,33,103,1)","rgba(51,109,141,1)","rgba(63,79,137,1)","rgba(43,143,138,1)","rgba(38,162,133,1)","rgba(65,70,135,1)","rgba(43,130,140,1)","rgba(58,97,139,1)","rgba(69,24,96,1)","rgba(43,129,140,1)","rgba(70,40,109,1)","rgba(70,37,107,1)","rgba(43,141,138,1)","rgba(68,5,86,1)","rgba(43,127,141,1)","rgba(43,126,141,1)","rgba(58,97,139,1)","rgba(60,92,138,1)","rgba(43,128,141,1)","rgba(55,103,140,1)","rgba(60,91,138,1)","rgba(40,157,135,1)","rgba(70,41,109,1)","rgba(53,107,140,1)","rgba(48,113,141,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(42,121,142,1)","rgba(70,34,104,1)","rgba(36,165,133,1)","rgba(43,123,141,1)","rgba(58,96,139,1)","rgba(63,80,137,1)","rgba(44,132,140,1)","rgba(43,143,138,1)","rgba(43,124,141,1)","rgba(69,13,90,1)","rgba(49,112,141,1)","rgba(42,148,137,1)","rgba(43,140,138,1)","rgba(42,121,142,1)","rgba(70,37,106,1)","rgba(69,21,95,1)","rgba(43,123,141,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(44,132,140,1)","rgba(44,118,142,1)","rgba(42,121,142,1)","rgba(70,31,101,1)","rgba(68,57,124,1)","rgba(68,6,86,1)","rgba(43,130,140,1)","rgba(63,79,137,1)","rgba(59,175,125,1)","rgba(70,38,107,1)","rgba(50,110,141,1)","rgba(45,117,142,1)","rgba(43,147,137,1)","rgba(64,77,136,1)","rgba(69,50,118,1)","rgba(64,75,136,1)","rgba(41,151,136,1)","rgba(64,73,136,1)","rgba(111,201,92,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(49,112,141,1)","rgba(40,156,135,1)","rgba(56,174,126,1)","rgba(55,102,140,1)","rgba(103,196,100,1)","rgba(43,146,137,1)","rgba(43,127,141,1)","rgba(48,113,141,1)","rgba(44,132,140,1)","rgba(63,79,137,1)","rgba(42,122,142,1)","rgba(46,115,141,1)","rgba(65,70,135,1)","rgba(70,39,108,1)","rgba(43,141,138,1)","rgba(54,173,126,1)","rgba(102,195,100,1)","rgba(68,58,125,1)","rgba(62,86,138,1)","rgba(43,128,141,1)","rgba(39,160,134,1)","rgba(43,123,142,1)","rgba(62,86,137,1)","rgba(41,154,135,1)","rgba(62,86,138,1)","rgba(43,126,141,1)","rgba(43,143,138,1)","rgba(51,109,141,1)","rgba(37,164,133,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(51,110,141,1)","rgba(51,110,141,1)","rgba(61,87,138,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(84,185,112,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(39,160,134,1)","rgba(43,145,137,1)","rgba(43,142,138,1)","rgba(49,111,141,1)","rgba(44,131,140,1)","rgba(47,115,141,1)","rgba(68,3,85,1)","rgba(63,176,123,1)","rgba(43,127,141,1)","rgba(69,22,95,1)","rgba(43,119,142,1)","rgba(69,52,120,1)","rgba(68,56,123,1)","rgba(70,25,97,1)","rgba(44,131,140,1)","rgba(43,129,140,1)","rgba(60,92,138,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(69,12,89,1)","rgba(42,121,142,1)","rgba(57,99,139,1)","rgba(47,114,141,1)","rgba(62,176,123,1)","rgba(61,87,138,1)","rgba(39,159,134,1)","rgba(70,41,110,1)","rgba(47,115,141,1)","rgba(46,115,141,1)","rgba(65,68,135,1)","rgba(69,50,118,1)","rgba(53,106,140,1)","rgba(42,150,136,1)","rgba(43,141,138,1)","rgba(43,129,140,1)","rgba(66,66,133,1)","rgba(62,84,137,1)","rgba(70,33,103,1)","rgba(48,113,141,1)","rgba(44,134,139,1)","rgba(54,104,140,1)","rgba(54,104,140,1)","rgba(44,136,139,1)","rgba(59,93,139,1)","rgba(41,154,135,1)","rgba(65,67,134,1)","rgba(57,98,139,1)","rgba(43,146,137,1)","rgba(66,65,132,1)","rgba(62,85,137,1)","rgba(43,127,141,1)","rgba(57,99,139,1)","rgba(45,117,142,1)","rgba(42,122,142,1)","rgba(39,158,134,1)","rgba(42,122,142,1)","rgba(43,125,141,1)","rgba(54,104,140,1)","rgba(62,86,138,1)","rgba(85,186,112,1)","rgba(44,118,142,1)","rgba(60,175,124,1)","rgba(42,121,142,1)","rgba(51,110,141,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(69,24,96,1)","rgba(66,65,132,1)","rgba(43,128,141,1)","rgba(43,128,141,1)","rgba(48,113,141,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(70,27,98,1)","rgba(49,112,141,1)","rgba(69,43,111,1)","rgba(42,122,142,1)","rgba(69,50,118,1)","rgba(43,129,140,1)","rgba(36,165,133,1)","rgba(41,154,135,1)","rgba(43,119,142,1)","rgba(50,110,141,1)","rgba(36,164,133,1)","rgba(62,83,137,1)","rgba(49,113,141,1)","rgba(62,86,138,1)","rgba(54,104,140,1)","rgba(61,88,138,1)","rgba(61,89,138,1)","rgba(45,117,142,1)","rgba(54,104,140,1)","rgba(43,144,137,1)","rgba(60,91,138,1)","rgba(59,94,139,1)","rgba(70,41,110,1)","rgba(42,148,137,1)","rgba(50,111,141,1)","rgba(44,135,139,1)","rgba(53,107,140,1)","rgba(43,143,138,1)","rgba(43,128,140,1)","rgba(56,101,140,1)","rgba(44,137,139,1)","rgba(41,154,135,1)","rgba(69,25,97,1)","rgba(43,128,140,1)","rgba(52,108,141,1)","rgba(52,108,141,1)","rgba(45,117,142,1)","rgba(43,123,141,1)","rgba(35,167,132,1)","rgba(43,147,137,1)","rgba(64,73,136,1)","rgba(69,18,92,1)","rgba(56,101,140,1)","rgba(43,144,137,1)","rgba(70,34,104,1)","rgba(57,99,139,1)","rgba(62,85,137,1)","rgba(49,112,141,1)","rgba(34,167,132,1)","rgba(64,76,136,1)","rgba(67,60,127,1)","rgba(39,158,134,1)","rgba(54,104,140,1)","rgba(43,123,142,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(58,95,139,1)","rgba(44,139,138,1)","rgba(44,133,140,1)","rgba(43,127,141,1)","rgba(44,136,139,1)","rgba(43,146,137,1)","rgba(63,81,137,1)","rgba(43,123,142,1)","rgba(64,73,136,1)","rgba(66,65,132,1)","rgba(63,81,137,1)","rgba(43,140,138,1)","rgba(43,126,141,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(56,101,140,1)","rgba(66,64,131,1)","rgba(69,42,111,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(70,41,110,1)","rgba(70,40,109,1)","rgba(72,180,119,1)","rgba(54,105,140,1)","rgba(65,70,135,1)","rgba(65,66,134,1)","rgba(65,68,135,1)","rgba(43,119,142,1)","rgba(43,129,140,1)","rgba(67,61,128,1)","rgba(42,147,137,1)","rgba(38,162,133,1)","rgba(44,137,139,1)","rgba(84,185,112,1)","rgba(43,141,138,1)","rgba(41,154,135,1)","rgba(35,166,132,1)","rgba(69,46,114,1)","rgba(69,22,95,1)","rgba(47,115,141,1)","rgba(41,152,136,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(65,70,135,1)","rgba(60,175,124,1)","rgba(42,148,137,1)","rgba(44,131,140,1)","rgba(47,171,129,1)","rgba(70,27,99,1)","rgba(44,135,139,1)","rgba(57,99,139,1)","rgba(59,93,138,1)","rgba(43,127,141,1)","rgba(54,105,140,1)","rgba(64,72,136,1)","rgba(43,127,141,1)","rgba(69,25,97,1)","rgba(44,135,139,1)","rgba(44,137,139,1)","rgba(43,139,138,1)","rgba(44,132,140,1)","rgba(37,163,133,1)","rgba(54,104,140,1)","rgba(48,114,141,1)","rgba(43,128,141,1)","rgba(118,206,86,1)","rgba(64,76,136,1)","rgba(43,141,138,1)","rgba(48,114,141,1)","rgba(69,17,92,1)","rgba(64,76,136,1)","rgba(69,17,92,1)","rgba(48,171,128,1)","rgba(37,162,133,1)","rgba(63,79,137,1)","rgba(58,97,139,1)","rgba(43,146,137,1)","rgba(44,134,139,1)","rgba(47,115,141,1)","rgba(43,146,137,1)","rgba(51,109,141,1)","rgba(70,26,98,1)","rgba(51,109,141,1)","rgba(43,128,140,1)","rgba(49,112,141,1)","rgba(42,148,136,1)","rgba(38,161,134,1)","rgba(37,163,133,1)","rgba(43,119,142,1)","rgba(43,130,140,1)","rgba(66,64,131,1)","rgba(59,93,139,1)","rgba(39,159,134,1)","rgba(49,113,141,1)","rgba(75,181,117,1)","rgba(53,106,140,1)","rgba(44,136,139,1)","rgba(69,53,120,1)","rgba(43,124,141,1)","rgba(69,25,97,1)","rgba(64,75,136,1)","rgba(54,104,140,1)","rgba(36,168,132,1)","rgba(64,76,136,1)","rgba(64,74,136,1)","rgba(43,142,138,1)","rgba(68,4,85,1)","rgba(70,34,104,1)","rgba(43,130,140,1)","rgba(42,147,137,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(64,76,136,1)","rgba(40,156,135,1)","rgba(42,121,142,1)","rgba(41,153,135,1)","rgba(46,116,141,1)","rgba(43,128,141,1)","rgba(41,153,135,1)","rgba(53,106,140,1)","rgba(38,161,134,1)","rgba(42,121,142,1)","rgba(69,11,89,1)","rgba(57,98,139,1)","rgba(40,156,135,1)","rgba(69,46,115,1)","rgba(60,90,138,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(49,112,141,1)","rgba(49,112,141,1)","rgba(67,59,126,1)","rgba(53,106,140,1)","rgba(54,105,140,1)","rgba(47,114,141,1)","rgba(64,75,136,1)","rgba(55,173,126,1)","rgba(58,97,139,1)","rgba(62,84,137,1)","rgba(47,114,141,1)","rgba(40,155,135,1)","rgba(60,92,138,1)","rgba(64,73,136,1)","rgba(62,84,137,1)","rgba(61,87,138,1)","rgba(61,88,138,1)","rgba(37,163,133,1)","rgba(48,113,141,1)","rgba(35,168,132,1)","rgba(38,161,134,1)","rgba(63,82,137,1)","rgba(42,149,136,1)","rgba(47,114,141,1)","rgba(62,83,137,1)","rgba(54,105,140,1)","rgba(54,105,140,1)","rgba(58,97,139,1)","rgba(64,75,136,1)","rgba(43,119,142,1)","rgba(44,132,140,1)","rgba(67,59,126,1)","rgba(63,81,137,1)","rgba(44,131,140,1)","rgba(51,110,141,1)","rgba(54,105,140,1)","rgba(55,103,140,1)","rgba(68,54,121,1)","rgba(68,7,87,1)","rgba(63,79,137,1)","rgba(52,107,140,1)","rgba(43,129,140,1)","rgba(68,56,124,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(51,109,141,1)","rgba(36,165,133,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(38,161,134,1)","rgba(63,78,136,1)","rgba(46,171,129,1)","rgba(68,57,125,1)","rgba(61,88,138,1)","rgba(54,105,140,1)","rgba(43,127,141,1)","rgba(43,140,138,1)","rgba(48,114,141,1)","rgba(50,110,141,1)","rgba(43,170,130,1)","rgba(42,122,142,1)","rgba(94,190,106,1)","rgba(64,74,136,1)","rgba(43,126,141,1)","rgba(57,98,139,1)","rgba(40,157,134,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(58,96,139,1)","rgba(69,48,116,1)","rgba(49,172,128,1)","rgba(53,106,140,1)","rgba(44,135,139,1)","rgba(44,134,139,1)","rgba(43,143,138,1)","rgba(40,155,135,1)","rgba(37,169,131,1)","rgba(65,70,135,1)","rgba(69,21,94,1)","rgba(64,73,136,1)","rgba(43,144,137,1)","rgba(68,55,122,1)","rgba(36,164,133,1)","rgba(43,130,140,1)","rgba(42,149,136,1)","rgba(53,107,140,1)","rgba(67,61,129,1)","rgba(69,23,96,1)","rgba(41,152,136,1)","rgba(46,116,142,1)","rgba(42,149,136,1)","rgba(61,88,138,1)","rgba(60,90,138,1)","rgba(40,155,135,1)","rgba(43,126,141,1)","rgba(43,146,137,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(56,101,140,1)","rgba(43,129,140,1)","rgba(42,151,136,1)","rgba(34,168,132,1)","rgba(43,129,140,1)","rgba(62,86,138,1)","rgba(88,187,110,1)","rgba(43,130,140,1)","rgba(36,166,133,1)","rgba(43,145,137,1)","rgba(34,168,132,1)","rgba(68,4,85,1)","rgba(62,83,137,1)","rgba(64,76,136,1)","rgba(61,88,138,1)","rgba(70,42,111,1)","rgba(69,46,114,1)","rgba(38,161,134,1)","rgba(39,169,131,1)","rgba(69,51,119,1)","rgba(36,165,133,1)","rgba(44,137,139,1)","rgba(43,141,138,1)","rgba(152,214,75,1)","rgba(42,149,136,1)","rgba(42,122,142,1)","rgba(56,101,140,1)","rgba(43,144,137,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(43,130,140,1)","rgba(60,91,138,1)","rgba(42,149,136,1)","rgba(69,46,114,1)","rgba(41,152,136,1)","rgba(36,164,133,1)","rgba(48,114,141,1)","rgba(40,155,135,1)","rgba(44,132,140,1)","rgba(49,172,128,1)","rgba(50,111,141,1)","rgba(42,148,137,1)","rgba(44,135,139,1)","rgba(50,172,128,1)","rgba(44,139,138,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(70,32,103,1)","rgba(39,160,134,1)","rgba(68,55,123,1)","rgba(69,24,96,1)","rgba(35,166,132,1)","rgba(70,26,98,1)","rgba(44,134,139,1)","rgba(43,119,142,1)","rgba(43,143,138,1)","rgba(44,139,138,1)","rgba(48,114,141,1)","rgba(46,116,141,1)","rgba(56,101,140,1)","rgba(69,18,93,1)","rgba(60,91,138,1)","rgba(67,61,128,1)","rgba(42,151,136,1)","rgba(57,99,139,1)","rgba(44,136,139,1)","rgba(57,98,139,1)","rgba(44,131,140,1)","rgba(70,30,101,1)","rgba(39,159,134,1)","rgba(69,51,119,1)","rgba(43,119,142,1)","rgba(57,99,139,1)","rgba(53,107,140,1)","rgba(42,122,142,1)","rgba(43,129,140,1)","rgba(69,14,90,1)","rgba(43,141,138,1)","rgba(43,123,141,1)","rgba(68,6,86,1)","rgba(66,65,132,1)","rgba(70,27,99,1)","rgba(59,94,139,1)","rgba(70,30,101,1)","rgba(70,31,101,1)","rgba(51,110,141,1)","rgba(42,150,136,1)","rgba(48,171,129,1)","rgba(41,153,136,1)","rgba(43,142,138,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(67,62,129,1)","rgba(42,122,142,1)","rgba(42,147,137,1)","rgba(43,144,137,1)","rgba(38,162,134,1)","rgba(49,112,141,1)","rgba(43,144,137,1)","rgba(45,117,142,1)","rgba(48,114,141,1)","rgba(62,86,138,1)","rgba(44,133,140,1)","rgba(42,149,136,1)","rgba(41,152,136,1)","rgba(65,67,134,1)","rgba(44,132,140,1)","rgba(42,121,142,1)","rgba(40,169,131,1)","rgba(63,81,137,1)","rgba(40,155,135,1)","rgba(53,106,140,1)","rgba(46,116,142,1)","rgba(41,152,136,1)","rgba(43,124,141,1)","rgba(52,107,140,1)","rgba(44,118,142,1)","rgba(43,123,141,1)","rgba(38,162,133,1)","rgba(69,19,93,1)","rgba(70,36,106,1)","rgba(42,151,136,1)","rgba(44,134,139,1)","rgba(43,147,137,1)","rgba(43,123,142,1)","rgba(57,98,139,1)","rgba(70,41,110,1)","rgba(42,150,136,1)","rgba(44,136,139,1)","rgba(60,91,138,1)","rgba(39,160,134,1)","rgba(70,28,99,1)","rgba(44,134,139,1)","rgba(83,185,113,1)","rgba(43,131,140,1)","rgba(69,12,89,1)","rgba(70,34,104,1)","rgba(64,76,136,1)","rgba(43,141,138,1)","rgba(42,147,137,1)","rgba(53,106,140,1)","rgba(43,139,138,1)","rgba(50,111,141,1)","rgba(43,130,140,1)","rgba(63,81,137,1)","rgba(39,158,134,1)","rgba(44,134,139,1)","rgba(61,87,138,1)","rgba(65,71,135,1)","rgba(70,36,105,1)","rgba(68,4,85,1)","rgba(69,13,90,1)","rgba(70,36,106,1)","rgba(63,81,137,1)","rgba(44,137,139,1)","rgba(43,129,140,1)","rgba(65,67,134,1)","rgba(44,131,140,1)","rgba(43,146,137,1)","rgba(69,48,116,1)","rgba(37,169,131,1)","rgba(57,99,139,1)","rgba(43,123,141,1)","rgba(43,142,138,1)","rgba(69,25,97,1)","rgba(70,27,99,1)","rgba(57,98,139,1)","rgba(62,176,123,1)","rgba(43,127,141,1)","rgba(43,119,142,1)","rgba(77,182,117,1)","rgba(69,19,93,1)","rgba(69,21,94,1)","rgba(56,101,140,1)","rgba(43,147,137,1)","rgba(43,124,141,1)","rgba(43,128,141,1)","rgba(39,158,134,1)","rgba(67,59,126,1)","rgba(44,131,140,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(43,119,142,1)","rgba(59,93,138,1)","rgba(64,77,136,1)","rgba(62,83,137,1)","rgba(65,68,135,1)","rgba(108,199,95,1)","rgba(64,75,136,1)","rgba(68,55,123,1)","rgba(43,123,142,1)","rgba(58,97,139,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(44,139,138,1)","rgba(42,120,142,1)","rgba(54,104,140,1)","rgba(42,122,142,1)","rgba(61,89,138,1)","rgba(58,97,139,1)","rgba(70,39,108,1)","rgba(42,150,136,1)","rgba(58,97,139,1)","rgba(37,169,131,1)","rgba(65,72,136,1)","rgba(43,123,141,1)","rgba(42,122,142,1)","rgba(43,144,137,1)","rgba(68,7,87,1)","rgba(70,27,99,1)","rgba(44,136,139,1)","rgba(43,129,140,1)","rgba(68,57,125,1)","rgba(69,50,118,1)","rgba(44,118,142,1)","rgba(56,100,140,1)","rgba(44,139,138,1)","rgba(63,79,136,1)","rgba(44,118,142,1)","rgba(43,141,138,1)","rgba(65,68,135,1)","rgba(42,150,136,1)","rgba(63,81,137,1)","rgba(44,137,139,1)","rgba(62,85,137,1)","rgba(65,69,135,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(44,136,139,1)","rgba(44,138,139,1)","rgba(53,107,140,1)","rgba(66,63,131,1)","rgba(63,81,137,1)","rgba(37,163,133,1)","rgba(41,154,135,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(70,41,110,1)","rgba(43,128,141,1)","rgba(58,95,139,1)","rgba(42,122,142,1)","rgba(43,119,142,1)","rgba(43,141,138,1)","rgba(50,111,141,1)","rgba(60,175,124,1)","rgba(43,144,137,1)","rgba(65,71,135,1)","rgba(50,111,141,1)","rgba(54,105,140,1)","rgba(69,17,92,1)","rgba(36,165,133,1)","rgba(65,177,122,1)","rgba(44,132,140,1)","rgba(44,137,139,1)","rgba(52,108,140,1)","rgba(61,89,138,1)","rgba(64,76,136,1)","rgba(68,2,84,1)","rgba(63,79,137,1)","rgba(39,159,134,1)","rgba(43,119,142,1)","rgba(69,24,97,1)","rgba(69,17,92,1)","rgba(61,89,138,1)","rgba(68,55,123,1)","rgba(43,126,141,1)","rgba(62,84,137,1)","rgba(43,123,141,1)","rgba(42,148,137,1)","rgba(43,129,140,1)","rgba(44,135,139,1)","rgba(42,120,142,1)","rgba(69,48,116,1)","rgba(63,82,137,1)","rgba(43,128,140,1)","rgba(60,90,138,1)","rgba(59,95,139,1)","rgba(65,70,135,1)","rgba(58,96,139,1)","rgba(44,135,139,1)","rgba(68,57,124,1)","rgba(70,179,120,1)","rgba(54,104,140,1)","rgba(43,123,141,1)","rgba(43,146,137,1)","rgba(48,114,141,1)","rgba(54,105,140,1)","rgba(43,128,140,1)","rgba(44,132,140,1)","rgba(68,2,85,1)","rgba(69,15,91,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(63,79,137,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(70,31,102,1)","rgba(49,112,141,1)","rgba(55,104,140,1)","rgba(43,128,141,1)","rgba(63,82,137,1)","rgba(70,34,104,1)","rgba(37,163,133,1)","rgba(80,183,115,1)","rgba(43,140,138,1)","rgba(43,130,140,1)","rgba(69,51,119,1)","rgba(65,71,135,1)","rgba(44,133,140,1)","rgba(46,115,141,1)","rgba(69,9,88,1)","rgba(44,134,139,1)","rgba(37,163,133,1)","rgba(115,204,88,1)","rgba(53,105,140,1)","rgba(69,8,88,1)","rgba(43,142,138,1)","rgba(42,122,142,1)","rgba(52,107,140,1)","rgba(57,99,139,1)","rgba(43,123,141,1)","rgba(69,51,118,1)","rgba(63,82,137,1)","rgba(74,180,118,1)","rgba(52,107,140,1)","rgba(43,127,141,1)","rgba(43,119,142,1)","rgba(69,45,114,1)","rgba(70,41,110,1)","rgba(43,145,137,1)","rgba(130,210,79,1)","rgba(69,11,89,1)","rgba(69,47,115,1)","rgba(62,85,137,1)","rgba(42,122,142,1)","rgba(39,159,134,1)","rgba(51,109,141,1)","rgba(52,108,141,1)","rgba(73,180,119,1)","rgba(44,135,139,1)","rgba(69,15,91,1)","rgba(58,97,139,1)","rgba(69,50,117,1)","rgba(44,118,142,1)","rgba(49,112,141,1)","rgba(44,131,140,1)","rgba(42,148,136,1)","rgba(44,131,140,1)","rgba(43,123,141,1)","rgba(38,160,134,1)","rgba(65,71,135,1)","rgba(70,42,111,1)","rgba(59,94,139,1)","rgba(54,104,140,1)","rgba(43,145,137,1)","rgba(53,107,140,1)","rgba(67,62,129,1)","rgba(41,155,135,1)","rgba(44,136,139,1)","rgba(54,105,140,1)","rgba(43,124,141,1)","rgba(43,126,141,1)","rgba(63,80,137,1)","rgba(64,75,136,1)","rgba(42,150,136,1)","rgba(43,143,138,1)","rgba(70,36,106,1)","rgba(61,88,138,1)","rgba(65,67,134,1)","rgba(43,140,138,1)","rgba(64,74,136,1)","rgba(56,101,140,1)","rgba(57,98,139,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(59,95,139,1)","rgba(37,164,133,1)","rgba(65,67,134,1)","rgba(70,41,110,1)","rgba(57,98,139,1)","rgba(44,135,139,1)","rgba(47,115,141,1)","rgba(62,83,137,1)","rgba(47,115,141,1)","rgba(51,109,141,1)","rgba(43,123,142,1)","rgba(61,87,138,1)","rgba(44,134,139,1)","rgba(43,125,141,1)","rgba(69,15,91,1)","rgba(52,108,140,1)","rgba(44,117,142,1)","rgba(65,177,122,1)","rgba(42,148,137,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(70,40,109,1)","rgba(44,138,139,1)","rgba(70,37,107,1)","rgba(64,77,136,1)","rgba(69,20,94,1)","rgba(40,157,134,1)","rgba(63,81,137,1)","rgba(40,156,135,1)","rgba(66,65,132,1)","rgba(43,125,141,1)","rgba(50,111,141,1)","rgba(57,99,139,1)","rgba(42,151,136,1)","rgba(70,26,98,1)","rgba(69,22,95,1)","rgba(74,180,118,1)","rgba(43,145,137,1)","rgba(57,99,139,1)","rgba(70,30,101,1)","rgba(42,149,136,1)","rgba(43,126,141,1)","rgba(59,93,139,1)","rgba(43,141,138,1)","rgba(79,183,116,1)","rgba(54,105,140,1)","rgba(55,104,140,1)","rgba(43,143,138,1)","rgba(53,107,140,1)","rgba(139,212,78,1)","rgba(68,178,121,1)","rgba(43,145,137,1)","rgba(44,132,140,1)","rgba(68,55,122,1)","rgba(40,155,135,1)","rgba(44,118,142,1)","rgba(68,58,125,1)","rgba(42,120,142,1)","rgba(53,106,140,1)","rgba(77,182,117,1)","rgba(44,134,139,1)","rgba(37,162,133,1)","rgba(43,144,137,1)","rgba(47,114,141,1)","rgba(70,32,102,1)","rgba(68,2,85,1)","rgba(64,78,136,1)","rgba(56,100,139,1)","rgba(69,43,112,1)","rgba(46,116,141,1)","rgba(69,42,111,1)","rgba(50,111,141,1)","rgba(58,97,139,1)","rgba(68,1,84,1)","rgba(43,119,142,1)","rgba(43,124,141,1)","rgba(60,90,138,1)","rgba(54,105,140,1)","rgba(70,31,102,1)","rgba(43,142,138,1)","rgba(69,18,92,1)","rgba(44,132,140,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(42,122,142,1)","rgba(57,98,139,1)","rgba(64,74,136,1)","rgba(59,94,139,1)","rgba(53,106,140,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(60,92,138,1)","rgba(60,91,138,1)","rgba(51,109,141,1)","rgba(52,108,140,1)","rgba(60,91,138,1)","rgba(43,143,138,1)","rgba(43,128,141,1)","rgba(66,65,132,1)","rgba(69,42,111,1)","rgba(41,152,136,1)","rgba(70,28,100,1)","rgba(44,137,139,1)","rgba(61,88,138,1)","rgba(43,124,141,1)","rgba(70,30,101,1)","rgba(52,108,140,1)","rgba(60,90,138,1)","rgba(43,123,141,1)","rgba(65,69,135,1)","rgba(43,141,138,1)","rgba(40,169,131,1)","rgba(44,135,139,1)","rgba(37,163,133,1)","rgba(69,17,92,1)","rgba(60,90,138,1)","rgba(43,143,138,1)","rgba(50,111,141,1)","rgba(41,153,136,1)","rgba(53,107,140,1)","rgba(61,87,138,1)","rgba(47,114,141,1)","rgba(59,95,139,1)","rgba(41,154,135,1)","rgba(59,95,139,1)","rgba(42,148,136,1)","rgba(40,155,135,1)","rgba(66,177,122,1)","rgba(43,126,141,1)","rgba(60,92,138,1)","rgba(68,6,86,1)","rgba(70,35,105,1)","rgba(41,152,136,1)","rgba(51,109,141,1)","rgba(69,13,90,1)","rgba(53,106,140,1)","rgba(41,170,130,1)","rgba(50,111,141,1)","rgba(41,153,135,1)","rgba(44,138,139,1)","rgba(57,99,139,1)","rgba(54,104,140,1)","rgba(69,45,114,1)","rgba(44,133,140,1)","rgba(53,106,140,1)","rgba(42,149,136,1)","rgba(42,120,142,1)","rgba(43,119,142,1)","rgba(44,139,138,1)","rgba(44,133,140,1)","rgba(42,122,142,1)","rgba(70,42,111,1)","rgba(69,24,97,1)","rgba(69,44,113,1)","rgba(111,201,92,1)","rgba(68,54,122,1)","rgba(70,35,105,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(68,4,86,1)","rgba(44,139,138,1)","rgba(61,175,124,1)","rgba(70,35,105,1)","rgba(66,64,131,1)","rgba(70,32,102,1)","rgba(65,66,134,1)","rgba(51,109,141,1)","rgba(65,70,135,1)","rgba(41,153,135,1)","rgba(62,86,138,1)","rgba(44,138,139,1)","rgba(39,158,134,1)","rgba(42,148,137,1)","rgba(63,82,137,1)","rgba(43,141,138,1)","rgba(45,117,142,1)","rgba(64,73,136,1)","rgba(59,93,139,1)","rgba(43,123,142,1)","rgba(40,156,135,1)","rgba(60,91,138,1)","rgba(61,86,138,1)","rgba(51,109,141,1)","rgba(57,99,139,1)","rgba(42,121,142,1)","rgba(62,83,137,1)","rgba(44,136,139,1)","rgba(59,95,139,1)","rgba(44,136,139,1)","rgba(65,68,135,1)","rgba(42,148,137,1)","rgba(63,81,137,1)","rgba(57,98,139,1)","rgba(89,188,109,1)","rgba(64,72,136,1)","rgba(64,73,136,1)","rgba(39,159,134,1)","rgba(57,99,139,1)","rgba(43,144,137,1)","rgba(42,121,142,1)","rgba(35,166,132,1)","rgba(47,115,141,1)","rgba(43,128,141,1)","rgba(45,117,142,1)","rgba(43,126,141,1)","rgba(98,193,104,1)","rgba(43,140,138,1)","rgba(48,113,141,1)","rgba(70,30,101,1)","rgba(50,111,141,1)","rgba(70,31,101,1)","rgba(44,138,139,1)","rgba(58,97,139,1)","rgba(43,125,141,1)","rgba(71,179,120,1)","rgba(70,35,105,1)","rgba(70,27,98,1)","rgba(38,162,133,1)","rgba(70,33,103,1)","rgba(53,107,140,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(53,106,140,1)","rgba(44,136,139,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(43,140,138,1)","rgba(62,84,137,1)","rgba(43,145,137,1)","rgba(69,51,119,1)","rgba(69,43,112,1)","rgba(42,151,136,1)","rgba(43,145,137,1)","rgba(52,108,141,1)","rgba(43,119,142,1)","rgba(69,25,97,1)","rgba(63,82,137,1)","rgba(53,106,140,1)","rgba(43,125,141,1)","rgba(43,119,142,1)","rgba(52,108,140,1)","rgba(44,134,139,1)","rgba(42,148,137,1)","rgba(44,135,139,1)","rgba(44,133,140,1)","rgba(44,135,139,1)","rgba(58,96,139,1)","rgba(53,106,140,1)","rgba(61,89,138,1)","rgba(44,136,139,1)","rgba(64,75,136,1)","rgba(43,145,137,1)","rgba(48,113,141,1)","rgba(64,75,136,1)","rgba(64,177,123,1)","rgba(44,134,139,1)","rgba(74,181,118,1)","rgba(37,164,133,1)","rgba(61,86,138,1)","rgba(64,77,136,1)","rgba(38,161,134,1)","rgba(69,51,119,1)","rgba(55,102,140,1)","rgba(43,128,141,1)","rgba(56,101,140,1)","rgba(55,102,140,1)","rgba(43,142,138,1)","rgba(43,125,141,1)","rgba(52,172,127,1)","rgba(39,160,134,1)","rgba(55,102,140,1)","rgba(44,118,142,1)","rgba(42,122,142,1)","rgba(42,151,136,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(41,154,135,1)","rgba(63,79,137,1)","rgba(64,72,136,1)","rgba(52,107,140,1)","rgba(43,143,138,1)","rgba(44,132,140,1)","rgba(65,67,134,1)","rgba(43,125,141,1)","rgba(57,98,139,1)","rgba(70,26,98,1)","rgba(43,145,137,1)","rgba(70,31,102,1)","rgba(43,146,137,1)","rgba(45,117,142,1)","rgba(34,168,132,1)","rgba(54,104,140,1)","rgba(43,126,141,1)","rgba(69,50,118,1)","rgba(43,144,137,1)","rgba(54,105,140,1)","rgba(36,165,133,1)","rgba(48,113,141,1)","rgba(42,150,136,1)","rgba(43,128,141,1)","rgba(49,112,141,1)","rgba(43,123,142,1)","rgba(43,124,141,1)","rgba(46,116,141,1)","rgba(62,85,137,1)","rgba(69,52,120,1)","rgba(44,131,140,1)","rgba(59,93,138,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(43,126,141,1)","rgba(46,116,141,1)","rgba(70,27,99,1)","rgba(42,121,142,1)","rgba(37,169,131,1)","rgba(43,129,140,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(38,161,134,1)","rgba(66,66,133,1)","rgba(60,92,138,1)","rgba(41,154,135,1)","rgba(44,135,139,1)","rgba(41,153,135,1)","rgba(69,24,96,1)","rgba(38,161,134,1)","rgba(51,109,141,1)","rgba(69,24,96,1)","rgba(43,129,140,1)","rgba(69,20,94,1)","rgba(43,146,137,1)","rgba(70,33,103,1)","rgba(46,116,141,1)","rgba(53,106,140,1)","rgba(39,159,134,1)","rgba(42,121,142,1)","rgba(62,86,137,1)","rgba(43,130,140,1)","rgba(64,75,136,1)","rgba(47,115,141,1)","rgba(63,82,137,1)","rgba(70,36,106,1)","rgba(41,153,135,1)","rgba(70,41,110,1)","rgba(40,157,135,1)","rgba(42,148,137,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(44,136,139,1)","rgba(70,32,103,1)","rgba(51,109,141,1)","rgba(35,167,132,1)","rgba(68,55,123,1)","rgba(67,60,127,1)","rgba(43,140,138,1)","rgba(69,15,91,1)","rgba(43,141,138,1)","rgba(66,66,133,1)","rgba(69,25,97,1)","rgba(48,114,141,1)","rgba(45,117,142,1)","rgba(39,160,134,1)","rgba(42,121,142,1)","rgba(43,144,137,1)","rgba(56,102,140,1)","rgba(47,114,141,1)","rgba(57,99,139,1)","rgba(69,49,117,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(43,125,141,1)","rgba(43,139,138,1)","rgba(70,35,105,1)","rgba(56,102,140,1)","rgba(43,125,141,1)","rgba(40,156,135,1)","rgba(64,75,136,1)","rgba(66,63,130,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(53,107,140,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(112,202,92,1)","rgba(43,140,138,1)","rgba(65,67,134,1)","rgba(45,117,142,1)","rgba(44,133,140,1)","rgba(111,201,93,1)","rgba(85,186,112,1)","rgba(65,69,135,1)","rgba(66,65,132,1)","rgba(69,14,90,1)","rgba(43,147,137,1)","rgba(43,123,141,1)","rgba(43,130,140,1)","rgba(69,21,94,1)","rgba(43,123,141,1)","rgba(40,157,135,1)","rgba(43,127,141,1)","rgba(59,94,139,1)","rgba(62,86,138,1)","rgba(64,76,136,1)","rgba(81,184,114,1)","rgba(51,110,141,1)","rgba(67,62,129,1)","rgba(50,111,141,1)","rgba(65,67,134,1)","rgba(43,127,141,1)","rgba(45,117,142,1)","rgba(68,3,85,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(65,72,136,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(44,132,140,1)","rgba(42,150,136,1)","rgba(42,149,136,1)","rgba(43,127,141,1)","rgba(69,47,115,1)","rgba(57,98,139,1)","rgba(75,181,118,1)","rgba(66,63,130,1)","rgba(55,103,140,1)","rgba(43,128,141,1)","rgba(44,139,138,1)","rgba(68,5,86,1)","rgba(44,118,142,1)","rgba(70,32,103,1)","rgba(43,124,141,1)","rgba(56,101,140,1)","rgba(68,7,87,1)","rgba(43,130,140,1)","rgba(34,168,132,1)","rgba(43,143,138,1)","rgba(54,105,140,1)","rgba(57,98,139,1)","rgba(42,120,142,1)","rgba(44,134,139,1)","rgba(42,121,142,1)","rgba(70,33,103,1)","rgba(46,116,142,1)","rgba(44,133,140,1)","rgba(70,27,99,1)","rgba(69,22,95,1)","rgba(50,111,141,1)","rgba(44,133,140,1)","rgba(69,51,118,1)","rgba(53,107,140,1)","rgba(43,127,141,1)","rgba(51,109,141,1)","rgba(58,96,139,1)","rgba(44,133,140,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(48,114,141,1)","rgba(66,63,130,1)","rgba(44,139,138,1)","rgba(45,117,142,1)","rgba(53,106,140,1)","rgba(44,133,140,1)","rgba(58,97,139,1)","rgba(39,159,134,1)","rgba(63,78,136,1)","rgba(64,74,136,1)","rgba(96,192,105,1)","rgba(121,208,83,1)","rgba(43,147,137,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(43,125,141,1)","rgba(44,134,139,1)","rgba(46,115,141,1)","rgba(47,115,141,1)","rgba(42,148,136,1)","rgba(52,107,140,1)","rgba(42,151,136,1)","rgba(67,178,121,1)","rgba(69,19,93,1)","rgba(87,187,110,1)","rgba(43,119,142,1)","rgba(42,148,137,1)","rgba(49,112,141,1)","rgba(48,113,141,1)","rgba(43,145,137,1)","rgba(43,144,137,1)","rgba(42,122,142,1)","rgba(43,127,141,1)","rgba(42,121,142,1)","rgba(43,123,141,1)","rgba(42,151,136,1)","rgba(43,127,141,1)","rgba(42,122,142,1)","rgba(49,112,141,1)","rgba(45,116,142,1)","rgba(59,95,139,1)","rgba(43,125,141,1)","rgba(52,108,140,1)","rgba(38,161,134,1)","rgba(48,113,141,1)","rgba(37,163,133,1)","rgba(52,107,140,1)","rgba(49,113,141,1)","rgba(48,113,141,1)","rgba(43,125,141,1)","rgba(68,56,123,1)","rgba(70,31,102,1)","rgba(69,13,90,1)","rgba(51,109,141,1)","rgba(38,161,134,1)","rgba(60,91,138,1)","rgba(44,131,140,1)","rgba(67,61,128,1)","rgba(49,171,128,1)","rgba(43,129,140,1)","rgba(69,12,89,1)","rgba(42,122,142,1)","rgba(70,26,98,1)","rgba(44,137,139,1)","rgba(70,30,101,1)","rgba(42,149,136,1)","rgba(43,128,141,1)","rgba(35,166,133,1)","rgba(44,139,138,1)","rgba(58,96,139,1)","rgba(46,116,142,1)","rgba(43,140,138,1)","rgba(61,87,138,1)","rgba(43,119,142,1)","rgba(80,183,115,1)","rgba(42,120,142,1)","rgba(68,56,123,1)","rgba(43,142,138,1)","rgba(39,158,134,1)","rgba(69,9,88,1)","rgba(43,145,137,1)","rgba(43,143,138,1)","rgba(57,100,139,1)","rgba(43,127,141,1)","rgba(114,204,89,1)","rgba(69,21,95,1)","rgba(49,112,141,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(115,204,88,1)","rgba(43,141,138,1)","rgba(39,158,134,1)","rgba(37,164,133,1)","rgba(63,83,137,1)","rgba(53,106,140,1)","rgba(41,153,136,1)","rgba(61,86,138,1)","rgba(59,93,138,1)","rgba(69,44,112,1)","rgba(43,140,138,1)","rgba(53,106,140,1)","rgba(62,86,137,1)","rgba(64,78,136,1)","rgba(107,199,96,1)","rgba(42,120,142,1)","rgba(52,172,127,1)","rgba(44,118,142,1)","rgba(62,84,137,1)","rgba(69,50,118,1)","rgba(42,122,142,1)","rgba(70,39,108,1)","rgba(69,19,93,1)","rgba(43,126,141,1)","rgba(42,148,137,1)","rgba(53,105,140,1)","rgba(49,112,141,1)","rgba(40,156,135,1)","rgba(43,139,138,1)","rgba(44,137,139,1)","rgba(43,128,140,1)","rgba(36,165,133,1)","rgba(44,135,139,1)","rgba(35,167,132,1)","rgba(68,58,125,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(61,89,138,1)","rgba(43,143,138,1)","rgba(69,51,119,1)","rgba(41,152,136,1)","rgba(40,156,135,1)","rgba(42,149,136,1)","rgba(65,70,135,1)","rgba(41,154,135,1)","rgba(55,102,140,1)","rgba(69,49,117,1)","rgba(44,132,140,1)","rgba(39,159,134,1)","rgba(43,140,138,1)","rgba(44,135,139,1)","rgba(65,67,134,1)","rgba(45,117,142,1)","rgba(41,152,136,1)","rgba(65,72,136,1)","rgba(43,130,140,1)","rgba(64,73,136,1)","rgba(50,110,141,1)","rgba(42,147,137,1)","rgba(43,130,140,1)","rgba(40,169,131,1)","rgba(56,100,139,1)","rgba(69,13,90,1)","rgba(45,117,142,1)","rgba(68,57,125,1)","rgba(66,63,130,1)","rgba(42,120,142,1)","rgba(67,59,127,1)","rgba(44,137,139,1)","rgba(43,129,140,1)","rgba(61,89,138,1)","rgba(53,107,140,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(66,63,130,1)","rgba(65,71,135,1)","rgba(43,145,137,1)","rgba(43,127,141,1)","rgba(43,143,138,1)","rgba(47,115,141,1)","rgba(44,133,140,1)","rgba(69,48,116,1)","rgba(56,101,140,1)","rgba(68,55,123,1)","rgba(53,106,140,1)","rgba(70,36,105,1)","rgba(43,128,141,1)","rgba(42,147,137,1)","rgba(50,111,141,1)","rgba(39,159,134,1)","rgba(43,130,140,1)","rgba(51,109,141,1)","rgba(42,120,142,1)","rgba(51,109,141,1)","rgba(44,138,139,1)","rgba(63,80,137,1)","rgba(44,137,139,1)","rgba(43,130,140,1)","rgba(44,131,140,1)","rgba(61,88,138,1)","rgba(62,85,137,1)","rgba(62,83,137,1)","rgba(67,61,129,1)","rgba(65,72,136,1)","rgba(69,44,113,1)","rgba(69,13,90,1)","rgba(36,164,133,1)","rgba(40,156,135,1)","rgba(43,140,138,1)","rgba(44,132,140,1)","rgba(43,142,138,1)","rgba(63,176,123,1)","rgba(69,11,89,1)","rgba(67,62,129,1)","rgba(44,133,140,1)","rgba(42,150,136,1)","rgba(43,144,137,1)","rgba(64,76,136,1)","rgba(47,114,141,1)","rgba(35,166,132,1)","rgba(68,4,86,1)","rgba(41,152,136,1)","rgba(43,141,138,1)","rgba(44,136,139,1)","rgba(43,125,141,1)","rgba(43,142,138,1)","rgba(43,128,141,1)","rgba(60,92,138,1)","rgba(70,39,108,1)","rgba(46,116,141,1)","rgba(42,148,137,1)","rgba(56,101,140,1)","rgba(53,106,140,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(47,115,141,1)","rgba(122,209,81,1)","rgba(39,159,134,1)","rgba(58,174,125,1)","rgba(43,146,137,1)","rgba(51,172,128,1)","rgba(62,86,138,1)","rgba(36,165,133,1)","rgba(50,172,128,1)","rgba(43,125,141,1)","rgba(43,145,137,1)","rgba(58,97,139,1)","rgba(43,127,141,1)","rgba(63,81,137,1)","rgba(44,135,139,1)","rgba(43,124,141,1)","rgba(61,89,138,1)","rgba(55,103,140,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(59,93,138,1)","rgba(43,146,137,1)","rgba(43,139,138,1)","rgba(62,85,137,1)","rgba(70,38,107,1)","rgba(59,95,139,1)","rgba(56,101,140,1)","rgba(54,104,140,1)","rgba(57,99,139,1)","rgba(69,51,118,1)","rgba(43,127,141,1)","rgba(43,144,137,1)","rgba(52,108,141,1)","rgba(43,124,141,1)","rgba(43,142,138,1)","rgba(83,185,113,1)","rgba(59,175,125,1)","rgba(70,29,100,1)","rgba(44,136,139,1)","rgba(64,73,136,1)","rgba(70,36,106,1)","rgba(70,34,104,1)","rgba(44,133,140,1)","rgba(52,108,140,1)","rgba(48,114,141,1)","rgba(43,119,142,1)","rgba(43,130,140,1)","rgba(54,105,140,1)","rgba(65,67,134,1)","rgba(43,128,141,1)","rgba(42,149,136,1)","rgba(43,146,137,1)","rgba(55,103,140,1)","rgba(54,105,140,1)","rgba(40,169,131,1)","rgba(61,87,138,1)","rgba(69,50,118,1)","rgba(69,45,113,1)","rgba(53,106,140,1)","rgba(43,126,141,1)","rgba(43,127,141,1)","rgba(70,179,120,1)","rgba(69,23,96,1)","rgba(70,30,101,1)","rgba(44,133,140,1)","rgba(69,23,96,1)","rgba(52,109,141,1)","rgba(44,118,142,1)","rgba(54,104,140,1)","rgba(50,110,141,1)","rgba(49,112,141,1)","rgba(43,123,141,1)","rgba(34,168,132,1)","rgba(36,164,133,1)","rgba(43,128,141,1)","rgba(45,170,129,1)","rgba(70,32,102,1)","rgba(43,146,137,1)","rgba(40,155,135,1)","rgba(42,120,142,1)","rgba(61,89,138,1)","rgba(62,86,138,1)","rgba(57,99,139,1)","rgba(58,174,125,1)","rgba(44,138,139,1)","rgba(50,111,141,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(50,110,141,1)","rgba(55,103,140,1)","rgba(42,120,142,1)","rgba(64,73,136,1)","rgba(67,59,126,1)","rgba(47,115,141,1)","rgba(58,97,139,1)","rgba(69,43,111,1)","rgba(62,86,138,1)","rgba(43,127,141,1)","rgba(40,156,135,1)","rgba(47,115,141,1)","rgba(43,127,141,1)","rgba(66,66,133,1)","rgba(43,127,141,1)","rgba(68,54,121,1)","rgba(64,74,136,1)","rgba(58,97,139,1)","rgba(43,127,141,1)","rgba(43,139,138,1)","rgba(43,146,137,1)","rgba(70,40,109,1)","rgba(36,166,133,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(63,79,137,1)","rgba(71,179,119,1)","rgba(56,102,140,1)","rgba(59,93,139,1)","rgba(68,54,121,1)","rgba(51,109,141,1)","rgba(65,177,122,1)","rgba(64,75,136,1)","rgba(66,66,133,1)","rgba(112,202,92,1)","rgba(55,102,140,1)","rgba(65,71,135,1)","rgba(43,129,140,1)","rgba(42,122,142,1)","rgba(42,150,136,1)","rgba(43,128,141,1)","rgba(41,153,135,1)","rgba(44,134,139,1)","rgba(70,28,99,1)","rgba(44,137,139,1)","rgba(46,116,141,1)","rgba(48,171,128,1)","rgba(62,176,123,1)","rgba(68,6,86,1)","rgba(70,33,103,1)","rgba(34,167,132,1)","rgba(63,80,137,1)","rgba(52,108,140,1)","rgba(69,21,94,1)","rgba(44,134,139,1)","rgba(43,142,138,1)","rgba(44,136,139,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(49,112,141,1)","rgba(43,128,141,1)","rgba(43,146,137,1)","rgba(54,104,140,1)","rgba(62,86,138,1)","rgba(43,130,140,1)","rgba(59,95,139,1)","rgba(65,71,135,1)","rgba(42,122,142,1)","rgba(67,62,129,1)","rgba(46,116,142,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(44,133,140,1)","rgba(69,24,97,1)","rgba(44,134,139,1)","rgba(55,102,140,1)","rgba(70,36,106,1)","rgba(68,2,84,1)","rgba(51,109,141,1)","rgba(43,139,138,1)","rgba(62,84,137,1)","rgba(41,153,135,1)","rgba(38,169,131,1)","rgba(59,94,139,1)","rgba(48,114,141,1)","rgba(41,152,136,1)","rgba(42,149,136,1)","rgba(60,92,138,1)","rgba(57,99,139,1)","rgba(53,106,140,1)","rgba(59,93,139,1)","rgba(43,126,141,1)","rgba(56,174,126,1)","rgba(45,117,142,1)","rgba(69,51,119,1)","rgba(54,105,140,1)","rgba(51,109,141,1)","rgba(65,72,136,1)","rgba(61,87,138,1)","rgba(40,156,135,1)","rgba(44,138,139,1)","rgba(62,83,137,1)","rgba(51,172,128,1)","rgba(43,119,142,1)","rgba(49,112,141,1)","rgba(42,120,142,1)","rgba(43,129,140,1)","rgba(68,53,120,1)","rgba(69,25,97,1)","rgba(68,53,121,1)","rgba(43,139,138,1)","rgba(43,127,141,1)","rgba(70,30,101,1)","rgba(58,97,139,1)","rgba(44,137,139,1)","rgba(44,134,139,1)","rgba(43,144,137,1)","rgba(69,22,95,1)","rgba(68,56,123,1)","rgba(44,131,140,1)","rgba(43,130,140,1)","rgba(44,133,140,1)","rgba(70,34,104,1)","rgba(43,127,141,1)","rgba(66,65,132,1)","rgba(47,115,141,1)","rgba(63,78,136,1)","rgba(48,113,141,1)","rgba(70,31,102,1)","rgba(59,93,138,1)","rgba(47,115,141,1)","rgba(58,96,139,1)","rgba(44,136,139,1)","rgba(65,68,135,1)","rgba(43,124,141,1)","rgba(52,108,141,1)","rgba(70,37,107,1)","rgba(44,132,140,1)","rgba(44,138,139,1)","rgba(70,36,105,1)","rgba(46,116,142,1)","rgba(42,122,142,1)","rgba(43,128,141,1)","rgba(62,83,137,1)","rgba(44,136,139,1)","rgba(64,74,136,1)","rgba(61,87,138,1)","rgba(50,111,141,1)","rgba(40,157,134,1)","rgba(56,101,140,1)","rgba(43,147,137,1)","rgba(59,93,139,1)","rgba(70,37,106,1)","rgba(42,120,142,1)","rgba(69,43,112,1)","rgba(49,112,141,1)","rgba(59,94,139,1)","rgba(43,124,141,1)","rgba(44,136,139,1)","rgba(42,120,142,1)","rgba(63,79,137,1)","rgba(43,126,141,1)","rgba(48,113,141,1)","rgba(38,161,134,1)","rgba(49,112,141,1)","rgba(37,163,133,1)","rgba(42,151,136,1)","rgba(44,136,139,1)","rgba(68,57,125,1)","rgba(62,83,137,1)","rgba(69,43,111,1)","rgba(70,33,103,1)","rgba(69,46,115,1)","rgba(55,103,140,1)","rgba(68,2,84,1)","rgba(69,16,91,1)","rgba(43,141,138,1)","rgba(44,139,138,1)","rgba(54,105,140,1)","rgba(44,131,140,1)","rgba(65,70,135,1)","rgba(59,93,139,1)","rgba(65,72,136,1)","rgba(37,163,133,1)","rgba(68,53,121,1)","rgba(57,98,139,1)","rgba(63,79,137,1)","rgba(42,151,136,1)","rgba(49,112,141,1)","rgba(44,134,139,1)","rgba(59,93,138,1)","rgba(44,132,140,1)","rgba(43,145,137,1)","rgba(42,120,142,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(64,73,136,1)","rgba(39,158,134,1)","rgba(69,48,116,1)","rgba(68,58,125,1)","rgba(41,153,135,1)","rgba(70,32,102,1)","rgba(55,103,140,1)","rgba(68,54,122,1)","rgba(69,47,115,1)","rgba(63,79,137,1)","rgba(45,117,142,1)","rgba(68,1,84,1)","rgba(43,124,141,1)","rgba(43,123,141,1)","rgba(44,137,139,1)","rgba(62,83,137,1)","rgba(53,106,140,1)","rgba(60,90,138,1)","rgba(44,118,142,1)","rgba(47,115,141,1)","rgba(44,131,140,1)","rgba(69,53,120,1)","rgba(68,58,125,1)","rgba(43,140,138,1)","rgba(57,99,139,1)","rgba(61,87,138,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(62,83,137,1)","rgba(44,134,139,1)","rgba(69,45,114,1)","rgba(115,204,89,1)","rgba(70,40,109,1)","rgba(43,146,137,1)","rgba(43,147,137,1)","rgba(50,111,141,1)","rgba(49,113,141,1)","rgba(58,96,139,1)","rgba(43,119,142,1)","rgba(66,66,133,1)","rgba(43,143,138,1)","rgba(55,173,126,1)","rgba(69,52,120,1)","rgba(69,21,95,1)","rgba(45,117,142,1)","rgba(64,78,136,1)","rgba(43,125,141,1)","rgba(44,138,139,1)","rgba(59,94,139,1)","rgba(47,115,141,1)","rgba(43,140,138,1)","rgba(70,36,106,1)","rgba(43,130,140,1)","rgba(70,34,104,1)","rgba(62,86,137,1)","rgba(64,74,136,1)","rgba(43,145,137,1)","rgba(62,84,137,1)","rgba(56,101,140,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(44,118,142,1)","rgba(48,114,141,1)","rgba(42,148,136,1)","rgba(43,125,141,1)","rgba(49,113,141,1)","rgba(43,146,137,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(56,174,126,1)","rgba(43,127,141,1)","rgba(41,154,135,1)","rgba(69,14,91,1)","rgba(70,29,100,1)","rgba(65,71,135,1)","rgba(43,127,141,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(59,93,139,1)","rgba(47,115,141,1)","rgba(44,133,140,1)","rgba(69,47,115,1)","rgba(40,155,135,1)","rgba(44,134,139,1)","rgba(57,99,139,1)","rgba(42,121,142,1)","rgba(43,144,137,1)","rgba(60,92,138,1)","rgba(43,142,138,1)","rgba(55,102,140,1)","rgba(64,74,136,1)","rgba(55,103,140,1)","rgba(42,148,137,1)","rgba(51,172,127,1)","rgba(43,147,137,1)","rgba(38,160,134,1)","rgba(68,7,87,1)","rgba(59,94,139,1)","rgba(68,57,125,1)","rgba(51,110,141,1)","rgba(38,161,134,1)","rgba(64,75,136,1)","rgba(62,85,137,1)","rgba(52,108,140,1)","rgba(64,176,123,1)","rgba(41,152,136,1)","rgba(48,114,141,1)","rgba(62,85,137,1)","rgba(70,40,109,1)","rgba(68,5,86,1)","rgba(63,82,137,1)","rgba(58,97,139,1)","rgba(67,61,128,1)","rgba(43,127,141,1)","rgba(39,169,131,1)","rgba(41,152,136,1)","rgba(43,142,138,1)","rgba(54,173,126,1)","rgba(44,134,139,1)","rgba(44,132,140,1)","rgba(69,22,95,1)","rgba(43,130,140,1)","rgba(44,131,140,1)","rgba(50,111,141,1)","rgba(43,145,137,1)","rgba(43,130,140,1)","rgba(70,32,103,1)","rgba(43,119,142,1)","rgba(44,138,139,1)","rgba(43,145,137,1)","rgba(44,137,139,1)","rgba(42,121,142,1)","rgba(42,120,142,1)","rgba(59,93,139,1)","rgba(42,121,142,1)","rgba(43,126,141,1)","rgba(59,93,138,1)","rgba(42,148,137,1)","rgba(43,147,137,1)","rgba(43,124,141,1)","rgba(39,159,134,1)","rgba(42,122,142,1)","rgba(43,129,140,1)","rgba(44,133,140,1)","rgba(43,140,138,1)","rgba(38,169,131,1)","rgba(49,112,141,1)","rgba(70,27,98,1)","rgba(43,124,141,1)","rgba(44,137,139,1)","rgba(37,162,133,1)","rgba(58,96,139,1)","rgba(42,120,142,1)","rgba(47,114,141,1)","rgba(68,2,85,1)","rgba(46,116,142,1)","rgba(54,105,140,1)","rgba(69,51,119,1)","rgba(62,176,123,1)","rgba(54,104,140,1)","rgba(61,87,138,1)","rgba(69,15,91,1)","rgba(44,137,139,1)","rgba(43,123,141,1)","rgba(49,171,128,1)","rgba(69,42,111,1)","rgba(42,120,142,1)","rgba(43,142,138,1)","rgba(35,167,132,1)","rgba(44,136,139,1)","rgba(44,118,142,1)","rgba(55,103,140,1)","rgba(42,120,142,1)","rgba(47,114,141,1)","rgba(62,83,137,1)","rgba(44,135,139,1)","rgba(70,42,111,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(49,112,141,1)","rgba(46,116,142,1)","rgba(49,112,141,1)","rgba(50,111,141,1)","rgba(55,102,140,1)","rgba(60,90,138,1)","rgba(66,63,130,1)","rgba(41,154,135,1)","rgba(64,77,136,1)","rgba(37,162,133,1)","rgba(44,134,139,1)","rgba(63,80,137,1)","rgba(50,111,141,1)","rgba(53,106,140,1)","rgba(50,110,141,1)","rgba(58,97,139,1)","rgba(55,103,140,1)","rgba(69,10,88,1)","rgba(69,22,95,1)","rgba(69,43,112,1)","rgba(52,107,140,1)","rgba(41,153,135,1)","rgba(44,118,142,1)","rgba(85,186,112,1)","rgba(41,153,136,1)","rgba(55,103,140,1)","rgba(40,157,135,1)","rgba(43,124,141,1)","rgba(43,139,138,1)","rgba(41,152,136,1)","rgba(42,151,136,1)","rgba(43,119,142,1)","rgba(60,90,138,1)","rgba(50,111,141,1)","rgba(45,117,142,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(34,168,132,1)","rgba(44,135,139,1)","rgba(42,120,142,1)","rgba(67,62,129,1)","rgba(43,124,141,1)","rgba(42,151,136,1)","rgba(37,162,133,1)","rgba(43,125,141,1)","rgba(43,142,138,1)","rgba(43,140,138,1)","rgba(43,141,138,1)","rgba(43,143,138,1)","rgba(69,52,120,1)","rgba(43,140,138,1)","rgba(43,146,137,1)","rgba(66,64,131,1)","rgba(74,180,118,1)","rgba(54,105,140,1)","rgba(67,61,129,1)","rgba(53,107,140,1)","rgba(43,140,138,1)","rgba(35,167,132,1)","rgba(43,126,141,1)","rgba(48,171,129,1)","rgba(53,107,140,1)","rgba(68,1,84,1)","rgba(41,154,135,1)","rgba(56,102,140,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(68,58,126,1)","rgba(56,101,140,1)","rgba(44,135,139,1)","rgba(42,121,142,1)","rgba(68,55,123,1)","rgba(56,100,139,1)","rgba(63,81,137,1)","rgba(43,126,141,1)","rgba(41,154,135,1)","rgba(43,125,141,1)","rgba(49,172,128,1)","rgba(68,57,124,1)","rgba(65,69,135,1)","rgba(67,63,130,1)","rgba(64,75,136,1)","rgba(42,122,142,1)","rgba(35,167,132,1)","rgba(137,211,78,1)","rgba(58,97,139,1)","rgba(50,111,141,1)","rgba(43,127,141,1)","rgba(64,76,136,1)","rgba(59,95,139,1)","rgba(43,145,137,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(43,123,141,1)","rgba(44,139,138,1)","rgba(42,121,142,1)","rgba(66,65,132,1)","rgba(58,96,139,1)","rgba(58,96,139,1)","rgba(43,124,141,1)","rgba(50,110,141,1)","rgba(68,3,85,1)","rgba(68,55,123,1)","rgba(48,114,141,1)","rgba(56,101,140,1)","rgba(41,153,136,1)","rgba(60,91,138,1)","rgba(36,164,133,1)","rgba(43,141,138,1)","rgba(57,99,139,1)","rgba(44,131,140,1)","rgba(51,110,141,1)","rgba(43,125,141,1)","rgba(65,67,134,1)","rgba(42,120,142,1)","rgba(62,83,137,1)","rgba(56,100,139,1)","rgba(40,156,135,1)","rgba(44,132,140,1)","rgba(54,104,140,1)","rgba(43,129,140,1)","rgba(44,139,138,1)","rgba(52,108,140,1)","rgba(37,163,133,1)","rgba(65,72,135,1)","rgba(66,66,133,1)","rgba(63,80,137,1)","rgba(43,144,137,1)","rgba(43,126,141,1)","rgba(65,70,135,1)","rgba(52,108,140,1)","rgba(69,47,115,1)","rgba(64,78,136,1)","rgba(41,153,136,1)","rgba(68,55,122,1)","rgba(55,102,140,1)","rgba(43,125,141,1)","rgba(44,135,139,1)","rgba(54,105,140,1)","rgba(39,160,134,1)","rgba(57,99,139,1)","rgba(69,18,93,1)","rgba(44,136,139,1)","rgba(62,86,138,1)","rgba(40,157,135,1)","rgba(56,100,139,1)","rgba(56,100,139,1)","rgba(53,106,140,1)","rgba(60,92,138,1)","rgba(69,47,115,1)","rgba(53,106,140,1)","rgba(70,30,101,1)","rgba(64,73,136,1)","rgba(57,98,139,1)","rgba(53,106,140,1)","rgba(64,74,136,1)","rgba(40,156,135,1)","rgba(69,9,88,1)","rgba(69,9,88,1)","rgba(70,26,98,1)","rgba(44,137,139,1)","rgba(62,86,137,1)","rgba(42,121,142,1)","rgba(44,138,139,1)","rgba(49,112,141,1)","rgba(59,95,139,1)","rgba(69,24,96,1)","rgba(47,114,141,1)","rgba(69,50,118,1)","rgba(55,102,140,1)","rgba(57,99,139,1)","rgba(44,118,142,1)","rgba(43,130,140,1)","rgba(61,87,138,1)","rgba(49,111,141,1)","rgba(44,138,139,1)","rgba(43,141,138,1)","rgba(43,140,138,1)","rgba(46,115,141,1)","rgba(63,80,137,1)","rgba(68,1,84,1)","rgba(43,119,142,1)","rgba(62,83,137,1)","rgba(69,50,118,1)","rgba(58,97,139,1)","rgba(36,165,133,1)","rgba(45,116,142,1)","rgba(58,96,139,1)","rgba(43,126,141,1)","rgba(70,36,106,1)","rgba(69,8,87,1)","rgba(54,105,140,1)","rgba(54,105,140,1)","rgba(43,141,138,1)","rgba(35,166,133,1)","rgba(57,99,139,1)","rgba(69,51,119,1)","rgba(44,131,140,1)","rgba(49,112,141,1)","rgba(56,101,140,1)","rgba(55,103,140,1)","rgba(59,94,139,1)","rgba(42,149,136,1)","rgba(42,149,136,1)","rgba(44,134,139,1)","rgba(41,151,136,1)","rgba(47,114,141,1)","rgba(49,113,141,1)","rgba(60,90,138,1)","rgba(43,143,138,1)","rgba(43,144,137,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(56,100,139,1)","rgba(43,129,140,1)","rgba(50,111,141,1)","rgba(67,60,128,1)","rgba(43,123,141,1)","rgba(69,49,117,1)","rgba(62,86,138,1)","rgba(63,176,123,1)","rgba(37,164,133,1)","rgba(43,141,138,1)","rgba(70,28,99,1)","rgba(44,118,142,1)","rgba(43,123,141,1)","rgba(65,71,135,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(59,94,139,1)","rgba(61,87,138,1)","rgba(56,102,140,1)","rgba(42,150,136,1)","rgba(64,77,136,1)","rgba(60,91,138,1)","rgba(57,99,139,1)","rgba(70,31,101,1)","rgba(46,116,142,1)","rgba(51,109,141,1)","rgba(53,107,140,1)","rgba(45,116,142,1)","rgba(67,61,129,1)","rgba(43,127,141,1)","rgba(43,127,141,1)","rgba(45,171,129,1)","rgba(67,59,127,1)","rgba(43,127,141,1)","rgba(63,176,123,1)","rgba(65,70,135,1)","rgba(39,159,134,1)","rgba(42,151,136,1)","rgba(48,114,141,1)","rgba(69,50,118,1)","rgba(64,77,136,1)","rgba(102,195,100,1)","rgba(66,63,130,1)","rgba(70,29,100,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(43,130,140,1)","rgba(43,129,140,1)","rgba(43,141,138,1)","rgba(69,45,113,1)","rgba(44,135,139,1)","rgba(41,154,135,1)","rgba(44,131,140,1)","rgba(43,129,140,1)","rgba(50,111,141,1)","rgba(98,193,103,1)","rgba(40,155,135,1)","rgba(93,190,107,1)","rgba(52,107,140,1)","rgba(53,106,140,1)","rgba(40,155,135,1)","rgba(43,146,137,1)","rgba(70,29,100,1)","rgba(42,150,136,1)","rgba(43,129,140,1)","rgba(70,26,98,1)","rgba(58,97,139,1)","rgba(71,179,120,1)","rgba(69,8,87,1)","rgba(43,142,138,1)","rgba(44,138,139,1)","rgba(64,76,136,1)","rgba(44,134,139,1)","rgba(48,114,141,1)","rgba(42,148,137,1)","rgba(43,124,141,1)","rgba(43,139,138,1)","rgba(69,12,89,1)","rgba(43,128,141,1)","rgba(68,58,126,1)","rgba(41,154,135,1)","rgba(42,148,137,1)","rgba(40,157,134,1)","rgba(57,98,139,1)","rgba(43,140,138,1)","rgba(68,2,84,1)","rgba(52,172,127,1)","rgba(113,202,91,1)","rgba(43,128,141,1)","rgba(42,120,142,1)","rgba(42,122,142,1)","rgba(42,149,136,1)","rgba(51,109,141,1)","rgba(43,144,137,1)","rgba(57,99,139,1)","rgba(51,109,141,1)","rgba(43,119,142,1)","rgba(51,172,127,1)","rgba(59,93,138,1)","rgba(50,110,141,1)","rgba(61,88,138,1)","rgba(150,214,75,1)","rgba(69,13,90,1)","rgba(43,123,141,1)","rgba(52,107,140,1)","rgba(50,172,128,1)","rgba(44,134,139,1)","rgba(57,99,139,1)","rgba(40,155,135,1)","rgba(41,154,135,1)","rgba(42,120,142,1)","rgba(64,76,136,1)","rgba(49,112,141,1)","rgba(43,127,141,1)","rgba(44,138,139,1)","rgba(53,107,140,1)","rgba(43,143,138,1)","rgba(67,62,129,1)","rgba(43,142,138,1)","rgba(43,119,142,1)","rgba(42,120,142,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(56,102,140,1)","rgba(54,104,140,1)","rgba(69,45,113,1)","rgba(70,36,105,1)","rgba(43,147,137,1)","rgba(66,64,131,1)","rgba(60,92,138,1)","rgba(68,4,85,1)","rgba(69,44,113,1)","rgba(43,142,138,1)","rgba(61,89,138,1)","rgba(43,143,138,1)","rgba(52,108,140,1)","rgba(37,162,133,1)","rgba(42,150,136,1)","rgba(43,147,137,1)","rgba(44,138,139,1)","rgba(68,3,85,1)","rgba(69,44,112,1)","rgba(43,123,141,1)","rgba(43,127,141,1)","rgba(44,135,139,1)","rgba(35,168,132,1)","rgba(56,101,140,1)","rgba(44,139,138,1)","rgba(52,108,140,1)","rgba(52,107,140,1)","rgba(50,110,141,1)","rgba(56,101,140,1)","rgba(44,137,139,1)","rgba(69,17,92,1)","rgba(44,133,140,1)","rgba(65,72,136,1)","rgba(58,95,139,1)","rgba(65,70,135,1)","rgba(50,111,141,1)","rgba(65,68,135,1)","rgba(44,133,140,1)","rgba(54,104,140,1)","rgba(70,35,105,1)","rgba(57,98,139,1)","rgba(43,130,140,1)","rgba(35,167,132,1)","rgba(50,110,141,1)","rgba(58,174,125,1)","rgba(60,90,138,1)","rgba(43,140,138,1)","rgba(44,135,139,1)","rgba(43,126,141,1)","rgba(43,128,140,1)","rgba(60,91,138,1)","rgba(39,159,134,1)","rgba(56,101,140,1)","rgba(64,78,136,1)","rgba(59,95,139,1)","rgba(39,160,134,1)","rgba(52,108,140,1)","rgba(44,138,139,1)","rgba(39,158,134,1)","rgba(50,110,141,1)","rgba(44,133,140,1)","rgba(44,131,140,1)","rgba(43,130,140,1)","rgba(43,127,141,1)","rgba(47,115,141,1)","rgba(69,18,92,1)","rgba(43,124,141,1)","rgba(58,97,139,1)","rgba(48,113,141,1)","rgba(43,141,138,1)","rgba(60,90,138,1)","rgba(53,106,140,1)","rgba(38,160,134,1)","rgba(64,78,136,1)","rgba(42,122,142,1)","rgba(70,36,106,1)","rgba(42,120,142,1)","rgba(65,71,135,1)","rgba(55,103,140,1)","rgba(50,111,141,1)","rgba(70,179,120,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(53,106,140,1)","rgba(64,73,136,1)","rgba(43,144,137,1)","rgba(60,93,138,1)","rgba(46,116,142,1)","rgba(69,43,112,1)","rgba(53,106,140,1)","rgba(60,91,138,1)","rgba(69,51,119,1)","rgba(44,134,139,1)","rgba(44,136,139,1)","rgba(70,39,108,1)","rgba(41,153,136,1)","rgba(68,53,120,1)","rgba(82,184,114,1)","rgba(43,145,137,1)","rgba(57,98,139,1)","rgba(57,99,139,1)","rgba(109,200,94,1)","rgba(44,135,139,1)","rgba(43,142,138,1)","rgba(68,5,86,1)","rgba(38,161,134,1)","rgba(44,138,139,1)","rgba(67,62,130,1)","rgba(42,120,142,1)","rgba(42,121,142,1)","rgba(43,123,142,1)","rgba(44,132,140,1)","rgba(70,29,100,1)","rgba(40,157,135,1)","rgba(58,96,139,1)","rgba(43,142,138,1)","rgba(39,159,134,1)","rgba(42,122,142,1)","rgba(57,99,139,1)","rgba(70,31,102,1)","rgba(60,91,138,1)","rgba(58,96,139,1)","rgba(61,88,138,1)","rgba(51,172,127,1)","rgba(62,84,137,1)","rgba(67,59,127,1)","rgba(57,99,139,1)","rgba(42,122,142,1)","rgba(42,150,136,1)","rgba(68,55,123,1)","rgba(109,200,94,1)","rgba(51,110,141,1)","rgba(53,106,140,1)","rgba(60,90,138,1)","rgba(69,15,91,1)","rgba(44,135,139,1)","rgba(56,174,126,1)","rgba(43,123,141,1)","rgba(44,118,142,1)","rgba(44,132,140,1)","rgba(62,83,137,1)","rgba(49,112,141,1)","rgba(43,125,141,1)","rgba(64,76,136,1)","rgba(64,73,136,1)","rgba(66,64,131,1)","rgba(53,107,140,1)","rgba(44,118,142,1)","rgba(43,126,141,1)","rgba(36,164,133,1)","rgba(43,124,141,1)","rgba(42,148,137,1)","rgba(69,10,88,1)","rgba(49,112,141,1)","rgba(52,107,140,1)","rgba(69,23,96,1)","rgba(42,170,130,1)","rgba(70,27,99,1)","rgba(44,139,138,1)","rgba(51,109,141,1)","rgba(44,118,142,1)","rgba(46,116,142,1)","rgba(62,85,137,1)","rgba(43,126,141,1)","rgba(43,126,141,1)","rgba(43,129,140,1)","rgba(47,115,141,1)","rgba(46,116,142,1)","rgba(47,115,141,1)","rgba(56,101,140,1)","rgba(118,206,85,1)","rgba(59,93,139,1)","rgba(50,111,141,1)","rgba(43,126,141,1)","rgba(35,167,132,1)","rgba(43,145,137,1)","rgba(42,149,136,1)","rgba(42,121,142,1)","rgba(40,155,135,1)","rgba(44,137,139,1)","rgba(62,83,137,1)","rgba(66,66,133,1)","rgba(59,95,139,1)","rgba(46,116,141,1)","rgba(43,119,142,1)","rgba(64,77,136,1)","rgba(60,92,138,1)","rgba(44,131,140,1)","rgba(52,107,140,1)","rgba(69,11,89,1)","rgba(63,80,137,1)","rgba(43,141,138,1)","rgba(70,34,104,1)","rgba(68,178,121,1)","rgba(88,187,110,1)","rgba(57,98,139,1)","rgba(54,173,126,1)","rgba(69,42,111,1)","rgba(70,34,104,1)","rgba(43,128,141,1)","rgba(57,99,139,1)","rgba(69,50,118,1)","rgba(68,55,122,1)","rgba(44,138,139,1)","rgba(69,22,95,1)","rgba(43,124,141,1)","rgba(41,152,136,1)","rgba(70,26,98,1)","rgba(62,86,137,1)","rgba(51,110,141,1)","rgba(42,149,136,1)","rgba(51,109,141,1)","rgba(63,80,137,1)","rgba(51,110,141,1)","rgba(44,132,140,1)","rgba(45,117,142,1)","rgba(44,134,139,1)","rgba(42,149,136,1)","rgba(43,129,140,1)","rgba(43,124,141,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(110,201,93,1)","rgba(44,137,139,1)","rgba(60,90,138,1)","rgba(43,119,142,1)","rgba(43,143,138,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(53,106,140,1)","rgba(63,78,136,1)","rgba(44,139,138,1)","rgba(59,94,139,1)","rgba(43,128,141,1)","rgba(41,152,136,1)","rgba(54,105,140,1)","rgba(43,140,138,1)","rgba(41,153,135,1)","rgba(63,78,136,1)","rgba(43,143,138,1)","rgba(70,39,108,1)","rgba(36,165,133,1)","rgba(47,114,141,1)","rgba(43,128,141,1)","rgba(63,82,137,1)","rgba(66,65,132,1)","rgba(55,103,140,1)","rgba(70,37,106,1)","rgba(43,125,141,1)","rgba(63,80,137,1)","rgba(66,66,133,1)","rgba(44,132,140,1)","rgba(43,146,137,1)","rgba(61,89,138,1)","rgba(68,57,124,1)","rgba(68,57,125,1)","rgba(43,141,138,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(44,118,142,1)","rgba(54,104,140,1)","rgba(64,76,136,1)","rgba(69,48,116,1)","rgba(68,4,86,1)","rgba(70,28,100,1)","rgba(57,98,139,1)","rgba(43,128,141,1)","rgba(44,139,138,1)","rgba(46,116,141,1)","rgba(70,32,102,1)","rgba(44,133,140,1)","rgba(69,16,91,1)","rgba(43,123,141,1)","rgba(49,112,141,1)","rgba(43,143,138,1)","rgba(50,111,141,1)","rgba(53,106,140,1)","rgba(47,115,141,1)","rgba(45,117,142,1)","rgba(44,136,139,1)","rgba(56,100,140,1)","rgba(58,97,139,1)","rgba(94,190,106,1)","rgba(44,135,139,1)","rgba(43,140,138,1)","rgba(46,116,142,1)","rgba(69,51,119,1)","rgba(40,157,135,1)","rgba(43,141,138,1)","rgba(42,121,142,1)","rgba(43,124,141,1)","rgba(43,128,140,1)","rgba(44,136,139,1)","rgba(43,141,138,1)","rgba(70,40,109,1)","rgba(43,119,142,1)","rgba(43,126,141,1)","rgba(63,78,136,1)","rgba(44,134,139,1)","rgba(44,133,140,1)","rgba(49,112,141,1)","rgba(44,118,142,1)","rgba(59,94,139,1)","rgba(51,110,141,1)","rgba(38,161,134,1)","rgba(70,31,101,1)","rgba(70,39,108,1)","rgba(52,108,140,1)","rgba(68,2,84,1)","rgba(53,106,140,1)","rgba(70,37,106,1)","rgba(69,25,97,1)","rgba(43,126,141,1)","rgba(44,132,140,1)","rgba(44,135,139,1)","rgba(66,63,130,1)","rgba(43,130,140,1)","rgba(43,130,140,1)","rgba(42,147,137,1)","rgba(73,180,119,1)","rgba(67,62,129,1)","rgba(43,125,141,1)","rgba(42,150,136,1)","rgba(43,126,141,1)","rgba(60,91,138,1)","rgba(48,114,141,1)","rgba(68,54,121,1)","rgba(68,8,87,1)","rgba(76,182,117,1)","rgba(43,141,138,1)","rgba(43,141,138,1)","rgba(63,79,137,1)","rgba(43,119,142,1)","rgba(44,135,139,1)","rgba(44,134,139,1)","rgba(69,8,87,1)","rgba(52,108,141,1)","rgba(64,76,136,1)","rgba(41,154,135,1)","rgba(43,124,141,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(43,130,140,1)","rgba(43,129,140,1)","rgba(69,17,92,1)","rgba(43,125,141,1)","rgba(43,144,138,1)","rgba(59,93,139,1)","rgba(57,99,139,1)","rgba(63,78,136,1)","rgba(69,45,113,1)","rgba(44,135,139,1)","rgba(38,160,134,1)","rgba(70,29,100,1)","rgba(50,111,141,1)","rgba(48,114,141,1)","rgba(43,143,138,1)","rgba(69,25,97,1)","rgba(67,59,126,1)","rgba(64,73,136,1)","rgba(68,54,122,1)","rgba(55,103,140,1)","rgba(44,133,140,1)","rgba(67,177,121,1)","rgba(43,129,140,1)","rgba(44,135,139,1)","rgba(49,112,141,1)","rgba(70,30,101,1)","rgba(41,153,136,1)","rgba(46,116,142,1)","rgba(43,141,138,1)","rgba(50,111,141,1)","rgba(68,7,87,1)","rgba(40,169,131,1)","rgba(52,107,140,1)","rgba(70,32,103,1)","rgba(43,126,141,1)","rgba(51,109,141,1)","rgba(47,115,141,1)","rgba(44,132,140,1)","rgba(43,129,140,1)","rgba(43,126,141,1)","rgba(64,74,136,1)","rgba(65,70,135,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(46,116,142,1)","rgba(40,157,135,1)","rgba(48,113,141,1)","rgba(66,177,122,1)","rgba(57,98,139,1)","rgba(64,76,136,1)","rgba(53,106,140,1)","rgba(46,116,141,1)","rgba(43,129,140,1)","rgba(47,115,141,1)","rgba(55,102,140,1)","rgba(43,131,140,1)","rgba(46,116,141,1)","rgba(42,122,142,1)","rgba(39,160,134,1)","rgba(44,134,139,1)","rgba(34,168,132,1)","rgba(61,89,138,1)","rgba(38,161,134,1)","rgba(62,86,137,1)","rgba(55,102,140,1)","rgba(43,124,141,1)","rgba(69,15,91,1)","rgba(61,90,138,1)","rgba(49,172,128,1)","rgba(65,68,135,1)","rgba(69,47,115,1)","rgba(44,139,138,1)","rgba(63,82,137,1)","rgba(67,59,127,1)","rgba(38,160,134,1)","rgba(57,98,139,1)","rgba(46,115,141,1)","rgba(43,124,141,1)","rgba(54,104,140,1)","rgba(56,101,140,1)","rgba(42,122,142,1)","rgba(68,4,86,1)","rgba(68,6,87,1)","rgba(63,82,137,1)","rgba(51,109,141,1)","rgba(66,65,132,1)","rgba(40,156,135,1)","rgba(47,114,141,1)","rgba(68,3,85,1)","rgba(68,4,85,1)","rgba(44,133,140,1)","rgba(65,67,134,1)","rgba(67,58,126,1)","rgba(66,64,131,1)","rgba(54,104,140,1)","rgba(69,11,89,1)","rgba(69,23,96,1)","rgba(66,64,132,1)","rgba(43,130,140,1)","rgba(59,93,138,1)","rgba(56,101,140,1)","rgba(43,140,138,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(65,68,135,1)","rgba(50,111,141,1)","rgba(66,65,132,1)","rgba(44,131,140,1)","rgba(64,73,136,1)","rgba(43,140,138,1)","rgba(61,89,138,1)","rgba(44,136,139,1)","rgba(52,107,140,1)","rgba(69,22,95,1)","rgba(50,111,141,1)","rgba(44,117,142,1)","rgba(43,122,142,1)","rgba(43,143,138,1)","rgba(75,181,117,1)","rgba(70,26,98,1)","rgba(43,124,141,1)","rgba(69,22,95,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(53,107,140,1)","rgba(43,142,138,1)","rgba(51,109,141,1)","rgba(42,121,142,1)","rgba(42,149,136,1)","rgba(42,122,142,1)","rgba(68,53,120,1)","rgba(43,126,141,1)","rgba(62,83,137,1)","rgba(42,122,142,1)","rgba(44,137,139,1)","rgba(44,134,139,1)","rgba(69,178,120,1)","rgba(43,126,141,1)","rgba(55,104,140,1)","rgba(44,131,140,1)","rgba(42,120,142,1)","rgba(69,48,116,1)","rgba(58,97,139,1)","rgba(68,56,123,1)","rgba(43,123,141,1)","rgba(43,145,137,1)","rgba(68,178,121,1)","rgba(44,131,140,1)","rgba(69,9,88,1)","rgba(63,81,137,1)","rgba(43,129,140,1)","rgba(68,58,125,1)","rgba(60,90,138,1)","rgba(60,92,138,1)","rgba(43,124,141,1)","rgba(39,159,134,1)","rgba(66,63,130,1)","rgba(63,81,137,1)","rgba(43,128,141,1)","rgba(69,22,95,1)","rgba(43,125,141,1)","rgba(51,109,141,1)","rgba(43,127,141,1)","rgba(69,15,91,1)","rgba(63,82,137,1)","rgba(56,101,140,1)","rgba(42,120,142,1)","rgba(43,125,141,1)","rgba(53,106,140,1)","rgba(38,161,134,1)","rgba(58,96,139,1)","rgba(70,32,102,1)","rgba(62,85,137,1)","rgba(43,128,141,1)","rgba(58,96,139,1)","rgba(44,136,139,1)","rgba(69,43,111,1)","rgba(45,117,142,1)","rgba(69,44,113,1)","rgba(42,120,142,1)","rgba(42,120,142,1)","rgba(61,88,138,1)","rgba(50,111,141,1)","rgba(58,96,139,1)","rgba(70,42,111,1)","rgba(69,9,88,1)","rgba(69,46,115,1)","rgba(44,137,139,1)","rgba(48,113,141,1)","rgba(54,105,140,1)","rgba(46,116,141,1)","rgba(37,163,133,1)","rgba(52,108,141,1)","rgba(68,4,86,1)","rgba(69,9,88,1)","rgba(44,138,139,1)","rgba(41,153,135,1)","rgba(48,114,141,1)","rgba(45,117,142,1)","rgba(44,118,142,1)","rgba(59,94,139,1)","rgba(59,94,139,1)","rgba(69,13,90,1)","rgba(51,109,141,1)","rgba(57,99,139,1)","rgba(61,87,138,1)","rgba(44,137,139,1)","rgba(53,106,140,1)","rgba(67,59,126,1)","rgba(44,131,140,1)","rgba(42,121,142,1)","rgba(69,50,118,1)","rgba(68,2,84,1)","rgba(54,105,140,1)","rgba(64,74,136,1)","rgba(43,127,141,1)","rgba(66,63,130,1)","rgba(70,39,108,1)","rgba(51,110,141,1)","rgba(44,137,139,1)","rgba(43,127,141,1)","rgba(44,132,140,1)","rgba(68,55,122,1)","rgba(42,121,142,1)","rgba(40,157,135,1)","rgba(58,97,139,1)","rgba(36,164,133,1)","rgba(43,119,142,1)","rgba(57,98,139,1)","rgba(43,140,138,1)","rgba(66,63,130,1)","rgba(42,151,136,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(53,106,140,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(63,82,137,1)","rgba(64,77,136,1)","rgba(50,111,141,1)","rgba(44,138,139,1)","rgba(49,113,141,1)","rgba(41,154,135,1)","rgba(46,115,141,1)","rgba(40,157,135,1)","rgba(68,8,87,1)","rgba(64,176,123,1)","rgba(43,127,141,1)","rgba(43,124,141,1)","rgba(61,89,138,1)","rgba(44,135,139,1)","rgba(50,110,141,1)","rgba(51,109,141,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(46,116,142,1)","rgba(56,100,139,1)","rgba(42,121,142,1)","rgba(68,57,124,1)","rgba(42,148,137,1)","rgba(43,124,141,1)","rgba(69,14,90,1)","rgba(44,134,139,1)","rgba(61,86,138,1)","rgba(48,114,141,1)","rgba(113,203,90,1)","rgba(41,152,136,1)","rgba(70,31,102,1)","rgba(69,45,114,1)","rgba(69,21,94,1)","rgba(39,158,134,1)","rgba(43,142,138,1)","rgba(44,133,140,1)","rgba(43,124,141,1)","rgba(42,120,142,1)","rgba(59,94,139,1)","rgba(55,103,140,1)","rgba(60,90,138,1)","rgba(44,135,139,1)","rgba(49,112,141,1)","rgba(66,63,130,1)","rgba(43,125,141,1)","rgba(43,125,141,1)","rgba(44,137,139,1)","rgba(58,96,139,1)","rgba(41,154,135,1)","rgba(69,46,114,1)","rgba(61,87,138,1)","rgba(51,109,141,1)","rgba(43,141,138,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(39,158,134,1)","rgba(43,128,140,1)","rgba(69,14,90,1)","rgba(62,84,137,1)","rgba(57,99,139,1)","rgba(43,129,140,1)","rgba(68,6,87,1)","rgba(59,95,139,1)","rgba(44,137,139,1)","rgba(57,98,139,1)","rgba(48,113,141,1)","rgba(43,130,140,1)","rgba(69,44,113,1)","rgba(44,139,138,1)","rgba(55,103,140,1)","rgba(40,156,135,1)","rgba(44,135,139,1)","rgba(43,127,141,1)","rgba(60,91,138,1)","rgba(55,103,140,1)","rgba(44,137,139,1)","rgba(66,66,133,1)","rgba(48,113,141,1)","rgba(40,155,135,1)","rgba(49,112,141,1)","rgba(45,117,142,1)","rgba(43,144,137,1)","rgba(42,120,142,1)","rgba(69,46,114,1)","rgba(57,99,139,1)","rgba(70,28,99,1)","rgba(43,127,141,1)","rgba(69,18,93,1)","rgba(43,142,138,1)","rgba(42,151,136,1)","rgba(42,120,142,1)","rgba(69,18,93,1)","rgba(43,129,140,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(44,134,139,1)","rgba(70,28,99,1)","rgba(44,131,140,1)","rgba(55,103,140,1)","rgba(43,145,137,1)","rgba(59,95,139,1)","rgba(46,116,141,1)","rgba(50,110,141,1)","rgba(63,82,137,1)","rgba(63,81,137,1)","rgba(61,89,138,1)","rgba(60,91,138,1)","rgba(55,103,140,1)","rgba(35,166,132,1)","rgba(68,4,86,1)","rgba(50,110,141,1)","rgba(43,142,138,1)","rgba(52,108,141,1)","rgba(68,56,123,1)","rgba(70,29,100,1)","rgba(69,19,93,1)","rgba(43,143,138,1)","rgba(41,154,135,1)","rgba(67,59,126,1)","rgba(68,55,122,1)","rgba(62,83,137,1)","rgba(43,143,138,1)","rgba(70,40,109,1)","rgba(54,104,140,1)","rgba(44,138,139,1)","rgba(47,114,141,1)","rgba(64,77,136,1)","rgba(68,58,126,1)","rgba(69,24,97,1)","rgba(56,100,139,1)","rgba(70,37,106,1)","rgba(47,114,141,1)","rgba(66,65,132,1)","rgba(48,113,141,1)","rgba(69,12,89,1)","rgba(57,99,139,1)","rgba(43,125,141,1)","rgba(43,140,138,1)","rgba(55,103,140,1)","rgba(46,116,142,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(58,97,139,1)","rgba(59,94,139,1)","rgba(42,122,142,1)","rgba(66,66,133,1)","rgba(52,172,127,1)","rgba(43,144,137,1)","rgba(45,117,142,1)","rgba(65,69,135,1)","rgba(61,89,138,1)","rgba(68,56,124,1)","rgba(41,153,135,1)","rgba(42,121,142,1)","rgba(44,134,139,1)","rgba(43,143,138,1)","rgba(48,113,141,1)","rgba(52,108,140,1)","rgba(44,137,139,1)","rgba(44,133,140,1)","rgba(65,69,135,1)","rgba(64,73,136,1)","rgba(43,124,141,1)","rgba(69,14,90,1)","rgba(58,97,139,1)","rgba(42,122,142,1)","rgba(43,123,141,1)","rgba(61,87,138,1)","rgba(65,70,135,1)","rgba(44,131,140,1)","rgba(42,122,142,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(59,94,139,1)","rgba(42,122,142,1)","rgba(69,44,112,1)","rgba(42,151,136,1)","rgba(44,135,139,1)","rgba(70,33,104,1)","rgba(43,119,142,1)","rgba(69,23,96,1)","rgba(43,143,138,1)","rgba(43,141,138,1)","rgba(41,152,136,1)","rgba(43,143,138,1)","rgba(46,116,142,1)","rgba(59,95,139,1)","rgba(69,48,116,1)","rgba(43,128,140,1)","rgba(64,73,136,1)","rgba(44,135,139,1)","rgba(39,158,134,1)","rgba(60,175,124,1)","rgba(41,153,136,1)","rgba(49,113,141,1)","rgba(43,123,142,1)","rgba(41,154,135,1)","rgba(37,169,131,1)","rgba(43,147,137,1)","rgba(56,102,140,1)","rgba(42,121,142,1)","rgba(44,133,140,1)","rgba(64,73,136,1)","rgba(42,148,136,1)","rgba(41,153,136,1)","rgba(52,107,140,1)","rgba(40,157,135,1)","rgba(43,143,138,1)","rgba(43,127,141,1)","rgba(54,104,140,1)","rgba(43,145,137,1)","rgba(68,54,122,1)","rgba(44,133,140,1)","rgba(54,105,140,1)","rgba(67,60,127,1)","rgba(43,127,141,1)","rgba(51,109,141,1)","rgba(62,85,137,1)","rgba(43,144,137,1)","rgba(43,140,138,1)","rgba(43,126,141,1)","rgba(44,138,139,1)","rgba(51,110,141,1)","rgba(61,89,138,1)","rgba(62,83,137,1)","rgba(52,107,140,1)","rgba(55,103,140,1)","rgba(48,113,141,1)","rgba(63,80,137,1)","rgba(43,130,140,1)","rgba(67,59,127,1)","rgba(43,130,140,1)","rgba(69,18,92,1)","rgba(97,192,104,1)","rgba(52,108,141,1)","rgba(78,182,116,1)","rgba(41,153,135,1)","rgba(43,124,141,1)","rgba(58,97,139,1)","rgba(42,122,142,1)","rgba(60,90,138,1)","rgba(64,72,136,1)","rgba(57,99,139,1)","rgba(63,82,137,1)","rgba(54,105,140,1)","rgba(61,88,138,1)","rgba(57,98,139,1)","rgba(36,166,133,1)","rgba(54,105,140,1)","rgba(43,147,137,1)","rgba(69,46,114,1)","rgba(44,137,139,1)","rgba(42,120,142,1)","rgba(55,103,140,1)","rgba(43,124,141,1)","rgba(66,65,132,1)","rgba(62,85,137,1)","rgba(69,42,111,1)","rgba(64,78,136,1)","rgba(43,141,138,1)","rgba(115,204,88,1)","rgba(45,117,142,1)","rgba(70,29,100,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(70,30,101,1)","rgba(68,3,85,1)","rgba(43,146,137,1)","rgba(54,104,140,1)","rgba(37,169,131,1)","rgba(58,97,139,1)","rgba(69,48,116,1)","rgba(54,105,140,1)","rgba(41,152,136,1)","rgba(68,57,124,1)","rgba(44,131,140,1)","rgba(69,18,93,1)","rgba(43,126,141,1)","rgba(37,164,133,1)","rgba(49,112,141,1)","rgba(42,149,136,1)","rgba(41,152,136,1)","rgba(69,25,97,1)","rgba(54,104,140,1)","rgba(54,105,140,1)","rgba(63,82,137,1)","rgba(44,137,139,1)","rgba(43,124,141,1)","rgba(40,155,135,1)","rgba(70,179,120,1)","rgba(68,56,123,1)","rgba(56,100,139,1)","rgba(64,73,136,1)","rgba(43,143,138,1)","rgba(42,121,142,1)","rgba(57,98,139,1)","rgba(69,19,93,1)","rgba(42,150,136,1)","rgba(38,160,134,1)","rgba(69,20,94,1)","rgba(69,44,113,1)","rgba(57,100,139,1)","rgba(70,36,106,1)","rgba(70,35,105,1)","rgba(49,113,141,1)","rgba(42,148,137,1)","rgba(39,159,134,1)","rgba(43,126,141,1)","rgba(68,56,124,1)","rgba(45,117,142,1)","rgba(67,59,126,1)","rgba(43,170,130,1)","rgba(64,77,136,1)","rgba(57,99,139,1)","rgba(43,139,138,1)","rgba(69,16,91,1)","rgba(44,138,139,1)","rgba(61,87,138,1)","rgba(61,88,138,1)","rgba(43,119,142,1)","rgba(43,145,137,1)","rgba(44,139,138,1)","rgba(42,148,137,1)","rgba(43,127,141,1)","rgba(46,115,141,1)","rgba(39,159,134,1)","rgba(42,150,136,1)","rgba(43,147,137,1)","rgba(114,203,89,1)","rgba(44,133,140,1)","rgba(65,67,134,1)","rgba(56,101,140,1)","rgba(43,142,138,1)","rgba(37,162,133,1)","rgba(38,160,134,1)","rgba(57,98,139,1)","rgba(50,111,141,1)","rgba(42,150,136,1)","rgba(66,64,131,1)","rgba(70,28,100,1)","rgba(39,159,134,1)","rgba(44,138,139,1)","rgba(43,124,141,1)","rgba(69,17,92,1)","rgba(73,180,119,1)","rgba(35,166,133,1)","rgba(68,55,122,1)","rgba(69,11,89,1)","rgba(61,87,138,1)","rgba(43,139,138,1)","rgba(43,127,141,1)","rgba(43,129,140,1)","rgba(56,100,140,1)","rgba(69,20,94,1)","rgba(58,96,139,1)","rgba(43,145,137,1)","rgba(34,168,132,1)","rgba(43,146,137,1)","rgba(40,156,135,1)","rgba(44,137,139,1)","rgba(34,168,132,1)","rgba(44,133,140,1)","rgba(67,62,129,1)","rgba(64,78,136,1)","rgba(60,90,138,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(69,10,89,1)","rgba(65,69,135,1)","rgba(63,79,137,1)","rgba(44,136,139,1)","rgba(69,20,94,1)","rgba(54,105,140,1)","rgba(63,81,137,1)","rgba(39,158,134,1)","rgba(70,34,104,1)","rgba(38,161,134,1)","rgba(68,1,84,1)","rgba(44,131,140,1)","rgba(70,38,108,1)","rgba(42,150,136,1)","rgba(59,94,139,1)","rgba(43,130,140,1)","rgba(69,20,94,1)","rgba(44,131,140,1)","rgba(49,111,141,1)","rgba(36,164,133,1)","rgba(62,85,137,1)","rgba(70,37,106,1)","rgba(44,134,139,1)","rgba(38,161,134,1)","rgba(43,126,141,1)","rgba(42,120,142,1)","rgba(69,23,96,1)","rgba(43,145,137,1)","rgba(44,138,139,1)","rgba(43,125,141,1)","rgba(43,140,138,1)","rgba(201,222,61,1)","rgba(38,161,134,1)","rgba(46,116,142,1)","rgba(43,146,137,1)","rgba(42,122,142,1)","rgba(61,89,138,1)","rgba(59,93,139,1)","rgba(44,132,140,1)","rgba(42,148,137,1)","rgba(68,54,121,1)","rgba(54,173,127,1)","rgba(44,131,140,1)","rgba(42,121,142,1)","rgba(43,127,141,1)","rgba(70,35,105,1)","rgba(43,143,138,1)","rgba(69,10,88,1)","rgba(54,105,140,1)","rgba(69,25,97,1)","rgba(41,154,135,1)","rgba(157,215,73,1)","rgba(70,34,104,1)","rgba(62,86,138,1)","rgba(61,88,138,1)","rgba(64,75,136,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(43,126,141,1)","rgba(38,169,131,1)","rgba(70,40,109,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(52,109,141,1)","rgba(67,59,127,1)","rgba(64,76,136,1)","rgba(58,96,139,1)","rgba(43,141,138,1)","rgba(43,141,138,1)","rgba(67,62,129,1)","rgba(43,129,140,1)","rgba(50,110,141,1)","rgba(44,137,139,1)","rgba(58,96,139,1)","rgba(95,191,105,1)","rgba(40,155,135,1)","rgba(69,15,91,1)","rgba(55,103,140,1)","rgba(65,69,135,1)","rgba(43,129,140,1)","rgba(43,129,140,1)","rgba(68,54,122,1)","rgba(43,140,138,1)","rgba(52,107,140,1)","rgba(69,25,97,1)","rgba(70,36,106,1)","rgba(160,215,73,1)","rgba(113,203,90,1)","rgba(63,80,137,1)","rgba(58,97,139,1)","rgba(48,114,141,1)","rgba(68,55,123,1)","rgba(38,161,134,1)","rgba(46,116,142,1)","rgba(50,111,141,1)","rgba(41,153,135,1)","rgba(52,107,140,1)","rgba(44,139,138,1)","rgba(41,152,136,1)","rgba(49,112,141,1)","rgba(43,128,140,1)","rgba(65,72,136,1)","rgba(52,108,140,1)","rgba(62,83,137,1)","rgba(44,139,138,1)","rgba(44,118,142,1)","rgba(43,146,137,1)","rgba(53,107,140,1)","rgba(44,135,139,1)","rgba(44,133,140,1)","rgba(40,156,135,1)","rgba(66,66,133,1)","rgba(43,146,137,1)","rgba(57,98,139,1)","rgba(41,153,135,1)","rgba(65,70,135,1)","rgba(43,130,140,1)","rgba(70,30,101,1)","rgba(70,41,110,1)","rgba(44,131,140,1)","rgba(44,138,139,1)","rgba(44,118,142,1)","rgba(69,24,96,1)","rgba(49,112,141,1)","rgba(42,147,137,1)","rgba(43,125,141,1)","rgba(42,121,142,1)","rgba(55,102,140,1)","rgba(35,167,132,1)","rgba(48,114,141,1)","rgba(44,118,142,1)","rgba(62,84,137,1)","rgba(42,122,142,1)","rgba(44,135,139,1)","rgba(72,180,119,1)","rgba(44,118,142,1)","rgba(69,52,120,1)","rgba(44,131,140,1)","rgba(44,136,139,1)","rgba(43,119,142,1)","rgba(43,124,141,1)","rgba(44,170,130,1)","rgba(43,124,141,1)","rgba(43,142,138,1)","rgba(36,164,133,1)","rgba(69,24,97,1)","rgba(43,124,141,1)","rgba(42,150,136,1)","rgba(44,138,139,1)","rgba(43,146,137,1)","rgba(48,171,128,1)","rgba(43,147,137,1)","rgba(44,138,139,1)","rgba(44,139,138,1)","rgba(43,129,140,1)","rgba(39,158,134,1)","rgba(42,148,137,1)","rgba(42,149,136,1)","rgba(43,144,137,1)","rgba(43,139,138,1)","rgba(68,5,86,1)","rgba(62,84,137,1)","rgba(44,138,139,1)","rgba(43,142,138,1)","rgba(43,141,138,1)","rgba(43,127,141,1)","rgba(52,172,127,1)","rgba(54,105,140,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(43,128,141,1)","rgba(42,120,142,1)","rgba(34,167,132,1)","rgba(39,158,134,1)","rgba(40,157,135,1)","rgba(42,121,142,1)","rgba(61,89,138,1)","rgba(43,119,142,1)","rgba(68,55,123,1)","rgba(62,85,137,1)","rgba(70,34,104,1)","rgba(40,157,134,1)","rgba(45,117,142,1)","rgba(64,74,136,1)","rgba(39,158,134,1)","rgba(43,142,138,1)","rgba(41,153,136,1)","rgba(70,37,107,1)","rgba(68,56,124,1)","rgba(41,151,136,1)","rgba(40,157,135,1)","rgba(38,161,134,1)","rgba(43,146,137,1)","rgba(44,134,139,1)","rgba(44,134,139,1)","rgba(42,151,136,1)","rgba(49,112,141,1)","rgba(44,118,142,1)","rgba(43,128,140,1)","rgba(44,134,139,1)","rgba(102,196,100,1)","rgba(41,154,135,1)","rgba(40,155,135,1)","rgba(44,132,140,1)","rgba(43,119,142,1)","rgba(44,139,138,1)","rgba(65,67,134,1)","rgba(44,136,139,1)","rgba(50,172,128,1)","rgba(43,119,142,1)","rgba(55,102,140,1)","rgba(61,89,138,1)","rgba(43,139,138,1)","rgba(69,19,93,1)","rgba(43,123,141,1)","rgba(49,112,141,1)","rgba(63,79,137,1)","rgba(65,70,135,1)","rgba(69,52,119,1)","rgba(63,82,137,1)","rgba(65,67,134,1)","rgba(69,19,93,1)","rgba(44,131,140,1)","rgba(63,82,137,1)","rgba(42,120,142,1)","rgba(69,25,97,1)","rgba(70,37,107,1)","rgba(43,119,142,1)","rgba(43,119,142,1)","rgba(53,106,140,1)","rgba(42,151,136,1)","rgba(46,116,141,1)","rgba(69,46,115,1)","rgba(70,37,106,1)","rgba(43,146,137,1)","rgba(37,162,133,1)","rgba(57,98,139,1)","rgba(55,102,140,1)","rgba(57,174,126,1)","rgba(66,64,131,1)","rgba(58,97,139,1)","rgba(67,62,130,1)","rgba(39,158,134,1)","rgba(58,97,139,1)","rgba(69,17,92,1)","rgba(39,159,134,1)","rgba(68,4,86,1)","rgba(68,58,125,1)","rgba(43,143,138,1)","rgba(69,23,96,1)","rgba(42,149,136,1)","rgba(70,29,100,1)","rgba(58,96,139,1)","rgba(69,50,118,1)","rgba(69,45,113,1)","rgba(70,27,99,1)","rgba(69,47,115,1)","rgba(69,22,95,1)","rgba(43,126,141,1)","rgba(45,117,142,1)","rgba(52,108,140,1)","rgba(66,65,132,1)","rgba(68,57,124,1)","rgba(70,33,103,1)","rgba(43,146,137,1)","rgba(50,110,141,1)","rgba(66,65,132,1)","rgba(64,72,136,1)","rgba(65,66,133,1)","rgba(65,177,122,1)","rgba(59,93,139,1)","rgba(70,31,102,1)","rgba(42,149,136,1)","rgba(70,36,105,1)","rgba(44,139,138,1)","rgba(69,178,121,1)","rgba(69,50,118,1)","rgba(67,59,126,1)","rgba(69,46,114,1)","rgba(70,42,111,1)","rgba(43,146,137,1)","rgba(163,216,72,1)","rgba(53,105,140,1)","rgba(55,103,140,1)","rgba(69,52,120,1)","rgba(60,91,138,1)","rgba(56,101,140,1)","rgba(52,108,140,1)","rgba(70,26,98,1)","rgba(59,93,139,1)","rgba(55,173,126,1)","rgba(66,66,133,1)","rgba(62,84,137,1)","rgba(50,111,141,1)","rgba(42,120,142,1)","rgba(44,138,139,1)","rgba(59,93,138,1)","rgba(42,150,136,1)","rgba(43,140,138,1)","rgba(67,61,128,1)","rgba(42,149,136,1)","rgba(74,181,118,1)","rgba(40,157,135,1)","rgba(65,68,135,1)","rgba(59,94,139,1)","rgba(38,162,133,1)","rgba(58,96,139,1)","rgba(70,30,101,1)","rgba(44,133,140,1)","rgba(38,161,134,1)","rgba(37,162,133,1)","rgba(63,81,137,1)","rgba(90,188,109,1)","rgba(38,162,134,1)","rgba(57,98,139,1)","rgba(44,137,139,1)","rgba(44,132,140,1)","rgba(67,178,121,1)","rgba(44,135,139,1)","rgba(40,156,135,1)","rgba(41,153,136,1)","rgba(86,186,111,1)","rgba(67,60,127,1)","rgba(69,9,88,1)","rgba(70,29,100,1)","rgba(68,5,86,1)","rgba(70,29,100,1)","rgba(49,172,128,1)","rgba(65,67,134,1)","rgba(44,139,138,1)","rgba(43,146,137,1)","rgba(44,138,139,1)","rgba(46,115,141,1)","rgba(66,63,130,1)","rgba(44,135,139,1)","rgba(69,18,93,1)","rgba(43,126,141,1)","rgba(104,196,99,1)","rgba(43,140,138,1)","rgba(51,110,141,1)","rgba(59,175,125,1)","rgba(61,87,138,1)","rgba(69,49,117,1)","rgba(65,70,135,1)","rgba(54,173,126,1)","rgba(39,158,134,1)","rgba(42,170,130,1)","rgba(44,133,140,1)","rgba(36,164,133,1)","rgba(58,97,139,1)","rgba(61,89,138,1)","rgba(69,24,96,1)","rgba(64,73,136,1)","rgba(69,52,119,1)","rgba(44,139,138,1)","rgba(54,105,140,1)","rgba(66,66,133,1)","rgba(70,25,97,1)","rgba(70,42,111,1)","rgba(45,117,142,1)","rgba(64,75,136,1)","rgba(62,176,123,1)","rgba(42,151,136,1)","rgba(43,142,138,1)","rgba(41,169,130,1)","rgba(58,95,139,1)","rgba(43,124,141,1)","rgba(61,88,138,1)","rgba(49,112,141,1)","rgba(48,113,141,1)","rgba(65,70,135,1)","rgba(67,61,128,1)","rgba(70,32,103,1)","rgba(43,119,142,1)","rgba(62,83,137,1)","rgba(69,10,88,1)","rgba(42,150,136,1)","rgba(69,48,116,1)","rgba(58,96,139,1)","rgba(43,142,138,1)","rgba(58,97,139,1)","rgba(43,119,142,1)","rgba(57,100,139,1)","rgba(66,65,132,1)","rgba(43,143,138,1)","rgba(42,148,137,1)","rgba(43,146,137,1)","rgba(53,106,140,1)","rgba(34,168,132,1)","rgba(57,100,139,1)","rgba(40,155,135,1)","rgba(69,47,115,1)","rgba(69,46,114,1)","rgba(70,30,101,1)","rgba(43,119,142,1)","rgba(64,75,136,1)","rgba(42,150,136,1)","rgba(107,199,96,1)","rgba(70,27,99,1)","rgba(57,98,139,1)","rgba(99,194,102,1)","rgba(43,144,137,1)","rgba(40,155,135,1)","rgba(68,56,123,1)","rgba(58,97,139,1)","rgba(43,139,138,1)","rgba(58,96,139,1)","rgba(50,111,141,1)","rgba(47,171,129,1)","rgba(41,151,136,1)","rgba(38,161,134,1)","rgba(54,105,140,1)","rgba(55,103,140,1)","rgba(57,100,139,1)","rgba(37,162,133,1)","rgba(46,116,142,1)","rgba(68,57,124,1)","rgba(39,159,134,1)","rgba(36,164,133,1)","rgba(44,135,139,1)","rgba(40,155,135,1)","rgba(68,58,125,1)","rgba(68,58,125,1)","rgba(68,53,121,1)","rgba(69,20,94,1)","rgba(69,50,118,1)","rgba(44,136,139,1)","rgba(70,34,104,1)","rgba(62,85,137,1)","rgba(99,194,102,1)","rgba(40,156,135,1)","rgba(39,158,134,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(69,21,95,1)","rgba(42,122,142,1)","rgba(39,169,131,1)","rgba(44,136,139,1)","rgba(55,104,140,1)","rgba(43,124,141,1)","rgba(69,22,95,1)","rgba(68,4,85,1)","rgba(53,106,140,1)","rgba(53,106,140,1)","rgba(55,103,140,1)","rgba(43,147,137,1)","rgba(41,154,135,1)","rgba(54,104,140,1)","rgba(66,65,132,1)","rgba(39,160,134,1)","rgba(38,160,134,1)","rgba(43,126,141,1)","rgba(44,137,139,1)","rgba(68,5,86,1)","rgba(59,93,139,1)","rgba(41,154,135,1)","rgba(57,99,139,1)","rgba(44,131,140,1)","rgba(44,131,140,1)","rgba(62,83,137,1)","rgba(54,104,140,1)","rgba(44,131,140,1)","rgba(69,23,96,1)","rgba(69,51,119,1)","rgba(62,176,124,1)","rgba(42,170,130,1)","rgba(42,148,137,1)","rgba(43,122,142,1)","rgba(70,36,105,1)","rgba(42,148,137,1)","rgba(69,10,88,1)","rgba(66,63,130,1)","rgba(43,143,138,1)","rgba(70,36,106,1)","rgba(64,78,136,1)","rgba(70,41,110,1)","rgba(56,101,140,1)","rgba(69,51,119,1)","rgba(41,152,136,1)","rgba(59,95,139,1)","rgba(67,59,126,1)","rgba(37,163,133,1)","rgba(70,29,100,1)","rgba(70,35,104,1)","rgba(43,128,140,1)","rgba(42,122,142,1)","rgba(63,81,137,1)","rgba(69,15,91,1)","rgba(67,62,129,1)","rgba(70,35,105,1)","rgba(63,82,137,1)","rgba(57,99,139,1)","rgba(43,141,138,1)","rgba(44,138,139,1)","rgba(78,182,116,1)","rgba(43,141,138,1)","rgba(65,72,136,1)","rgba(57,98,139,1)","rgba(66,66,133,1)","rgba(43,129,140,1)","rgba(55,103,140,1)","rgba(68,4,86,1)","rgba(45,117,142,1)","rgba(65,67,134,1)","rgba(43,123,141,1)","rgba(43,144,137,1)","rgba(69,19,93,1)","rgba(66,63,130,1)","rgba(58,97,139,1)","rgba(59,93,138,1)","rgba(69,25,97,1)","rgba(70,37,106,1)","rgba(69,23,96,1)","rgba(43,126,141,1)","rgba(54,104,140,1)","rgba(57,99,139,1)","rgba(65,70,135,1)","rgba(43,123,141,1)","rgba(59,94,139,1)","rgba(43,125,141,1)","rgba(44,131,140,1)","rgba(64,75,136,1)","rgba(42,148,137,1)","rgba(69,16,91,1)","rgba(66,64,131,1)","rgba(66,66,133,1)","rgba(41,152,136,1)","rgba(53,107,140,1)","rgba(70,35,105,1)","rgba(60,92,138,1)","rgba(64,76,136,1)","rgba(35,167,132,1)","rgba(56,100,139,1)","rgba(54,104,140,1)","rgba(44,133,140,1)","rgba(68,6,87,1)","rgba(67,60,128,1)","rgba(44,132,140,1)","rgba(43,141,138,1)","rgba(63,80,137,1)","rgba(53,106,140,1)","rgba(41,154,135,1)","rgba(43,142,138,1)","rgba(69,14,90,1)","rgba(60,91,138,1)","rgba(53,106,140,1)","rgba(55,102,140,1)","rgba(50,110,141,1)","rgba(69,17,92,1)","rgba(44,136,139,1)","rgba(43,124,141,1)","rgba(66,64,131,1)","rgba(64,76,136,1)","rgba(54,105,140,1)","rgba(61,88,138,1)","rgba(66,65,132,1)","rgba(69,48,116,1)","rgba(70,35,105,1)","rgba(67,63,130,1)","rgba(69,12,89,1)","rgba(67,62,130,1)","rgba(42,151,136,1)","rgba(62,86,138,1)","rgba(49,112,141,1)","rgba(80,183,115,1)","rgba(44,134,139,1)","rgba(64,76,136,1)","rgba(69,14,90,1)","rgba(47,114,141,1)","rgba(69,46,114,1)","rgba(69,16,91,1)","rgba(66,64,131,1)","rgba(64,72,136,1)","rgba(44,134,139,1)","rgba(43,126,141,1)","rgba(59,94,139,1)","rgba(41,154,135,1)","rgba(69,49,117,1)","rgba(69,8,87,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(70,27,99,1)","rgba(67,61,128,1)","rgba(44,131,140,1)","rgba(40,155,135,1)","rgba(65,68,135,1)","rgba(68,3,85,1)","rgba(58,97,139,1)","rgba(57,99,139,1)","rgba(47,114,141,1)","rgba(63,78,136,1)","rgba(65,67,134,1)","rgba(70,26,98,1)","rgba(57,99,139,1)","rgba(66,64,131,1)","rgba(68,54,122,1)","rgba(44,136,139,1)","rgba(41,152,136,1)","rgba(69,52,120,1)","rgba(64,74,136,1)","rgba(62,85,137,1)","rgba(43,122,142,1)","rgba(69,48,116,1)","rgba(90,188,109,1)","rgba(38,162,134,1)","rgba(49,112,141,1)","rgba(53,107,140,1)","rgba(37,162,133,1)","rgba(59,94,139,1)","rgba(69,43,112,1)","rgba(70,29,100,1)","rgba(51,110,141,1)","rgba(69,9,88,1)","rgba(48,114,141,1)","rgba(43,140,138,1)","rgba(42,148,136,1)","rgba(69,46,114,1)","rgba(42,149,136,1)","rgba(64,76,136,1)","rgba(40,157,135,1)","rgba(56,101,140,1)","rgba(38,162,133,1)","rgba(53,106,140,1)","rgba(69,20,94,1)","rgba(69,20,94,1)","rgba(42,122,142,1)","rgba(53,107,140,1)","rgba(61,87,138,1)","rgba(70,36,106,1)","rgba(64,77,136,1)","rgba(70,41,109,1)","rgba(43,129,140,1)","rgba(68,58,125,1)","rgba(44,137,139,1)","rgba(70,37,107,1)","rgba(68,178,121,1)","rgba(47,115,141,1)","rgba(44,138,139,1)","rgba(69,45,113,1)","rgba(44,139,138,1)","rgba(38,162,133,1)","rgba(44,118,142,1)","rgba(70,39,108,1)","rgba(43,123,141,1)","rgba(43,123,142,1)","rgba(63,79,137,1)","rgba(42,148,136,1)","rgba(82,184,114,1)","rgba(43,125,141,1)","rgba(44,132,140,1)","rgba(43,140,138,1)","rgba(51,109,141,1)","rgba(70,28,99,1)","rgba(60,90,138,1)","rgba(70,42,110,1)","rgba(53,106,140,1)","rgba(60,92,138,1)","rgba(70,38,107,1)","rgba(38,161,134,1)","rgba(60,91,138,1)","rgba(69,16,91,1)","rgba(60,92,138,1)","rgba(65,68,135,1)","rgba(53,106,140,1)","rgba(44,137,139,1)","rgba(59,94,139,1)","rgba(52,108,140,1)","rgba(62,84,137,1)","rgba(68,54,121,1)","rgba(64,73,136,1)","rgba(66,65,132,1)","rgba(64,72,136,1)","rgba(53,106,140,1)","rgba(70,38,107,1)","rgba(44,135,139,1)","rgba(62,176,124,1)","rgba(67,60,127,1)","rgba(69,22,95,1)","rgba(43,145,137,1)","rgba(44,136,139,1)","rgba(37,169,131,1)","rgba(70,30,101,1)","rgba(66,66,133,1)","rgba(70,32,102,1)","rgba(43,123,142,1)","rgba(43,127,141,1)","rgba(43,146,137,1)","rgba(70,37,106,1)","rgba(69,19,93,1)","rgba(64,76,136,1)","rgba(43,145,137,1)","rgba(61,89,138,1)","rgba(52,108,140,1)","rgba(62,84,137,1)","rgba(54,173,127,1)","rgba(68,56,123,1)","rgba(69,23,96,1)","rgba(63,80,137,1)","rgba(61,89,138,1)","rgba(70,29,100,1)","rgba(44,133,140,1)","rgba(36,165,133,1)","rgba(44,135,139,1)","rgba(42,150,136,1)","rgba(61,89,138,1)","rgba(51,109,141,1)","rgba(55,103,140,1)","rgba(69,12,89,1)","rgba(68,53,121,1)","rgba(42,150,136,1)","rgba(60,91,138,1)","rgba(67,62,129,1)","rgba(41,170,130,1)","rgba(70,38,108,1)","rgba(43,139,138,1)","rgba(40,156,135,1)","rgba(68,5,86,1)","rgba(61,87,138,1)","rgba(43,126,141,1)","rgba(64,78,136,1)","rgba(49,112,141,1)","rgba(37,163,133,1)","rgba(44,131,140,1)","rgba(70,28,100,1)","rgba(60,92,138,1)","rgba(35,167,132,1)","rgba(37,162,133,1)","rgba(44,118,142,1)","rgba(61,87,138,1)","rgba(68,3,85,1)","rgba(64,76,136,1)","rgba(69,22,95,1)","rgba(45,117,142,1)","rgba(70,27,99,1)","rgba(59,94,139,1)","rgba(70,31,102,1)","rgba(66,64,131,1)","rgba(42,121,142,1)","rgba(67,60,127,1)","rgba(59,95,139,1)","rgba(68,5,86,1)","rgba(68,53,120,1)","rgba(66,64,131,1)","rgba(70,40,109,1)","rgba(64,72,136,1)","rgba(43,125,141,1)","rgba(46,116,142,1)","rgba(63,82,137,1)","rgba(70,35,105,1)","rgba(65,67,134,1)","rgba(70,34,104,1)","rgba(69,50,118,1)","rgba(69,44,112,1)","rgba(70,28,99,1)","rgba(66,65,132,1)","rgba(70,37,106,1)","rgba(64,74,136,1)","rgba(70,40,109,1)","rgba(41,153,136,1)","rgba(43,143,138,1)","rgba(67,60,127,1)","rgba(43,119,142,1)","rgba(61,89,138,1)","rgba(42,121,142,1)","rgba(61,89,138,1)","rgba(44,118,142,1)","rgba(64,73,136,1)","rgba(65,67,134,1)","rgba(43,127,141,1)","rgba(67,178,121,1)","rgba(43,130,140,1)","rgba(43,145,137,1)","rgba(42,148,137,1)","rgba(38,162,134,1)","rgba(70,34,104,1)","rgba(43,147,137,1)","rgba(42,120,142,1)","rgba(43,126,141,1)","rgba(50,110,141,1)","rgba(70,28,100,1)","rgba(62,86,138,1)","rgba(59,93,139,1)","rgba(69,50,118,1)","rgba(59,93,139,1)","rgba(51,110,141,1)","rgba(61,89,138,1)","rgba(70,29,100,1)","rgba(58,97,139,1)","rgba(43,146,137,1)","rgba(70,32,103,1)","rgba(68,55,122,1)","rgba(42,121,142,1)","rgba(69,19,93,1)","rgba(37,163,133,1)","rgba(43,128,141,1)","rgba(62,83,137,1)","rgba(54,105,140,1)","rgba(42,121,142,1)","rgba(68,54,122,1)","rgba(63,80,137,1)","rgba(66,65,132,1)","rgba(53,106,140,1)","rgba(59,95,139,1)","rgba(59,94,139,1)","rgba(39,159,134,1)","rgba(55,103,140,1)","rgba(59,95,139,1)","rgba(60,92,138,1)","rgba(44,138,139,1)","rgba(43,119,142,1)","rgba(69,49,117,1)","rgba(40,157,135,1)","rgba(52,108,141,1)","rgba(100,194,101,1)","rgba(69,47,115,1)","rgba(63,81,137,1)","rgba(69,9,88,1)","rgba(60,91,138,1)","rgba(64,77,136,1)","rgba(48,113,141,1)","rgba(42,122,142,1)","rgba(61,88,138,1)","rgba(66,65,132,1)","rgba(67,61,128,1)","rgba(69,49,117,1)","rgba(63,176,123,1)","rgba(68,3,85,1)","rgba(64,73,136,1)","rgba(70,37,107,1)","rgba(63,81,137,1)","rgba(62,83,137,1)","rgba(64,77,136,1)","rgba(43,144,137,1)","rgba(57,98,139,1)","rgba(43,127,141,1)","rgba(70,29,100,1)","rgba(44,131,140,1)","rgba(43,131,140,1)","rgba(54,105,140,1)","rgba(43,144,137,1)","rgba(61,89,138,1)","rgba(43,141,138,1)","rgba(53,107,140,1)","rgba(70,32,102,1)","rgba(70,39,108,1)","rgba(50,111,141,1)","rgba(44,118,142,1)","rgba(57,99,139,1)","rgba(44,133,140,1)","rgba(69,17,92,1)","rgba(42,150,136,1)","rgba(70,30,101,1)","rgba(68,4,86,1)","rgba(43,123,141,1)","rgba(43,147,137,1)","rgba(53,106,140,1)","rgba(68,54,122,1)","rgba(70,32,102,1)","rgba(70,38,107,1)","rgba(69,13,90,1)","rgba(42,121,142,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(38,160,134,1)","rgba(70,30,101,1)","rgba(43,144,137,1)","rgba(43,125,141,1)","rgba(34,168,132,1)","rgba(68,54,122,1)","rgba(55,102,140,1)","rgba(44,133,140,1)","rgba(43,144,137,1)","rgba(51,109,141,1)","rgba(50,172,128,1)","rgba(43,140,138,1)","rgba(48,113,141,1)","rgba(42,121,142,1)","rgba(44,138,139,1)","rgba(44,136,139,1)","rgba(64,73,136,1)","rgba(69,12,89,1)","rgba(59,93,138,1)","rgba(64,73,136,1)","rgba(43,147,137,1)","rgba(52,107,140,1)","rgba(69,16,91,1)","rgba(60,91,138,1)","rgba(43,129,140,1)","rgba(70,29,100,1)","rgba(106,198,97,1)","rgba(79,183,115,1)","rgba(69,21,95,1)","rgba(65,67,134,1)","rgba(43,140,138,1)","rgba(54,104,140,1)","rgba(40,155,135,1)","rgba(66,64,131,1)","rgba(70,29,100,1)","rgba(43,139,138,1)","rgba(36,165,133,1)","rgba(41,154,135,1)","rgba(95,191,105,1)","rgba(67,62,129,1)","rgba(58,97,139,1)","rgba(67,60,127,1)","rgba(52,108,141,1)","rgba(37,164,133,1)","rgba(64,77,136,1)","rgba(67,61,128,1)","rgba(67,61,129,1)","rgba(66,65,132,1)","rgba(49,112,141,1)","rgba(50,111,141,1)","rgba(49,112,141,1)","rgba(68,57,125,1)","rgba(64,75,136,1)","rgba(62,86,138,1)","rgba(36,165,133,1)","rgba(56,100,140,1)","rgba(69,15,91,1)","rgba(65,70,135,1)","rgba(58,97,139,1)","rgba(38,160,134,1)","rgba(59,93,138,1)","rgba(64,78,136,1)","rgba(41,152,136,1)","rgba(70,39,108,1)","rgba(69,44,112,1)","rgba(67,59,126,1)","rgba(44,135,139,1)","rgba(65,72,136,1)","rgba(37,163,133,1)","rgba(69,21,94,1)","rgba(42,150,136,1)","rgba(39,158,134,1)","rgba(39,159,134,1)","rgba(70,42,111,1)","rgba(70,42,110,1)","rgba(38,162,133,1)","rgba(36,165,133,1)","rgba(40,156,135,1)","rgba(65,66,133,1)","rgba(57,98,139,1)","rgba(34,167,132,1)","rgba(69,52,120,1)","rgba(46,116,141,1)","rgba(42,150,136,1)","rgba(69,52,120,1)","rgba(44,133,140,1)","rgba(69,11,89,1)","rgba(50,111,141,1)","rgba(57,99,139,1)","rgba(63,81,137,1)","rgba(64,72,136,1)","rgba(69,16,91,1)","rgba(46,116,142,1)","rgba(40,169,131,1)","rgba(69,22,95,1)","rgba(64,74,136,1)","rgba(39,160,134,1)","rgba(39,158,134,1)","rgba(43,126,141,1)","rgba(68,1,84,1)","rgba(62,83,137,1)","rgba(67,61,128,1)","rgba(47,115,141,1)","rgba(68,54,122,1)","rgba(57,99,139,1)","rgba(69,11,89,1)","rgba(68,6,86,1)","rgba(65,72,136,1)","rgba(69,12,89,1)","rgba(68,4,86,1)","rgba(50,111,141,1)","rgba(63,79,137,1)","rgba(68,5,86,1)","rgba(70,38,107,1)","rgba(69,48,116,1)","rgba(42,149,136,1)","rgba(60,90,138,1)","rgba(63,78,136,1)","rgba(50,111,141,1)","rgba(61,90,138,1)","rgba(69,20,94,1)","rgba(60,90,138,1)","rgba(65,70,135,1)","rgba(45,117,142,1)","rgba(61,88,138,1)","rgba(65,72,136,1)","rgba(70,25,97,1)","rgba(68,178,121,1)","rgba(42,151,136,1)","rgba(43,126,141,1)","rgba(42,120,142,1)","rgba(43,142,138,1)","rgba(69,14,90,1)","rgba(43,124,141,1)","rgba(48,113,141,1)","rgba(56,100,139,1)","rgba(68,56,123,1)","rgba(55,103,140,1)","rgba(39,169,131,1)","rgba(44,138,139,1)","rgba(67,61,129,1)","rgba(37,163,133,1)","rgba(66,66,133,1)","rgba(69,20,94,1)","rgba(36,165,133,1)","rgba(69,16,91,1)","rgba(41,152,136,1)","rgba(43,146,137,1)","rgba(42,148,137,1)","rgba(42,121,142,1)","rgba(56,100,140,1)","rgba(43,130,140,1)","rgba(68,53,121,1)","rgba(66,64,131,1)","rgba(56,101,140,1)","rgba(60,92,138,1)","rgba(64,76,136,1)","rgba(69,12,89,1)","rgba(70,37,107,1)","rgba(44,138,139,1)","rgba(48,114,141,1)","rgba(43,130,140,1)","rgba(46,116,142,1)","rgba(65,70,135,1)","rgba(41,153,135,1)","rgba(58,97,139,1)","rgba(70,34,104,1)","rgba(44,132,140,1)","rgba(39,160,134,1)","rgba(56,102,140,1)","rgba(35,166,132,1)","rgba(63,79,137,1)","rgba(65,70,135,1)","rgba(44,118,142,1)","rgba(42,120,142,1)","rgba(70,39,108,1)","rgba(70,37,106,1)","rgba(65,70,135,1)","rgba(69,12,89,1)","rgba(69,23,96,1)","rgba(64,73,136,1)","rgba(69,16,91,1)","rgba(68,178,121,1)","rgba(43,142,138,1)","rgba(70,33,104,1)","rgba(43,123,141,1)","rgba(37,163,133,1)","rgba(43,130,140,1)","rgba(57,99,139,1)","rgba(70,30,101,1)","rgba(40,156,135,1)","rgba(67,62,129,1)","rgba(43,144,137,1)","rgba(48,114,141,1)","rgba(69,25,97,1)","rgba(70,33,104,1)","rgba(69,14,90,1)","rgba(43,144,137,1)","rgba(68,3,85,1)","rgba(65,71,135,1)","rgba(47,115,141,1)","rgba(56,102,140,1)","rgba(64,72,136,1)","rgba(64,75,136,1)","rgba(69,44,112,1)","rgba(70,32,103,1)","rgba(64,75,136,1)","rgba(44,134,139,1)","rgba(53,107,140,1)","rgba(50,111,141,1)","rgba(43,143,138,1)","rgba(44,137,139,1)","rgba(39,159,134,1)","rgba(68,2,85,1)","rgba(65,66,133,1)","rgba(68,7,87,1)","rgba(35,167,132,1)","rgba(64,75,136,1)","rgba(53,106,140,1)","rgba(35,167,132,1)","rgba(63,81,137,1)","rgba(70,39,108,1)","rgba(69,46,115,1)","rgba(69,43,112,1)","rgba(70,34,104,1)","rgba(70,36,106,1)","rgba(70,27,99,1)","rgba(63,79,137,1)","rgba(68,3,85,1)","rgba(69,24,96,1)","rgba(63,83,137,1)","rgba(65,69,135,1)","rgba(70,34,104,1)","rgba(59,95,139,1)","rgba(68,4,86,1)","rgba(44,136,139,1)","rgba(80,183,115,1)","rgba(64,75,136,1)","rgba(67,59,126,1)","rgba(39,169,131,1)","rgba(45,117,142,1)","rgba(69,44,112,1)","rgba(57,99,139,1)","rgba(61,90,138,1)","rgba(53,105,140,1)","rgba(69,12,89,1)","rgba(62,85,137,1)","rgba(70,28,100,1)","rgba(67,61,129,1)","rgba(67,60,128,1)","rgba(54,104,140,1)","rgba(68,57,124,1)","rgba(64,78,136,1)","rgba(65,69,135,1)","rgba(69,16,91,1)","rgba(63,79,137,1)","rgba(68,56,123,1)","rgba(69,13,90,1)","rgba(62,83,137,1)","rgba(42,149,136,1)","rgba(43,125,141,1)","rgba(70,29,100,1)","rgba(43,127,141,1)","rgba(67,60,127,1)","rgba(62,85,137,1)","rgba(64,76,136,1)","rgba(52,108,141,1)","rgba(54,173,127,1)","rgba(43,142,138,1)","rgba(43,124,141,1)","rgba(42,122,142,1)","rgba(42,148,137,1)","rgba(44,137,139,1)","rgba(48,113,141,1)","rgba(43,141,138,1)","rgba(55,103,140,1)","rgba(44,132,140,1)","rgba(65,69,135,1)","rgba(68,4,85,1)","rgba(64,73,136,1)","rgba(70,40,109,1)","rgba(61,87,138,1)","rgba(57,174,125,1)","rgba(43,143,138,1)","rgba(43,129,140,1)","rgba(48,114,141,1)","rgba(41,153,135,1)","rgba(69,25,97,1)","rgba(47,115,141,1)","rgba(45,117,142,1)","rgba(44,132,140,1)","rgba(70,31,102,1)","rgba(215,225,55,1)","rgba(42,149,136,1)","rgba(69,16,92,1)","rgba(42,120,142,1)","rgba(64,77,136,1)","rgba(69,11,89,1)","rgba(69,50,117,1)","rgba(65,67,134,1)","rgba(70,38,107,1)","rgba(68,53,120,1)","rgba(46,116,141,1)","rgba(107,198,96,1)","rgba(69,23,96,1)","rgba(43,129,140,1)","rgba(70,29,100,1)","rgba(44,136,139,1)","rgba(66,65,132,1)","rgba(61,88,138,1)","rgba(65,72,136,1)","rgba(43,129,140,1)","rgba(65,72,135,1)","rgba(67,59,126,1)","rgba(43,140,138,1)","rgba(43,128,141,1)","rgba(44,132,140,1)","rgba(70,42,111,1)","rgba(70,29,100,1)","rgba(68,1,84,1)","rgba(58,97,139,1)","rgba(43,147,137,1)","rgba(55,103,140,1)","rgba(36,166,133,1)","rgba(39,158,134,1)","rgba(69,11,89,1)","rgba(69,12,89,1)","rgba(69,14,90,1)","rgba(70,31,101,1)","rgba(41,152,136,1)","rgba(69,43,112,1)","rgba(69,24,97,1)","rgba(62,84,137,1)","rgba(64,74,136,1)","rgba(43,131,140,1)","rgba(38,169,131,1)","rgba(69,43,111,1)","rgba(57,99,139,1)","rgba(42,150,136,1)","rgba(67,60,127,1)","rgba(48,114,141,1)","rgba(44,170,130,1)","rgba(59,93,139,1)","rgba(39,158,134,1)","rgba(62,85,137,1)","rgba(69,44,113,1)","rgba(70,38,108,1)","rgba(70,26,98,1)","rgba(52,107,140,1)","rgba(62,84,137,1)","rgba(67,60,128,1)","rgba(63,83,137,1)","rgba(64,74,136,1)","rgba(70,41,110,1)","rgba(65,67,134,1)","rgba(65,69,135,1)","rgba(43,125,141,1)","rgba(64,74,136,1)","rgba(66,65,132,1)","rgba(64,74,136,1)","rgba(53,107,140,1)","rgba(69,46,114,1)","rgba(56,100,139,1)","rgba(65,67,134,1)","rgba(61,89,138,1)","rgba(43,123,141,1)","rgba(55,103,140,1)","rgba(67,63,130,1)","rgba(52,107,140,1)","rgba(64,72,136,1)","rgba(70,36,106,1)","rgba(58,96,139,1)","rgba(67,60,127,1)","rgba(65,71,135,1)","rgba(64,73,136,1)","rgba(69,20,94,1)","rgba(68,53,121,1)","rgba(68,53,120,1)","rgba(69,51,119,1)","rgba(68,57,125,1)","rgba(70,36,106,1)","rgba(68,58,125,1)","rgba(66,65,132,1)","rgba(58,96,139,1)","rgba(67,59,126,1)","rgba(61,88,138,1)","rgba(65,69,135,1)","rgba(69,52,119,1)","rgba(68,2,85,1)","rgba(69,52,120,1)","rgba(58,96,139,1)","rgba(69,16,91,1)","rgba(64,77,136,1)","rgba(44,133,140,1)","rgba(44,132,140,1)","rgba(34,168,132,1)","rgba(42,149,136,1)","rgba(48,113,141,1)","rgba(55,102,140,1)","rgba(43,129,140,1)","rgba(69,52,120,1)","rgba(67,62,129,1)","rgba(60,91,138,1)","rgba(40,157,135,1)","rgba(65,71,135,1)","rgba(62,84,137,1)","rgba(70,33,103,1)","rgba(69,178,120,1)","rgba(53,105,140,1)","rgba(47,114,141,1)","rgba(69,47,116,1)","rgba(68,56,123,1)","rgba(68,6,86,1)","rgba(64,74,136,1)","rgba(69,19,93,1)","rgba(70,40,109,1)","rgba(69,22,95,1)","rgba(66,63,130,1)","rgba(40,156,135,1)","rgba(63,81,137,1)","rgba(70,35,105,1)","rgba(69,52,120,1)","rgba(69,48,116,1)","rgba(39,159,134,1)","rgba(43,139,138,1)","rgba(46,116,142,1)","rgba(43,126,141,1)","rgba(65,67,134,1)","rgba(107,198,96,1)","rgba(43,145,137,1)","rgba(64,73,136,1)","rgba(70,29,100,1)","rgba(62,176,124,1)","rgba(35,166,133,1)","rgba(43,147,137,1)","rgba(70,37,106,1)","rgba(54,173,127,1)","rgba(42,149,136,1)","rgba(35,168,132,1)","rgba(65,71,135,1)","rgba(60,92,138,1)","rgba(55,103,140,1)","rgba(60,91,138,1)","rgba(70,38,107,1)","rgba(52,108,141,1)","rgba(60,91,138,1)","rgba(42,151,136,1)","rgba(65,70,135,1)","rgba(50,110,141,1)","rgba(68,6,87,1)","rgba(70,28,99,1)","rgba(51,109,141,1)","rgba(69,49,117,1)","rgba(70,37,107,1)","rgba(67,61,128,1)","rgba(70,35,105,1)","rgba(43,143,138,1)","rgba(49,112,141,1)","rgba(36,168,131,1)","rgba(41,152,136,1)","rgba(63,79,137,1)","rgba(43,143,138,1)","rgba(44,134,139,1)","rgba(42,149,136,1)","rgba(52,173,127,1)","rgba(73,180,119,1)","rgba(69,12,89,1)","rgba(55,103,140,1)","rgba(56,174,126,1)","rgba(39,158,134,1)","rgba(65,177,122,1)","rgba(42,150,136,1)","rgba(50,111,141,1)","rgba(35,168,132,1)","rgba(47,115,141,1)","rgba(43,144,137,1)","rgba(43,170,130,1)","rgba(69,25,97,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(43,128,141,1)","rgba(43,139,138,1)","rgba(43,142,138,1)","rgba(41,153,135,1)","rgba(36,164,133,1)","rgba(60,175,124,1)","rgba(44,133,140,1)","rgba(49,113,141,1)","rgba(42,121,142,1)","rgba(60,90,138,1)","rgba(64,77,136,1)","rgba(71,179,119,1)","rgba(41,153,136,1)","rgba(125,209,80,1)","rgba(44,136,139,1)","rgba(41,153,135,1)","rgba(36,165,133,1)","rgba(43,129,140,1)","rgba(69,43,112,1)","rgba(43,145,137,1)","rgba(43,123,141,1)","rgba(40,156,135,1)","rgba(43,126,141,1)","rgba(68,4,86,1)","rgba(69,14,90,1)","rgba(68,5,86,1)","rgba(41,153,135,1)","rgba(67,178,121,1)","rgba(44,135,139,1)","rgba(69,48,116,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(37,164,133,1)","rgba(89,188,110,1)","rgba(42,150,136,1)","rgba(47,171,129,1)","rgba(56,101,140,1)","rgba(67,59,126,1)","rgba(42,148,137,1)","rgba(44,118,142,1)","rgba(44,137,139,1)","rgba(42,121,142,1)","rgba(59,95,139,1)","rgba(69,13,90,1)","rgba(78,182,116,1)","rgba(38,160,134,1)","rgba(52,108,141,1)","rgba(91,189,108,1)","rgba(44,138,139,1)","rgba(43,139,138,1)","rgba(43,130,140,1)","rgba(71,179,120,1)","rgba(44,136,139,1)","rgba(69,25,97,1)","rgba(42,149,136,1)","rgba(55,103,140,1)","rgba(69,49,117,1)","rgba(43,140,138,1)","rgba(43,119,142,1)","rgba(43,126,141,1)","rgba(60,92,138,1)","rgba(44,134,139,1)","rgba(82,184,114,1)","rgba(69,43,112,1)","rgba(50,172,128,1)","rgba(59,175,125,1)","rgba(104,196,99,1)","rgba(90,188,109,1)","rgba(43,144,137,1)","rgba(42,150,136,1)","rgba(54,105,140,1)","rgba(62,83,137,1)","rgba(48,113,141,1)","rgba(43,119,142,1)","rgba(43,123,142,1)","rgba(59,93,139,1)","rgba(59,175,125,1)","rgba(94,191,106,1)","rgba(49,112,141,1)","rgba(91,189,108,1)","rgba(48,114,141,1)","rgba(39,158,134,1)","rgba(42,149,136,1)","rgba(39,158,134,1)","rgba(68,57,125,1)","rgba(75,181,118,1)","rgba(42,150,136,1)","rgba(42,122,142,1)","rgba(43,145,137,1)","rgba(41,153,136,1)","rgba(43,144,137,1)","rgba(77,182,117,1)","rgba(69,178,121,1)","rgba(58,96,139,1)","rgba(63,81,137,1)","rgba(42,147,137,1)","rgba(34,167,132,1)","rgba(100,194,102,1)","rgba(119,207,84,1)","rgba(41,152,136,1)","rgba(34,168,132,1)","rgba(56,101,140,1)","rgba(43,143,138,1)","rgba(67,59,126,1)","rgba(44,137,139,1)","rgba(65,71,135,1)","rgba(37,163,133,1)","rgba(69,44,113,1)","rgba(63,79,137,1)","rgba(58,97,139,1)","rgba(38,160,134,1)","rgba(42,151,136,1)","rgba(39,158,134,1)","rgba(97,192,104,1)","rgba(41,154,135,1)","rgba(35,166,133,1)","rgba(43,123,142,1)","rgba(65,68,135,1)","rgba(43,123,141,1)","rgba(63,80,137,1)","rgba(70,38,107,1)","rgba(38,162,134,1)","rgba(48,114,141,1)","rgba(68,55,123,1)","rgba(36,164,133,1)","rgba(42,120,142,1)","rgba(44,118,142,1)","rgba(69,8,87,1)","rgba(45,117,142,1)","rgba(60,92,138,1)","rgba(44,132,140,1)","rgba(38,161,134,1)","rgba(66,66,133,1)","rgba(41,153,136,1)","rgba(38,161,134,1)","rgba(69,23,96,1)","rgba(43,119,142,1)","rgba(43,125,141,1)","rgba(62,83,137,1)","rgba(70,40,109,1)","rgba(69,9,88,1)","rgba(43,125,141,1)","rgba(62,86,138,1)","rgba(64,73,136,1)","rgba(44,135,139,1)","rgba(61,88,138,1)","rgba(36,164,133,1)","rgba(44,132,140,1)","rgba(44,136,139,1)","rgba(53,106,140,1)","rgba(36,165,133,1)","rgba(62,86,138,1)","rgba(76,182,117,1)","rgba(56,101,140,1)","rgba(69,23,96,1)","rgba(70,40,109,1)","rgba(68,54,122,1)","rgba(67,62,129,1)","rgba(68,53,121,1)","rgba(41,170,130,1)","rgba(35,167,132,1)","rgba(70,26,98,1)","rgba(70,38,108,1)","rgba(83,185,113,1)","rgba(60,175,124,1)","rgba(45,117,142,1)","rgba(51,109,141,1)","rgba(56,101,140,1)","rgba(69,49,117,1)","rgba(44,170,130,1)","rgba(77,182,116,1)","rgba(46,116,142,1)","rgba(44,135,139,1)","rgba(68,2,85,1)","rgba(45,116,142,1)","rgba(36,166,133,1)","rgba(70,31,102,1)","rgba(44,134,139,1)","rgba(45,117,142,1)","rgba(44,138,139,1)","rgba(42,170,130,1)","rgba(57,98,139,1)","rgba(42,148,137,1)","rgba(42,120,142,1)","rgba(61,87,138,1)","rgba(69,20,94,1)","rgba(70,34,104,1)","rgba(60,90,138,1)","rgba(43,119,142,1)","rgba(69,21,95,1)","rgba(50,111,141,1)","rgba(62,84,137,1)","rgba(70,29,100,1)","rgba(69,48,116,1)","rgba(61,89,138,1)","rgba(68,56,123,1)","rgba(60,90,138,1)","rgba(44,137,139,1)","rgba(68,2,84,1)","rgba(64,74,136,1)","rgba(50,111,141,1)","rgba(91,189,108,1)","rgba(53,107,140,1)","rgba(69,46,114,1)","rgba(65,67,134,1)","rgba(68,58,125,1)","rgba(56,101,140,1)","rgba(37,163,133,1)","rgba(69,11,89,1)","rgba(41,151,136,1)","rgba(42,149,136,1)","rgba(52,108,140,1)","rgba(37,162,133,1)","rgba(68,55,123,1)","rgba(50,110,141,1)","rgba(65,70,135,1)","rgba(69,50,118,1)","rgba(43,147,137,1)","rgba(66,63,131,1)","rgba(45,116,142,1)","rgba(41,153,135,1)","rgba(69,45,113,1)","rgba(43,146,137,1)","rgba(62,85,137,1)","rgba(69,51,119,1)","rgba(64,76,136,1)","rgba(68,7,87,1)","rgba(66,66,133,1)","rgba(55,103,140,1)","rgba(47,171,129,1)","rgba(43,143,138,1)","rgba(66,65,132,1)","rgba(43,142,138,1)","rgba(43,145,137,1)","rgba(58,95,139,1)","rgba(81,184,114,1)","rgba(58,96,139,1)","rgba(68,55,123,1)","rgba(69,10,88,1)","rgba(63,81,137,1)","rgba(58,96,139,1)","rgba(44,170,129,1)","rgba(59,93,139,1)","rgba(66,66,133,1)","rgba(55,102,140,1)","rgba(45,170,129,1)","rgba(62,83,137,1)","rgba(40,156,135,1)","rgba(69,46,114,1)","rgba(70,30,101,1)","rgba(40,169,131,1)","rgba(43,123,142,1)","rgba(70,31,102,1)","rgba(43,126,141,1)","rgba(43,126,141,1)","rgba(43,125,141,1)","rgba(54,105,140,1)","rgba(56,101,140,1)","rgba(64,77,136,1)","rgba(68,54,122,1)","rgba(78,182,116,1)","rgba(68,55,123,1)","rgba(94,191,106,1)","rgba(54,173,127,1)","rgba(43,125,141,1)","rgba(44,135,139,1)","rgba(44,132,140,1)","rgba(38,160,134,1)","rgba(43,119,142,1)","rgba(72,179,119,1)","rgba(63,81,137,1)","rgba(42,149,136,1)","rgba(52,108,140,1)","rgba(68,5,86,1)","rgba(64,73,136,1)","rgba(70,30,101,1)","rgba(57,99,139,1)","rgba(39,158,134,1)","rgba(39,158,134,1)","rgba(39,160,134,1)","rgba(90,189,109,1)","rgba(43,139,138,1)","rgba(68,56,124,1)","rgba(58,97,139,1)","rgba(69,21,94,1)","rgba(44,136,139,1)","rgba(69,51,119,1)","rgba(64,72,136,1)","rgba(43,125,141,1)","rgba(69,25,97,1)","rgba(69,17,92,1)","rgba(45,117,142,1)","rgba(66,63,130,1)","rgba(96,192,104,1)","rgba(56,100,139,1)","rgba(41,153,135,1)","rgba(51,109,141,1)","rgba(43,126,141,1)","rgba(75,181,117,1)","rgba(43,145,137,1)","rgba(38,161,134,1)","rgba(61,175,124,1)","rgba(44,134,139,1)","rgba(56,101,140,1)","rgba(69,46,114,1)","rgba(42,120,142,1)","rgba(63,81,137,1)","rgba(43,145,137,1)","rgba(70,28,99,1)","rgba(69,24,96,1)","rgba(49,113,141,1)","rgba(60,92,138,1)","rgba(51,110,141,1)","rgba(46,116,141,1)","rgba(69,45,114,1)","rgba(43,144,137,1)","rgba(68,56,123,1)","rgba(78,182,116,1)","rgba(69,23,96,1)","rgba(48,113,141,1)","rgba(43,143,138,1)","rgba(49,112,141,1)","rgba(43,130,140,1)","rgba(49,113,141,1)","rgba(49,112,141,1)","rgba(44,134,139,1)","rgba(59,93,138,1)","rgba(53,106,140,1)","rgba(54,105,140,1)","rgba(61,87,138,1)","rgba(65,72,136,1)","rgba(57,99,139,1)","rgba(68,55,123,1)","rgba(44,135,139,1)","rgba(71,179,119,1)","rgba(43,120,142,1)","rgba(36,164,133,1)","rgba(42,151,136,1)","rgba(36,165,133,1)","rgba(43,130,140,1)","rgba(51,109,141,1)","rgba(43,145,137,1)","rgba(44,132,140,1)","rgba(42,149,136,1)","rgba(42,148,137,1)","rgba(118,206,86,1)","rgba(42,149,136,1)","rgba(34,167,132,1)","rgba(35,166,132,1)","rgba(69,21,94,1)","rgba(44,136,139,1)","rgba(44,135,139,1)","rgba(35,166,132,1)","rgba(44,136,139,1)","rgba(43,145,137,1)","rgba(43,119,142,1)","rgba(52,173,127,1)","rgba(38,161,134,1)","rgba(43,140,138,1)","rgba(70,27,98,1)","rgba(42,150,136,1)","rgba(89,188,109,1)","rgba(43,122,142,1)","rgba(65,69,135,1)","rgba(42,147,137,1)","rgba(42,150,136,1)","rgba(51,110,141,1)","rgba(62,85,137,1)","rgba(44,134,139,1)","rgba(44,135,139,1)","rgba(55,103,140,1)","rgba(42,121,142,1)","rgba(69,23,96,1)","rgba(70,36,106,1)","rgba(69,44,113,1)","rgba(69,23,96,1)","rgba(44,131,140,1)","rgba(44,118,142,1)","rgba(53,106,140,1)","rgba(42,121,142,1)","rgba(105,197,98,1)","rgba(43,147,137,1)","rgba(43,139,138,1)","rgba(44,132,140,1)","rgba(42,150,136,1)","rgba(53,173,127,1)","rgba(42,151,136,1)","rgba(70,34,104,1)","rgba(39,158,134,1)","rgba(42,121,142,1)","rgba(77,182,116,1)","rgba(43,145,137,1)","rgba(69,179,120,1)","rgba(61,89,138,1)","rgba(39,158,134,1)","rgba(53,105,140,1)","rgba(39,158,134,1)","rgba(44,137,139,1)","rgba(36,166,133,1)","rgba(69,19,93,1)","rgba(43,142,138,1)","rgba(44,138,139,1)","rgba(111,201,93,1)","rgba(65,69,135,1)","rgba(42,122,142,1)","rgba(43,139,138,1)","rgba(69,21,94,1)","rgba(69,43,111,1)","rgba(70,33,104,1)","rgba(73,180,119,1)","rgba(43,131,140,1)","rgba(66,65,132,1)","rgba(45,116,142,1)","rgba(43,145,137,1)","rgba(60,175,124,1)","rgba(43,142,138,1)","rgba(70,30,101,1)","rgba(41,154,135,1)","rgba(39,160,134,1)","rgba(40,155,135,1)","rgba(40,155,135,1)","rgba(69,21,95,1)","rgba(70,26,98,1)","rgba(44,134,139,1)","rgba(55,103,140,1)","rgba(36,164,133,1)","rgba(44,134,139,1)","rgba(79,183,115,1)","rgba(65,72,136,1)","rgba(97,192,104,1)","rgba(44,137,139,1)","rgba(74,180,118,1)","rgba(38,160,134,1)","rgba(43,146,137,1)","rgba(43,123,141,1)","rgba(67,61,128,1)","rgba(41,154,135,1)","rgba(43,125,141,1)","rgba(43,146,137,1)","rgba(44,131,140,1)","rgba(41,153,135,1)","rgba(68,53,121,1)","rgba(44,132,140,1)","rgba(41,154,135,1)","rgba(43,128,140,1)","rgba(93,190,107,1)","rgba(68,178,121,1)","rgba(67,178,121,1)","rgba(42,148,136,1)","rgba(43,144,137,1)","rgba(64,76,136,1)","rgba(35,167,132,1)","rgba(41,155,135,1)","rgba(63,79,137,1)","rgba(42,120,142,1)","rgba(40,156,135,1)","rgba(43,145,137,1)","rgba(43,131,140,1)","rgba(37,169,131,1)","rgba(68,1,84,1)","rgba(38,160,134,1)","rgba(65,177,122,1)","rgba(69,25,97,1)"]}},"hoveron":"points","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","_isNestedKey":false,"frame":null},{"x":[-2.5,-2.5,null,2.5,2.5],"y":[-3.65411872478483,76.7426839028378,null,-3.65411872478483,76.7426839028378],"text":["xintercept: -2.5","xintercept: -2.5",null,"xintercept: 2.5","xintercept: 2.5"],"type":"scatter","mode":"lines","line":{"width":1.88976377952756,"color":"rgba(190,190,190,1)","dash":"dash"},"hoveron":"points","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[-6],"y":[-4.44089209850063e-16],"name":"99_854ca33fd6ba3e0a6dcd5738f3d68ac3","type":"scatter","mode":"markers","opacity":0,"hoverinfo":"skip","showlegend":false,"marker":{"color":[0,1],"colorscale":[[0,"#440154"],[0.00334448160535116,"#440355"],[0.00668896321070235,"#440456"],[0.0100334448160535,"#440656"],[0.0133779264214047,"#450857"],[0.0167224080267559,"#450958"],[0.020066889632107,"#450B59"],[0.0234113712374582,"#450D5A"],[0.0267558528428094,"#450E5B"],[0.0301003344481605,"#45105B"],[0.0334448160535117,"#45115C"],[0.0367892976588629,"#45135D"],[0.040133779264214,"#45145E"],[0.0434782608695652,"#45155F"],[0.0468227424749164,"#451760"],[0.0501672240802676,"#451860"],[0.0535117056856187,"#451961"],[0.0568561872909699,"#461A62"],[0.0602006688963211,"#461B63"],[0.0635451505016722,"#461D64"],[0.0668896321070234,"#461E65"],[0.0702341137123745,"#461F65"],[0.0735785953177258,"#462066"],[0.0769230769230769,"#462167"],[0.0802675585284281,"#462268"],[0.0836120401337793,"#462369"],[0.0869565217391305,"#46246A"],[0.0903010033444816,"#46256B"],[0.0936454849498328,"#46266B"],[0.096989966555184,"#46276C"],[0.100334448160535,"#46286D"],[0.103678929765886,"#46296E"],[0.107023411371237,"#462A6F"],[0.110367892976589,"#452B70"],[0.11371237458194,"#452C70"],[0.117056856187291,"#452D71"],[0.120401337792642,"#452E72"],[0.123745819397993,"#452F73"],[0.127090301003345,"#453074"],[0.130434782608696,"#453175"],[0.133779264214047,"#453276"],[0.137123745819398,"#453377"],[0.140468227424749,"#453477"],[0.1438127090301,"#443578"],[0.147157190635451,"#443679"],[0.150501672240803,"#44367A"],[0.153846153846154,"#44377B"],[0.157190635451505,"#44387C"],[0.160535117056856,"#44397D"],[0.163879598662207,"#443A7D"],[0.167224080267559,"#433B7E"],[0.17056856187291,"#433C7F"],[0.173913043478261,"#433D80"],[0.177257525083612,"#433E81"],[0.180602006688963,"#433F82"],[0.183946488294314,"#424083"],[0.187290969899666,"#424184"],[0.190635451505017,"#424185"],[0.193979933110368,"#424285"],[0.197324414715719,"#414386"],[0.20066889632107,"#414487"],[0.204013377926421,"#414587"],[0.207357859531773,"#414687"],[0.210702341137124,"#414787"],[0.214046822742475,"#414888"],[0.217391304347826,"#404988"],[0.220735785953177,"#404A88"],[0.224080267558528,"#404A88"],[0.22742474916388,"#404B88"],[0.230769230769231,"#404C88"],[0.234113712374582,"#404D88"],[0.237458193979933,"#404E88"],[0.240802675585284,"#3F4F89"],[0.244147157190635,"#3F5089"],[0.247491638795987,"#3F5189"],[0.250836120401338,"#3F5289"],[0.254180602006689,"#3F5289"],[0.25752508361204,"#3E5389"],[0.260869565217391,"#3E5489"],[0.264214046822742,"#3E5589"],[0.267558528428094,"#3E568A"],[0.270903010033445,"#3D578A"],[0.274247491638796,"#3D588A"],[0.277591973244147,"#3D598A"],[0.280936454849498,"#3D598A"],[0.28428093645485,"#3C5A8A"],[0.287625418060201,"#3C5B8A"],[0.290969899665552,"#3C5C8A"],[0.294314381270903,"#3B5D8A"],[0.297658862876254,"#3B5E8B"],[0.301003344481605,"#3B5F8B"],[0.304347826086956,"#3A5F8B"],[0.307692307692308,"#3A608B"],[0.311036789297659,"#3A618B"],[0.31438127090301,"#39628B"],[0.317725752508361,"#39638B"],[0.321070234113712,"#38648B"],[0.324414715719064,"#38658C"],[0.327759197324415,"#38658C"],[0.331103678929766,"#37668C"],[0.334448160535117,"#37678C"],[0.337792642140468,"#36688C"],[0.341137123745819,"#36698C"],[0.344481605351171,"#356A8C"],[0.347826086956522,"#356B8C"],[0.351170568561873,"#346B8C"],[0.354515050167224,"#346C8D"],[0.357859531772575,"#336D8D"],[0.361204013377926,"#326E8D"],[0.364548494983278,"#326F8D"],[0.367892976588629,"#31708D"],[0.37123745819398,"#31718D"],[0.374581939799331,"#30718D"],[0.377926421404682,"#2F728D"],[0.381270903010033,"#2F738D"],[0.384615384615385,"#2E748E"],[0.387959866220736,"#2D758E"],[0.391304347826087,"#2C768E"],[0.394648829431438,"#2B778E"],[0.397993311036789,"#2B778E"],[0.40133779264214,"#2A788E"],[0.404682274247492,"#2A798E"],[0.408026755852843,"#2A7A8E"],[0.411371237458194,"#2B7B8E"],[0.414715719063545,"#2B7B8D"],[0.418060200668896,"#2B7C8D"],[0.421404682274248,"#2B7D8D"],[0.424749163879599,"#2B7E8D"],[0.42809364548495,"#2B7F8D"],[0.431438127090301,"#2B7F8D"],[0.434782608695652,"#2B808D"],[0.438127090301003,"#2B818C"],[0.441471571906354,"#2B828C"],[0.444816053511706,"#2B838C"],[0.448160535117057,"#2C838C"],[0.451505016722408,"#2C848C"],[0.454849498327759,"#2C858C"],[0.45819397993311,"#2C868B"],[0.461538461538462,"#2C878B"],[0.464882943143813,"#2C878B"],[0.468227424749164,"#2C888B"],[0.471571906354515,"#2C898B"],[0.474916387959866,"#2C8A8B"],[0.478260869565217,"#2C8B8B"],[0.481605351170569,"#2B8B8A"],[0.48494983277592,"#2B8C8A"],[0.488294314381271,"#2B8D8A"],[0.491638795986622,"#2B8E8A"],[0.494983277591973,"#2B8F8A"],[0.498327759197325,"#2B8F8A"],[0.501672240802676,"#2B9089"],[0.505016722408027,"#2B9189"],[0.508361204013378,"#2B9289"],[0.511705685618729,"#2B9389"],[0.51505016722408,"#2A9389"],[0.518394648829432,"#2A9489"],[0.521739130434783,"#2A9588"],[0.525083612040134,"#2A9688"],[0.528428093645485,"#2A9788"],[0.531772575250836,"#2A9788"],[0.535117056856187,"#299888"],[0.538461538461538,"#299987"],[0.54180602006689,"#299A87"],[0.545150501672241,"#299B87"],[0.548494983277592,"#289B87"],[0.551839464882943,"#289C87"],[0.555183946488294,"#289D87"],[0.558528428093645,"#279E86"],[0.561872909698997,"#279F86"],[0.565217391304348,"#27A086"],[0.568561872909699,"#26A086"],[0.57190635451505,"#26A186"],[0.575250836120401,"#26A285"],[0.578595317725753,"#25A385"],[0.581939799331104,"#25A485"],[0.585284280936455,"#24A485"],[0.588628762541806,"#24A585"],[0.591973244147157,"#23A684"],[0.595317725752508,"#23A784"],[0.59866220735786,"#22A884"],[0.602006688963211,"#24A884"],[0.605351170568562,"#27A983"],[0.608695652173913,"#2AAA82"],[0.612040133779264,"#2DAA81"],[0.615384615384615,"#2FAB81"],[0.618729096989967,"#32AC80"],[0.622073578595318,"#34AC7F"],[0.625418060200669,"#36AD7E"],[0.62876254180602,"#38AE7E"],[0.632107023411371,"#3AAE7D"],[0.635451505016722,"#3CAF7C"],[0.638795986622073,"#3EB07B"],[0.642140468227425,"#40B07B"],[0.645484949832776,"#42B17A"],[0.648829431438127,"#43B279"],[0.652173913043478,"#45B278"],[0.65551839464883,"#47B378"],[0.658862876254181,"#48B477"],[0.662207357859532,"#4AB476"],[0.665551839464883,"#4BB575"],[0.668896321070234,"#4DB675"],[0.672240802675585,"#4EB674"],[0.675585284280936,"#50B773"],[0.678929765886288,"#51B872"],[0.682274247491639,"#53B971"],[0.68561872909699,"#54B971"],[0.688963210702341,"#55BA70"],[0.692307692307692,"#57BB6F"],[0.695652173913043,"#58BB6E"],[0.698996655518395,"#59BC6D"],[0.702341137123746,"#5BBD6C"],[0.705685618729097,"#5CBD6C"],[0.709030100334448,"#5DBE6B"],[0.712374581939799,"#5EBF6A"],[0.71571906354515,"#5FBF69"],[0.719063545150502,"#61C068"],[0.722408026755853,"#62C167"],[0.725752508361204,"#63C166"],[0.729096989966555,"#64C266"],[0.732441471571906,"#65C365"],[0.735785953177258,"#66C464"],[0.739130434782609,"#67C463"],[0.74247491638796,"#69C562"],[0.745819397993311,"#6AC661"],[0.749163879598662,"#6BC660"],[0.752508361204013,"#6CC75F"],[0.755852842809365,"#6DC85E"],[0.759197324414716,"#6EC85D"],[0.762541806020067,"#6FC95C"],[0.765886287625418,"#70CA5B"],[0.769230769230769,"#71CB5A"],[0.77257525083612,"#72CB59"],[0.775919732441472,"#73CC58"],[0.779264214046823,"#74CD57"],[0.782608695652174,"#75CD56"],[0.785953177257525,"#76CE55"],[0.789297658862876,"#77CF54"],[0.792642140468227,"#78CF53"],[0.795986622073579,"#79D052"],[0.79933110367893,"#7AD151"],[0.802675585284281,"#7CD151"],[0.806020066889632,"#7FD250"],[0.809364548494983,"#81D250"],[0.812709030100335,"#84D34F"],[0.816053511705686,"#87D34F"],[0.819397993311037,"#89D34E"],[0.822742474916388,"#8CD44D"],[0.826086956521739,"#8ED44D"],[0.82943143812709,"#91D54C"],[0.832775919732441,"#93D54C"],[0.836120401337793,"#95D54B"],[0.839464882943144,"#98D64B"],[0.842809364548495,"#9AD64A"],[0.846153846153846,"#9DD74A"],[0.849498327759197,"#9FD749"],[0.852842809364548,"#A1D748"],[0.8561872909699,"#A3D848"],[0.859531772575251,"#A6D847"],[0.862876254180602,"#A8D947"],[0.866220735785953,"#AAD946"],[0.869565217391304,"#ACD946"],[0.872909698996655,"#AFDA45"],[0.876254180602007,"#B1DA44"],[0.879598662207358,"#B3DB44"],[0.882943143812709,"#B5DB43"],[0.88628762541806,"#B7DB42"],[0.889632107023411,"#BADC42"],[0.892976588628762,"#BCDC41"],[0.896321070234114,"#BEDC40"],[0.899665551839465,"#C0DD40"],[0.903010033444816,"#C2DD3F"],[0.906354515050167,"#C4DE3E"],[0.909698996655518,"#C6DE3E"],[0.91304347826087,"#C8DE3D"],[0.916387959866221,"#CBDF3C"],[0.919732441471572,"#CDDF3B"],[0.923076923076923,"#CFDF3B"],[0.926421404682274,"#D1E03A"],[0.929765886287625,"#D3E039"],[0.933110367892977,"#D5E038"],[0.936454849498328,"#D7E138"],[0.939799331103679,"#D9E137"],[0.94314381270903,"#DBE136"],[0.946488294314381,"#DDE235"],[0.949832775919732,"#DFE234"],[0.953177257525084,"#E1E233"],[0.956521739130435,"#E3E333"],[0.959866220735786,"#E5E332"],[0.963210702341137,"#E7E331"],[0.966555183946488,"#E9E430"],[0.969899665551839,"#EBE42F"],[0.973244147157191,"#EDE42E"],[0.976588628762542,"#EFE52D"],[0.979933110367893,"#F1E52C"],[0.983277591973244,"#F3E52B"],[0.986622073578595,"#F5E62A"],[0.989966555183946,"#F7E629"],[0.993311036789298,"#F9E627"],[0.996655518394649,"#FBE726"],[1,"#FDE725"]],"colorbar":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":2.06156048675734,"thickness":23.04,"title":"log10(baseMean)","titlefont":{"color":"rgba(0,0,0,1)","family":"","size":15.9402241594022},"tickmode":"array","ticktext":["1","2","3","4","5"],"tickvals":[0.0468507930397103,0.268885520937976,0.490920248836241,0.712954976734506,0.934989704632772],"tickfont":{"color":"rgba(0,0,0,1)","family":"","size":12.7521793275218},"ticklen":2,"len":0.5}},"xaxis":"x","yaxis":"y","frame":null}],"layout":{"margin":{"t":23.9701120797011,"r":7.97011207970112,"b":40.6475716064757,"l":40.6475716064757},"plot_bgcolor":"rgba(255,255,255,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":15.9402241594022},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-6.40187094298865,3.87894841824218],"tickmode":"array","ticktext":["-6","-4","-2","0","2"],"tickvals":[-6,-4,-2,0,2],"categoryorder":"array","categoryarray":["-6","-4","-2","0","2"],"nticks":null,"ticks":"outside","tickcolor":"rgba(77,77,77,1)","ticklen":3.98505603985056,"tickwidth":0.724555643609193,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":12.7521793275218},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":false,"gridcolor":null,"gridwidth":0,"zeroline":false,"anchor":"y","title":{"text":"log2FoldChange","font":{"color":"rgba(0,0,0,1)","family":"","size":15.9402241594022}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-3.65411872478483,76.7426839028378],"tickmode":"array","ticktext":["0","20","40","60"],"tickvals":[-4.44089209850063e-16,20,40,60],"categoryorder":"array","categoryarray":["0","20","40","60"],"nticks":null,"ticks":"outside","tickcolor":"rgba(77,77,77,1)","ticklen":3.98505603985056,"tickwidth":0.724555643609193,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":12.7521793275218},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":false,"gridcolor":null,"gridwidth":0,"zeroline":false,"anchor":"x","title":{"text":"-log10(padj)","font":{"color":"rgba(0,0,0,1)","family":"","size":15.9402241594022}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":"transparent","line":{"color":"rgba(77,77,77,1)","width":0.724555643609193,"linetype":"solid"},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":false,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":2.06156048675734,"font":{"color":"rgba(0,0,0,1)","family":"","size":12.7521793275218}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","showSendToCloud":false},"source":"A","attrs":{"76751b34d531":{"x":{},"y":{},"colour":{},"key":{},"type":"scatter"},"7675299891eb":{"xintercept":{}}},"cur_data":"76751b34d531","visdat":{"76751b34d531":["function (y) ","x"],"7675299891eb":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> ] --- name:ggsave # Saving plots .pull-left[ ```r ggsave(plot = p1, filename = "figs/volcanoplot_small.png", height = 4, width = 4, units = "in", dpi = 100) ``` <center><img src="figs/volcanoplot_small.png" width="60%" height="60%"></center> ] .pull-right[ ```r ggsave(plot = p1, filename = "figs/volcanoplot_large.png", height = 10, width = 10, units = "in", dpi = 300) ``` <center><img src="figs/volcanoplot_large.png" width="60%" height="60%"></center> ] --- # Exercise Complete the fifth section of the `practice_ggplot.Rmd` file: "Histogram". --- class:center, middle, inverse # References and Links --- <!-- 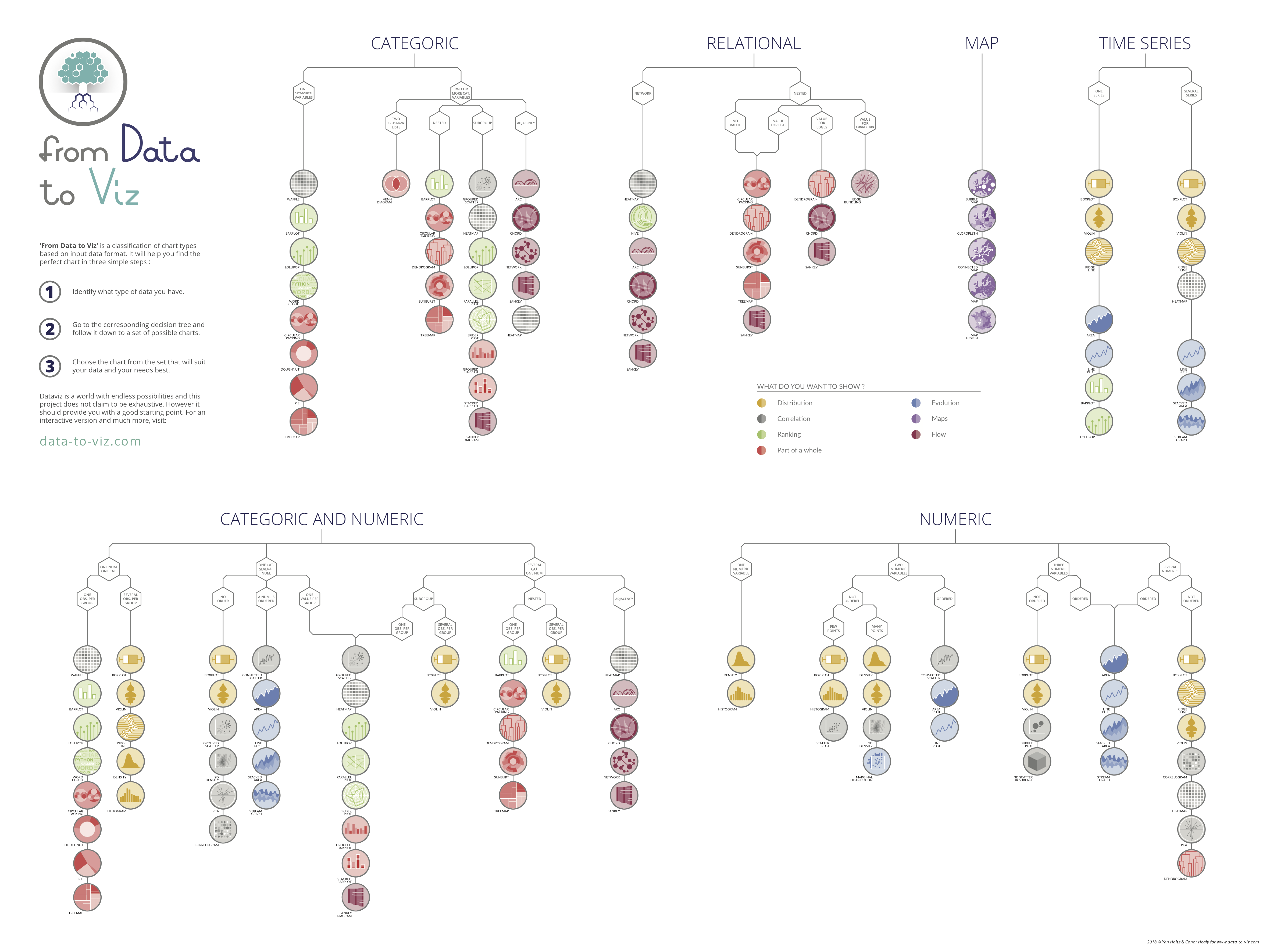 --> <center><img src="img/poster_big.png" width="85%" height="80%"><a href="https://www.data-to-viz.com/poster.html"><br>https://www.data-to-viz.com/poster.html</a></center> --- # Many, many ggplot extensions! Some examples at the [ggplot2 extensions gallery](https://exts.ggplot2.tidyverse.org/gallery/) <center><img src="img/ggextensiongallery.png" width="100%" height="100%"><a href="https://exts.ggplot2.tidyverse.org/gallery/"><br>Allison Horst</a></center> --- # Many, many themes and palettes/scales! We used themes from [`ggthemes`](https://yutannihilation.github.io/allYourFigureAreBelongToUs/ggthemes/) and [`hrbrthemes`](https://github.com/hrbrmstr/hrbrthemes) as well as built in themes, but there are many more: .pull-left[<center><img src="img/simpsonstheme.png" width="50%" height="50%"><a href="https://ryo-n7.github.io/2019-05-16-introducing-tvthemes-package/"><br>TV Themes</a></center> <center><img src="img/ggpomological.png" width="50%" height="50%"><a href="https://www.garrickadenbuie.com/project/ggpomological/"><br>ggpomological</a></center>] .pull-right[ <center><img src="img/bbplot.png" width="50%" height="50%"><a href="https://github.com/bbc/bbplot/"><br>bbplot for BBC themes</a></center> <center><img src="img/ggthemr.png" width="50%" height="50%"><a href="https://www.shanelynn.ie/themes-and-colours-for-r-ggplots-with-ggthemr/"><br>ggthemr</a></center> ] from ["Themes to improve your ggplot figures" by David Keyes](https://rfortherestofus.com/2019/08/themes-to-improve-your-ggplot-figures/) --- # R colors and palettes .pull-left[ <center><img src="img/ggplot2-colour-names.png" width="50%" height="50%"><a href="http://sape.inf.usi.ch/quick-reference/ggplot2/colour"><br>Built in R Colors</a></center> ] .pull-right[ <center><img src="img/wesanderson.png" width="60%" height="60%"><a href="https://www.datanovia.com/en/blog/ggplot-colors-best-tricks-you-will-love/"><br>wesanderson package</a></center> ] --- # Changing the order of names (levels) within a categorical variable * The default order of names within a categorical variable is alphanumeric * Such as `africa, americas, asia, europe` for the `four_regions` variable * Often we want a different order when making plots though. ## `factor` level variables Do this by making the categorical variable a `factor` level variable in R. * We can change the order of names within a `factor` level variable, and even rename the levels. * The `forcats` package makes this easy to do. See https://forcats.tidyverse.org/. --- # References <!-- to-do: check this slide --> - [ggplot cheatsheet](https://github.com/rstudio/cheatsheets/raw/master/data-visualization-2.1.pdf) - [ggplot2 package reference](https://ggplot2.tidyverse.org/reference/) - [ggplot2: Elegant Graphics for Data Analysis](https://ggplot2-book.org/) by Hadley Wickham - [*Data Visualizaton* online textbook by Kieran Healy](https://socviz.co/makeplot.html) - [R Graphics Cookbook](http://www.cookbook-r.com/Graphs/) by Winston Chang - [*R for Data Science* online textbook by Hadley Wickham](https://r4ds.had.co.nz/data-visualisation.html) - [*Introduction to Data Science* online textbook by Rafael A. Irizarry](https://rafalab.github.io/dsbook/ggplot2.html) Example plots and extensions: - [R Graph Gallery](https://www.r-graph-gallery.com/) - [ggplot2 extension gallery](https://exts.ggplot2.tidyverse.org/gallery/) - [All Your Figure Are Belong To Us](https://yutannihilation.github.io/allYourFigureAreBelongToUs/) - [from Data to Viz](https://www.data-to-viz.com/) - beautiful flowcharts to help you decide on a plot based on the variable type(s); check out their [poster](https://www.data-to-viz.com/poster.html) - [Top 50 ggplot2 Visualizations - The Master List (With Full R Code)](http://r-statistics.co/Top50-Ggplot2-Visualizations-MasterList-R-Code.html) OHSU class: - [CS 631 Data Visualization](https://www.ohsu.edu/school-of-medicine/csee/data-science) --- # Inspiration for this talk - [github/flipbookr](https://github.com/EvaMaeRey/flipbookr) - [Kieran Healy's rstudio::conf2020 data viz materials](https://github.com/rstudio-conf-2020/dataviz) --- class: left # Thank you! ## Contact info: - Jessica Minnier: _minnier@ohsu.edu_ - Meike Niederhausen: _niederha@ohsu.edu_ ## This workshop info: - Code for these slides are on github, with links to other course materials: [jminnier/berd_r_courses](https://github.com/jminnier/berd_r_courses) - The `.Rmd` file that generated the slides is on [github](https://github.com/jminnier/berd_r_courses/blob/master/04-ggplot/04_ggplot_slides.Rmd) and can be downloaded [here](https://jminnier-berd-r-courses.netlify.com/04-ggplot/04_ggplot_slides.Rmd), though you need to download the whole [R project](https://github.com/jminnier/berd_r_courses/archive/master.zip) to knit the file. - The project folder of examples can be downloaded at [github.com/jminnier/berd_ggplot_project](https://github.com/jminnier/berd_ggplot_project) & the solutions are in the `solns/` folder.